Abstract
Background
N6-Methyladenosine (m6A) and long non-coding RNAs (lncRNAs) are both crucial regulators in human cancer growth and metastasis. However, their regulation on cervical squamous cell carcinoma (CSCC) is largely unclear. The present study aimed to explore the role of m6A-associated lncRNAs in CSCC.
Methods
We screened the expression of methylation modification-related enzymes in CECC samples from TCGA. The qRT-PCR was used to detect METTL3 and lncRNA METTL4-2 expression. The biological activities of METTL3 in CSCC cells were evaluated by CCK-8, colony formation, transwell, wound healing, and xenograft tumor assays, respectively. The SRAMP tool was used to screen m6A modification sites of METTL4-2. Finally, the quantitative analysis of m6A modification was carried out by MeRIP.
Results
METTL3 expression was upregulated in CSCC cells and tissues. Biological function and function loss analysis indicated that METTL3 promoted the migration and proliferation of CSCC cells. In addition, METTL3 promoted CSCC tumor growth in vivo. Mechanically, METTL3 installed the m6A modification and enhanced METTL4-2 transcript stability to increase its expression. Meanwhile, the m6A “reader” YTHDF1 recognized METTL4-2 installed by METTL3 and facilitated the translation of METTL4-2.
Conclusions
In conclusion, our study highlights the function and mechanism of METTL3-induced METTL4-2 in CSCC. These findings support that METTL3-stabilized METTL4-2 promoted CSCC progression via a m6A-dependent modality, which provides new insights into therapeutic strategies for CSCC.
Similar content being viewed by others
Introduction
Cervical cancer (CC) is a human cancer with a high incidence rate of women, especially in developing countries [1]. In the report in 2018, it has been pointed out that CC accounts for about 3.2% of all new cancer cases and 3.3% of cancer deaths [2]. More importantly, the proportion of cervical squamous cell carcinoma (CSCC) has reached 90% of CC cases [3]. Studies have found a strong link between CC and human papillomavirus (HPV) infection [4]. Although the increase of HPV vaccination rate and the popularization of the HPV screening system have gradually accelerated the early diagnosis of CC and clarified the basis for diagnosis, the HPV infection rate is very high worldwide [5, 6]. Therefore, it is necessary to elucidate the molecular mechanism of CSCC occurrence and development, which may contribute to better study the therapeutic targets of CSCC.
Long non-coding RNAs (lncRNAs) refer to a special type of RNAs, with a length of 200 nt that do not encode proteins or have a limited protein-coding capacity [7]. Abnormal expression of lncRNAs has proposed to multiple biological functions, such as the initiation, metastasis, and epithelial-mesenchymal transformation (EMT) [8, 9]. In recent years, multiple studies have exhibited that lncRNAs are actually key determinants or diagnostic criteria of human cancer [10, 11]. Especially in CSCC, lncRNAs can also alter various malignant phenotypes [12, 13]. For example, the overexpressed lncRNA BLACAT1 in CSCC promoted cell invasion, proliferation, and migration to accelerate cancer progression [12, 14]. Therefore, lncRNA-mediated key regulation may greatly affect the progress of CSCC.
N6-Methyladenosine (m6A) is an epigenetic chemical modification existing in a variety of RNA molecules, which is the most abundant in eukaryotes [15]. In addition, m6A is a dynamic and reversible mode of RNA modification mainly mediated by RNA methyltransferases (m6A writers), RNA demethyltransferases (m6A erasers), and RNA methylation modification binding proteins (m6A readers) [16, 17]. METTL3 is a key catalyst for m6A modification [18], which display carcinogenic effects in breast cancer [19], endometrial cancer [20], and gastric cancer [21]. After different “readers” recognize gene m6A modification sites, cells will trigger different types of RNA metabolic effects, including mRNA structure, maturation, splicing, export, translation, and stability [22, 23]. YTH domain family 1 (YTHDF1) is the most functional “m6A reader” currently known, which can specifically recognize m6A-modified mRNAs and recruit multiple RNA-binding proteins to process mRNAs to improve translation efficiency [24]. For instance, the m6A reader YTHDF1 recognized the m6A residue on the CPCP1 3′‐UTR installed by METTL3 to facilitate CDCP1 translation in NSCLC [25]. In addition, growing evidence confirmed the regulation of m6A on EMT and metastasis of malignant tumors. For instance, m6A-related lncRNA RP11 triggered EMT of colorectal cancer through upregulation of Zeb1 [26]. However, the molecular mechanism of m6A on EMT and metastasis of CSCC has never been reported.
In our report, the role of METTL3 in the pathogenesis of CSCC was investigated. Data from this study confirmed that YTHDF1 recognized m6A-related lncRNA METTL4-2 by METTL3, improved the translation efficiency of METTL4-2 mRNA, and promoted its expression, thus promoting the EMT process of CSCC.
Materials and methods
Clinical tissues
Two group clinical patient samples: (1) 8 CSCC tissues and 8 corresponding normal tissues were collected from May 2019 and June 2020; (2) 8 CSCC metastatic tissue samples and 8 non-metastatic tissues were collected from October 2019 to May 2021. None of the patients had undergone preoperative chemoradiotherapy. The study protocol was approved by the Affiliated Tumor Hospital of Xinjiang Medical University. The written consent had obtained from each participant before the study.
Cell culture
The human CSCC cell lines (SiHa and C33A) and human normal cervical epithelial cell line (H8) were obtained from the Chinese Academy of Sciences Cell Bank (Shanghai, China) and American Type Culture Collection (Manassas, VA, USA). The cells were cultured in DMEM (Gibco, USA) containing 10% fetal bovine serum (Gibco, USA), 100 U/ml penicillin, and 100 mg/ml streptomycin. These cells were incubated in a 5% CO2 incubator at 37°C.
Cell transfection
All cell transfections were performed by Lipofectamine 2000 reagent (Invitrogen, USA), including Sh-NC, Sh-METTL3#1, Sh-METTL3#2; Vector-OE, METTL3-OE; lncRNA METTL4-2-OE; Sh-NC, Sh-YTHDF1#1; and Sh-YTHDF1#2; (GenePharma, Shanghai, China). Cells were harvested at 48 h post-transfection for the following experiments.
Animal experiment
Thirty-six BALB/c nude mice, 5-week-old, weighing about 18–20g, were fed on a 12-h light/dark schedule with free food and water. In order to conduct in vivo experiments, the lentivirus was customized by Shanghai Gene Pharma, including Sh-NC, Sh-METTL3#1, NC-OE, METTL3-OE, and lncRNA METTL4-2-OE. Subsequently, we subcutaneously injected C33A cells with adjusted gene expression into the axilla of mice. When a tumor mass appears (1 week), the tumor volume was measured with a vernier caliper every other week. Use the following formula to calculate: tumor volume (V) = (length × width2)/2. After 28 days, the mice were sacrificed and the tumors were isolated, measured the size and weight, and analyzed by a digital camera.
qRT-PCR
We use TRIZOL reagent (Thermo Fisher, USA) to extract total RNA from cells and tissues. For cDNA, reverse transcription was performed using the PrimeScript RT reagent Kit (TaKaRa, Otsu, Shiga, Japan). Gene expression was detected using a fluorescence quantitative PCR instrument (Analytick Jena A G, Germany). The reaction was conducted under the guidance of a fluorescent quantitative RT-PCR kit (SYBR Green, Bio-RAD, USA). The reaction conditions were described as 30 s denaturation at 94℃, followed by 35 cycles of denaturation at 95°C for 10 s, annealing at 58°C for 20 s, and extension at 72°C for 1 min. U6 and GAPDH were used as the internal control. The primer sequences were METTL3-F: 5′-CTATCTCCTGGCACTCGCAAGA-3′; METTL3-R: 5′-GCTTGAACCGTGCAACCACATC-3′; METTL4-2-F: 5′-TCCTAATAAGCCATTCCAGTCATT-3′; METTL4-2-R: 5′-TCTGCTCCTTCCTGCTATCT-3′; YTHDF1-F: 5′-GCACACAACCTCCATCTTCG-3′; YTHDF1-R: 5′-AACTGGTTCGCCCTCATTGT-3′; U6-F: 5′-CTCGCTTCGGCAGCACATATAC-3′; U6-R: 5′-AACGCTTCACGAATTTGCGTGTC-3′; GAPDH-F: 5′-GTCTCCTCTGACTTCAACAGCG-3′; and GAPDH-R: 5′-ACCACCCTGTTGCTGTAGCCAA-3′. All data were analyzed by adopting the 2−ΔΔCT method. ΔCT (test) = CT (target, test) − CT (internal control, test); ΔCT (calibrator) = CT (target, calibrator) − CT (internal control, calibrator); ΔΔCT = ΔCT (test) − ΔCT (calibrator).
Western blot
The cells were lysed using RIPA lysis buffer (Synthgene, Nanjing, China) and then centrifuged × 12,000 g for 20 min at 4°C. The concentration of protein in the supernatant was examined by the BCA protein Kit (Beyotime Biotechnology, China). Approximately 50 μg of protein was firstly placed on 10% SDS-PAGE and then shifted to a PVDF membrane. The proteins were incubated with the following primary antibodies at 4°C for 16 h: METTL3 (1:2000, Abcam, USA), E-cadherin, vimentin, N-cadherin, FN1 (1:1500, SBI, USA), and GAPDH (1:2000, Abcam, USA). GAPDH acts as a loading control. The membranes were washed with PBS for 3 times, and then corresponding secondary antibodies were added, followed by incubation for 1 h. Finally, the protein bands were quantified via ImageJ software (1.47V, NIH, USA).
Cell viability assay
Cell counting kit-8 assay (Beyotime, Shanghai, China) was used to evaluate the effect of transfection on the proliferation of CSCC cells. The cells were seeded in 96-well plates at a density of 2000 cells/well and then CCK8 reagent (10 μL) was added to each well. After incubation for 3 h, the absorbance was measured at 450-nm wavelength with an automatic microplate reader (MolecμLar Devices, Shanghai, China).
Colony formation assay
Briefly, C33A cells were grown in 6-well plates. Then, the cells (104 per plate) were seeded on plates and allowed to grow for 10–14 days. Then, the cells were fixed with methanol for 15 min and stained by 0.2% crystal violet. The colony number was manually counted and photographed.
Transwell assay
A transwell chamber (24-well chamber with an 8-µm pore) was used to detect the migration ability of C33A cells. The cells (1×104 cells/well) were seeded in the upper chamber with a serum-free medium. Subsequently, we added a medium containing 10% FBS to the lower chamber. After being incubated overnight, we fixed the cells with methanol and stained using crystal violet. Finally, cells were observed under an inverted microscope.
Wound healing assay
C33a cells were inoculated into 6-well culture plates and grew to confluence. We scraped the monolayer cells between two parallel straight lines in the middle of the culture plate with the tip of a sterile micropipette (0.5 mm) and washed culture plates with PBS to remove the floating cells. At 0 and 24 h after the scratch, the cell migration images were taken with an inverted microscope under the condition of 100 times magnification, respectively.
Immunohistochemistry (IHC)
The expression level of METTL3 in CSCC tissues was detected by immunohistochemistry. Briefly, the sections (5μm) were hydrated with conventional dewaxing and then rehydrated with alcohol. They were then incubated using primary antibody anti-METTL3 (1:100, Abcam, UK) overnight at 4°C and then incubated using a biotin-labeled secondary antibody (1:200, Abcam, UK) at 37°C for 30 min. Subsequently, the sections were counterstained using hematoxylin. Finally, observe and take pictures under a microscope.
HE staining
After mice were euthanized, CSCC tissues were collected and fixed. The sections (5μm) were stained using hematoxylin and eosin and dehydrated again (Olympus, Japan). Finally, histopathological characteristics were assessed under the microscope (CX31, Olympus, Japan).
m6A RNA methylation quantification
The total RNA m6A level of C33A cells was detected using the EpoQuikTM m6A RNA Methylation Quantification Kit (Colorimetric, No.#P-9005, Epigentek). In brief, the negative and diluted positive controls were then separately added to the assay wells with 200 ng of RNA and incubated for 90 min. The absorbance was monitored at 450-nm wavelength and the relative content of m6A was calculated by using the standard curve.
MeRIP-qRT-PCR
For quantification of m6A-modified METTL4-2 levels, methylated RNA immunoprecipitation was performed as previously reported [25]. In brief, total RNA was extracted from C33A cells by TRIzol (Invitrogen, CA, USA). One microgram of m6A antibody (ab151230; Abcam) was coupled to the mixture of Protein A beads (Thermo Fisher Scientific, Waltham, MA). Later, m6A RNA immunoprecipitation was performed using a GenSeq™ m6A RNA IP Kit and was eluted for m6A MeRIP library construction. Subsequently, beads were washed twice with RIP buffer and then resuspended in lysis buffer. After rotation and elution, the RNA enrichment was extracted using the PureLink RNA extraction kit (Ambion, TX, USA).
RNA stability
RNA stability was determined as previously described [27]. Briefly, cells were treated with 5 mg/ml actinomycin D (No.#A9415, Sigma) and then collected at different time points. RNA was then extracted and detected by qRT-PCR. The RNA levels at different times were calculated and normalized to GAPDH.
Statistical analyses
Data are presented as mean ± standard deviation (SD) of three independent experiments. GraphPad Prism 7.0 was used for data analysis. When comparing the two groups, Student’s t-test was used for statistical analysis. Besides, one-way ANOVA or two-way ANOVA was conducted followed by post hoc Dunnett’s test to calculate the mean difference between more than two groups. The Wilcoxon rank sum test was used to compare gene expression between CSCC tumor samples and paratumor samples in the TCGA dataset. P < 0.05 was considered statistically significant.
Results
METTL3 is upregulated in CSCC tissues and cells
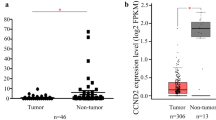
Plenty of researches have confirmed that m6A is highly variable and associated with tumorigenesis [28]. m6A affects the outcome of gene expression, this process of which includes localization, transcription, translation, and final decay, and can be jointly regulated by “writers,” “erasers,” and “readers” [29,30,31]. Based on this, we first searched the expression of m6A writers (such as METTL3, METTL14, and WTAP) and erasers (such as FTO and ALKBH5) through the TCGA database. The results found that METTL3 was overexpressed in CSCC tumors (Fig. 1A). qRT-PCR, western blot, and IHC also proved the above results (Fig. 1B–D). Consistently, METTL3 expression was significantly higher in CSCC cell lines (SiHa and C33A) than that in normal cervical epithelial cell lines (H8), especially C33A (Fig. 1E). Subsequently, METTL3 expression in non-metastatic and metastatic tissues was detected by qRT-qPCR and western blot. METTL3 levels were obviously upregulated in metastatic tissues (Fig. 1F, G). These results suggested that METTL3 was overexpressed in CSCC tissues, especially in metastatic tissues.
METTL3 is upregulated in CSCC tissues and cells. A Expression levels of m6A writers (METTL3, METTL14, and WTAP) and erasers (FTO and ALKBH5) were detected in the TCGA database. B qRT-PCR detected METTL3 expression in CSCC tissues and normal tissues (n=8). C The protein expression of METTL3 was detected by western blot in CSCC metastatic tissues (T) and non-metastatic tissues (N) (n=3). D Immunohistochemistry (IHC) staining was used to reveal METTL3 protein expression in CSCC tissues (n=3). E The mRNA level of METTL3 in CSCC cell lines (SiHa and C33A) and normal cervical epithelial cells (H8) was detected by qRT-PCR (n=3). F METTL3 expression in CSCC metastatic tissues and non-metastatic tissues was detected by qRT-PCR and G western blot (n=3). **p < 0.01; ***p < 0.001
Abnormal expression of METTL3 affects the progression of CSCC
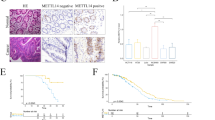
To further reveal the potential effect of METTL3 on CSCC cells, we studied the biological functions of METTL3 in C33A cells. The knockdown and overexpression efficiency of METTL3 was measured by RT-qPCR. METTL3 expression was significantly reduced in Sh-METTL3#1/#2 transfected cells. However, METTL3 level was significantly increased in METTL3 OE transfected cells (Fig. 2A). Based on CCK8 and clone formation experiments, the knockdown of METTL3 significantly attenuated cell growth, whereas overexpression of METTL3 remarkably increased cell growth (Fig. 2B, C). In further transwell and wound healing assays, METTL3 knockdown significantly attenuated the migration and invasion ability of cells, whereas METTL3 overexpression significantly increased the migration and invasion ability (Fig. 2D, E). Furthermore, METTL3 deficiency induced downregulation of vimentin, N-cadherin, FN1, and upregulation of E-cadherin, while overexpression of METTL3 had the opposite effects (Fig. 2F). All these results elucidated that the abnormal expression of METTL3 in CSCC cells affects the progression of CSCC in vitro.
Abnormal expression of METTL3 affects the progression of CSCC. A METTL3 expression was examined using qRT-PCR in C33A cells transfected with sh-NC, Sh-METTL3#1, Sh-METTL3#2, and METTL3 OE (n=3). B The proliferation of C33A cells was detected with a CCK-8 kit (n=3). C Colony formation assays (n=3). Cell migration and invasion were detected by D transwell assay and E wound healing assay (n=3). F The levels of E-cadherin, N-cadherin, vimentin, and FN1 in C33A cells were detected by western blot (n=3). **p < 0.01; ***p < 0.001
Next, to further verify the effect of METTL3 on the CSCC progression in vivo, we firstly knocked down or overexpressed METTL3 in C33A cells. Subsequently, we subcutaneously injected these C33A cells into BALB/c nude mice. The data showed that compared with control xenografts, the knockdown of METTL3 slowed tumor growth, whereas the upregulation of METTL3 promoted tumor growth (Fig. 3A–C). The HE stained of CSCC tissue slides also revealed that compared with the controls, overexpression of METTL3 increased the malignant morphology of the denser tumor, while knockdown of METTL3 reduced the malignant morphology of the looser tumor (Fig. 3D). In addition, IHC staining and qRT-PCR showed that knockdown of METTL3 significantly reduced METTL3 protein and mRNA levels, whereas overexpression of METTL3 significantly increased METTL3 protein and mRNA levels (Fig. 3E, F). These results suggested that abnormal expression of METTL3 affects the progression of CSCC in vivo.
Abnormal expression of METTL3 affects the growth of CSCC tumors. A Typical CSCC tumors from mice after subcutaneous injection of C33A cells with overexpression or knockdown of METTL3 (n=5). B, C The volume and weight of CSCC subcutaneous tumors in mice after different treatments (n=5). D HE staining of tumor tissues (n=5). E, F METTL3 expression in CSCC tumor tissue was determined by IHC staining and qRT-PCR, respectively (n=5). *p<0.05, **p<0.01, ***p<0.001
METTL3 upregulates lncRNA METTL4-2 expression
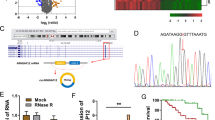
Recent advance in tumor epigenetic regulation has revealed the involvement of m6A in lncRNAs [26, 32]. We wanted to know whether m6A was associated with lncRNAs in CSCC. Therefore, in this study, we selected three CSCC tissues and matched normal tissues for lncRNA sequencing (Supplementary Table 1). We selected 6 upregulated lncRNAs with fold change over 200, among which MIR9-3HG [33] and KCNMB2-AS1 [34] were related to the development of CC, while the latter is stabilized by m6A modification to promote the growth of CC as a competitive endogenous RNA. In addition, the most differentially overexpressed lncRNA-NPHS2-6 has been studied in our previous work (data not published). Therefore, in this study, lncRNA-METTL4-2 that ranked second in fold change was selected to investigate the potential mechanism in CSCC. In the sequence-based RNA adenosine methylation site predictor (SRAMP) tool, we found that there were many m6A modification sites on METTL4-2 (Fig. 4A). MeRIP-qRT-PCR assay indicated that METTL4-2 was enriched in cells treated with anti-m6A antibody rather than IgG (Fig. 4B). Colorimetric quantificational analysis found that knockdown of METTL3 reduced the modification of m6A in CSCC cells (Fig. 4C). Next, whether METTL3 regulated METTL4-2 expression in CSCC cells was detected. It was found that METTL3 silencing significantly reduced the expression of METTL4-2 (Fig. 4D). In addition, MeRIP-qPCR demonstrated that the knockdown of METTL3 downregulated the m6A modification of METTL4-2 (Fig. 4E). In summary, these results suggested that METTL3 could upregulate METTL4-2 expression.
METTL3 enhances the stability of lncRNA METTL4-2 transcript and upregulates its expression. A The SRAMP tool was used to screen m6A modification sites of METTL4-2. B The enrichment of METTL4-2 followed by MeRIP-qRT-PCR assay in C33A cells (n=3). C m6A modification in C33A cells was conducted by colorimetric quantificational analysis (n=3). D The effect of METTL3 silencing on METTL4-2 expression was detected by qRT-PCR (n=3). E MeRIP‐qPCR demonstrated the m6A modification of METTL4-2 in C33A cells with METTL3 knockdown transfection (n=3). *p<0.05, **p<0.01, ***p<0.001
YTHDF1 enhances the stability of METTL4-2 via m6A modification
Epigenetic m6A marks A have been reported to be recognized mainly by “readers” [29,30,31]. Therefore, we detected the expression of m6A “reader” (such as YTHDC1, YTHDF1, and IGF2BP) through the TCGA database. The result found that the YTHDF1 level was higher in the CSCC tumors than the normal tissues (Fig. 5A). Subsequent clinical samples showed that YTHDF1 expression was overexpressed in CSCC tissues, especially in the metastatic tissues (Fig. 5B, C). Then, we studied the effect of YTHDF1 on METTL4-2 in CSCC cells. MeRIP-qPCR assay revealed that METTL4-2 was abundantly enriched by the YTHDF1 antibody (Fig. 5D). Next, we downregulated the level of YTHDF1 and qRT-PCR was used to detect knockdown efficiency (Fig. 5E). RT-qPCR results showed that METTL4-2 expression was inhibited after YTHDF1 knockdown (Fig. 5F). RNA stability analysis found that YTHDF1 knockdown reduced the stability of METTL4-2 (Fig. 5G). These results clearly suggested that YTHDF1 enhanced the stability of METTL4-2 via m6A modification.
YTHDF1 enhanced the stability of METTL4-2 via m6A modification. A The expression of m6A “reader” (such as YTHDC1, YTHDF1, and IGF2BP) was detected by the TCGA database. B qRT-PCR detected YTHDF1 expression in CSCC tissues and normal tissues (n=8). C qRT-PCR detected YTHDF1 expression in CSCC metastatic tissues and non-metastatic tissues (n=8). D RIP assay was performed in CSCC cells using an anti-YTHDF1 antibody, followed by the qRT-PCR analysis of METTL4-2 enrichment (n=3). E The transfection efficiency of YTHDF1 silencing by qRT-PCR (n=3). F The effect of YTHDF1 silencing on METTL4-2 expression was detected by qRT-PCR (n=3). G RNA stability analysis of METTL4-2 in treated C33A cells (n=3). *p < 0.05; **p < 0.01; ***p < 0.001
METTL3 regulates METTL4-2 expression and promotes the progression of CSCC
Functional complementation experiments were performed to evaluate the regulatory effect of METTL3 on METTL4-2. We firstly constructed C33A cells transfected with Sh-NC, lncRNA METTL4-2 OE, Sh-METTL3#1, and lncRNA METTL4-2 OE+Sh-METTL3#1 treatment, respectively. The CCK8 assay, colony formation, transwell assay, and wound healing showed that upregulated METTL4-2 significantly increased the cell proliferation, migration, and invasion, while downregulation of METTL3 significantly reduced cell proliferation, migration, and invasion. Importantly, simultaneous knockdown of METTL3 while overexpression of METTL4-2 neutralized the above effects (Fig. 6A–D). Furthermore, overexpression of METTL4-2 induced downregulation of vimentin, N-cadherin, and FN1 and upregulation of E-cadherin, while knockdown of METTL3 induced upregulation of vimentin, N-cadherin, and FN1 and downregulation of E-cadherin. Consistently, during overexpression of METTL4-2, simultaneous knockdown of METTL3 neutralized the above effects to EMT-associated factors (Fig. 6E). METTL3 could regulate the expression of METTL4-2 and promote EMT in CSCC cells. To further confirm this conclusion in vivo, we subcutaneously injected C33A cells with adjusted gene expression into the axilla of mice. The results were consistent with the trend of tumor growth in vitro experiments (Fig. 6F–H). In summary, we confirmed that METTL3 could regulate METTL4-2 expression and promote the progression of CSCC in vivo and in vitro.
Functional complementarity experiments demonstrated that METTL3 regulates METTL4-2 expression in CSCC progression. We firstly constructed C33A cells transfected with Sh-NC, lncRNA METTL4-2 OE, Sh-METTL3#1, and lncRNA METTL4-2 OE+Sh-METTL3#1 treatment, respectively. In animal experiments, we inoculated C33A cells performed with the above treatments subcutaneously at the armpits of mice. A Cell viability of C33A cells was measured by CCK-8 assay (n=3). B Colony formation assays of C33A cells (n=3). C The invasion of C33A cells was assessed by transwell assay (n=3). D The migration of C33A cells of indicated treatment was assessed by wound healing assay (n=3). E The levels of E-cadherin, N-cadherin, vimentin, and FN1 in C33A cells were detected by western blot (n=3). F Typical CSCC tumors from mice after subcutaneous injection of C33A cells with different treatments (n=4). G Tumor volume was measured every week by growth curve (n=4). H Tumor weight was measured at the end of experiments (at the 5 weeks) (n=4). *p < 0.05; **p < 0.01; ***p < 0.001
Discussion
Growing evidence suggests that lncRNAs are closely related to the pathogenesis of CSCC [12, 35]. With the rapid development of epigenetics and molecular biology, m6A-related lncRNA regulation resulting in self-renewal of tumor cells has gradually attracted the attention of researchers [36, 37]. Here, we proposed an interesting highlight that m6A is involved in the pathogenesis of CSCC together with lncRNAs.
Nowadays, m6A has made significant progress in the regulation of various stages of the RNA life cycle. M6A methylase complex is encoded by several “writers” where METTL3 localized in the catalytic core [23]. Increasing evidence supports the concept that METTL3 serves as a special mediator in tumorigenesis. The dual effects of METTL3 are inhibiting or promoting m6A modification in tumors [38]. For instance, Li et al. found that the deletion of METTL3 might promote the proliferation, migration, and invasion of renal small cell carcinoma [39]. However, Lin et al. observed that the knockdown of METTL3 dramatically inhibited liver cancer cell growth and EMT [40]. The above evidence suggested that METTL3 may be a novel marker for the tumorigenesis, development, and survival of cancer. Interestingly, METTL3 expression was upregulated in clinic CSCC tissues. Further cell studies confirmed that METTL3 overexpression in CSCC cells promoted cell proliferation, migration, and invasion, while deletion of METTL3 resulted in the opposite results. Overexpression of METTL3 could effectively promote subcutaneous tumor growth in nude mice, while knockdown of METTL3 could inhibit subcutaneous tumor growth. In addition, a series of studies have presented that METTL3 is associated with the occurrence and lung metastasis of liver cancer [41], colorectal cancer [27], gastric cancer [42], bladder cancer [43], and glioblastoma [44]. In this study, our clinical study also found METTL3 was upregulated in metastatic CSCC tissues. These data indicated that METTL3 could drive CSCC tumorigenesis and neoplasm metastasis.
It is well known that METTL3-mediated m6A affects targeted mRNA or miRNA, involved in EMT and metastasis of cancer [42]. For example, METTL3 preferentially recognized the m6A residues of CPCP1 and promoted its translation [25]. However, there are few studies on m6A-modified lncRNAs in the cancer field. In NSCLC, METTL3-correlated m6A modification enhanced the stability of lncRNA ABHD11-AS1 transcript to upregulate its expression [45]. Consistent with the above conclusions, our study first selected lncRNA METTL4-2 as the research object through lncRNA sequencing. Moreover, for the regulation of lncRNA METTL4-2, we found that METTL3 could preferentially install the m6A-modified site of METTL4-2 and enhance its transcript stability, thereby upregulating METTL4-2 expression. This finding is an interesting highlight that the METTL3-m6A-METTL4-2 axis could modulate the tumorigenesis and metastasis of CSCC.
RNA methyltransferase determines the abundance of m6A methylation modification after gene transcription, while the ultimate direction of m6A-modified abnormal mRNA in cells is determined by RNA methylation-modified binding protein. YTHDF1 is the most effective “m6A reader” known at present, which can specifically recognize m6A-modified mRNA and recruit multiple RNA-binding proteins to process the mRNA to improve translation efficiency and stability [24]. For instance, YTHDF1 recognized and bound to YY1 and MDM2 through m6A modification, which further promoted YY1 and MDM2 translation, thereby inhibiting p53 activation [46]. However, the regulatory role of YTHDF1 in tumor-related lncRNA translation is rarely studied. After our careful research, YTHDF1 was significantly upregulated in CSCC tumors based on the TCGA database. Clinical samples also showed that YTHDF1 expression was significantly increased in CSCC tissues, especially in the metastatic tissues. Further RIP assay revealed that YTHDF1 promoted the methylation modification of METTL4-2. RNA stability analysis found that YTHDF1 knockdown reduced the stability of METTL4-2. These results clearly suggested that YTHDF1 enhanced the stability of METTL4-2 via m6A modification.
Conclusion
Our study firstly demonstrated that YTHDF1 recognized METTL3-mediated m6A methylation of lncRNA METTL4-2 and improved the translation efficiency of METTL4-2, which thus promoted the EMT process of CSCC cells. In total, we investigated the functional roles and molecular mechanisms of METTL3 in CSCC. Besides, by highlighting the crucial role of METTL3, our discovery may pave the way for developing new therapeutic strategies against CSCC.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Shrestha AD, Neupane D, Vedsted P, et al. Cervical cancer prevalence, incidence and mortality in low and middle income countries: a systematic review. Asian Pac J Cancer Prev. 2018;19:319–24.
Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Groves IJ, Coleman N. Pathogenesis of human papillomavirus-associated mucosal disease. J Pathol. 2015;235:527–38.
Wentzensen N, Schiffman M, Palmer T, et al. Triage of HPV positive women in cervical cancer screening. J Clin Virol. 2016;76(Suppl 1):S49-s55.
Ronco G, Dillner J, Elfström KM, et al. Efficacy of HPV-based screening for prevention of invasive cervical cancer: follow-up of four European randomised controlled trials. Lancet. 2014;383:524–32.
de Martel C, Plummer M, Vignat J, et al. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer. 2017;141:664–70.
Raj A, Rinn JL. Illuminating genomic dark matter with RNA imaging. Cold Spring Harb Perspect Biol. 2019;11:a032094.
Wu Q, Xiang S, Ma J, et al. Long non-coding RNA CASC15 regulates gastric cancer cell proliferation, migration and epithelial mesenchymal transition by targeting CDKN1A and ZEB1. Mol Oncol. 2018;12:799–813.
Huang Y, Xiang B, Liu Y, et al. LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett. 2018;437:56–66.
Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–19.
Spizzo R, Almeida MI, Colombatti A, et al. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–87.
Cheng H, Tian J, Wang C, et al. LncRNA BLACAT1 is upregulated in cervical squamous cell carcinoma (CSCC) and predicts poor survival. Reprod Sci. 2020;27:585–91.
Liu H, Zhu C, Xu Z, et al. lncRNA PART1 and MIR17HG as ΔNp63α direct targets regulate tumor progression of cervical squamous cell carcinoma. Cancer Sci. 2020;111:4129–41.
Wang CH, Li YH, Tian HL, et al. Long non-coding RNA BLACAT1 promotes cell proliferation, migration and invasion in cervical cancer through activation of Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:3002–9.
Yue Y, Liu J, Cui X, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10.
Lin S, Choe J, Du P, et al. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335–45.
Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–99.
Wang X, Huang J, Zou T, et al. Human m(6)A writers: two subunits, 2 roles. RNA Biol. 2017;14:300–4.
Cai X, Wang X, Cao C, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–9.
Liu J, Eckert MA, Harada BT, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20:1074–83.
Lin S, Liu J, Jiang W, et al. METTL3 promotes the proliferation and mobility of gastric cancer cells. Open Med (Wars). 2019;14:25–31.
Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18:103.
Lan Q, Liu PY, Haase J, et al. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019;79:1285–92.
Edupuganti RR, Geiger S, Lindeboom RGH, et al. N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol. 2017;24:870–8.
Yang F, Jin H, Que B, et al. Dynamic m(6)A mRNA methylation reveals the role of METTL3-m(6)A-CDCP1 signaling axis in chemical carcinogenesis. Oncogene. 2019;38:4755–72.
Wu Y, Yang X, Chen Z, et al. m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019;18:87.
Li T, Hu PS, Zuo Z, et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112.
Pan Y, Ma P, Liu Y, et al. Multiple functions of m(6)A RNA methylation in cancer. J Hematol Oncol. 2018;11:48.
Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–200.
Wang CX, Cui GS, Liu X, et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018;16:e2004880.
Ping XL, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89.
Zheng ZQ, Li ZX, Zhou GQ, et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019;79:4612–26.
Yang H, Wang Z, Wang Z. Long noncoding RNA KCNMB2-AS1 increases ROCK1 expression by sponging microRNA-374a-3p to facilitate the progression of non-small-cell lung cancer. Cancer Manag Res. 2020;12:12679–95.
Zhang Y, Wang D, Wu D, et al. Long noncoding RNA KCNMB2-AS1 stabilized by N(6)-methyladenosine modification promotes cervical cancer growth through acting as a competing endogenous RNA. Cell Transplant. 2020;29:963689720964382.
Zhang Y, Xu H. LncRNA FAL1 upregulates SOX4 by downregulating miR-449a to promote the migration and invasion of cervical squamous cell carcinoma (CSCC) Cells. Reprod Sci. 2020;27:935–9.
Hu X, Peng WX, Zhou H, et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27:1782–94.
Hu Y, Ouyang Z, Sui X, et al. Oocyte competence is maintained by m(6)A methyltransferase KIAA1429-mediated RNA metabolism during mouse follicular development. Cell Death Differ. 2020;27:2468–83.
He L, Li J, Wang X, et al. The dual role of N6-methyladenosine modification of RNAs is involved in human cancers. J Cell Mol Med. 2018;22:4630–9.
Li X, Tang J, Huang W, et al. The M6A methyltransferase METTL3: acting as a tumor suppressor in renal cell carcinoma. Oncotarget. 2017;8:96103–16.
Lin X, Chai G, Wu Y, et al. RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat Commun. 2019;10:2065.
Fustin JM, Doi M, Yamaguchi Y, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806.
Yue B, Song C, Yang L, et al. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18:142.
Han J, Wang JZ, Yang X, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110.
Cui Q, Shi H, Ye P, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–34.
Xue L, Li J, Lin Y, et al. m(6) A transferase METTL3-induced lncRNA ABHD11-AS1 promotes the Warburg effect of non-small-cell lung cancer. J Cell Physiol. 2021;236:2649–58.
Zhao T, Sun D, Zhao M, et al. N(6)-methyladenosine mediates arsenite-induced human keratinocyte transformation by suppressing p53 activation. Environ Pollut. 2020;259:113908.
Acknowledgements
Not applicable.
Funding
This work was supported by Open Project of the State Key Laboratory of Prevention and Treatment of High Incidence Diseases in Central Asia (SKL-HIDCA-2021-18).
Author information
Authors and Affiliations
Contributions
GS and FL designed the study and drafted the manuscript. YW, YH, and GA were responsible for the collection and analysis of the experimental data. JY, CM, and CL revised the manuscript critically for important intellectual content. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Affiliated Tumor Hospital of Xinjiang Medical University, China. Signed written informed consents were obtained from the patients and/or guardians.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Table 1.
The top 6 lncRNAs from lncRNAs sequencing of CSCC tissues and matched normal tissues.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Shen, G., Li, F., Wang, Y. et al. New insights on the interaction between m6A modification and non-coding RNA in cervical squamous cell carcinoma. World J Surg Onc 21, 25 (2023). https://doi.org/10.1186/s12957-023-02907-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12957-023-02907-z