Abstract
Background
Papillary thyroid carcinoma (PTC) is the most common endocrine malignancy cancer among the malignancies of thyroid. Despite of wide usages of proteomics in PTC, the profile of acetylated proteins in PTC remains unsettled, which is helpful for understanding the carcinogenesis mechanism and identifying useful biomarkers for PTC.
Methods
The surgically removed specimens of cancer tissues (Ca-T) and adjacent normal tissues (Ca-N) from 10 female patients pathological diagnosed as PTC (TNM stage III) were enrolled in the study. After preparing the pooled extracts of the whole proteins and the acetylated proteins from 10 cases, TMT labeling and LC/MS/MS methods were applied to the assays of global proteomics and acetylated proteomics separately. Bioinformatics analysis, including KEGG, gene ontology (GO) and hierarchical clustering were performed. Some differentially expressed proteins (DEPs) and differentially expressed acetylated proteins (DEAPs) were validated by individual Western blots.
Results
Controlled with the normal tissues adjacent to the lesions, 147 out of 1923 identified proteins in tumor tissues were considered as DEPs in global proteomics, including 78 up-regulated and 69 down-regulated ones, while 57 out of 311 identified acetylated proteins in tumor tissues were DEAPs in acetylated proteomics, including 32 up-regulated and 25 down-regulated, respectively. The top 3 up- and down-regulated DEPs were fibronectin 1, KRT1B protein and chitinase-3-like protein 1, as well as keratin, type I cytoskeletal 16, A-gamma globin Osilo variant and Huntingtin interacting protein-1. The top 3 up- and down-regulated DEAPs were ribosomal protein L18a-like protein, alpha-1-acid glycoprotein 2 and eukaryotic peptide chain release factor GTP-binding subunit ERF3A, as well as trefoil factor 3, thyroglobulin and histone H2B. Functional GO annotation and KEGG pathway analysis based on the DEPs and DEAPs showed completely different changing pictures. Contrary to the top 10 up- and -down regulated DEPs, most of which were addressed in PTC and other types of carcinomas, changes of the majority DEAPs were not mentioned in the literatures.
Conclusions
Taken the profiling of the global and acetylated proteomics together will provide more broad view of protein alterations on the carcinogenesis and new direction for selecting biomarker for diagnosis of PTC.
Similar content being viewed by others
Background
Thyroid carcinoma is the most common endocrine cancer among the malignancies of head and neck [1,2,3], which accounts for about 1% of all kinds of carcinoma and comprises 91.5% of whole endocrine cancers. Recent statistics shows that the incidence of papillary thyroid carcinomas (PTC) is increasing in the past years and accounts for 80% of all thyroid cancers [4, 5]. Timely diagnosis and appropriate treatments are critical for raising long-term survival rate of PTC patients. Cytological examination and fine-needle aspiration, sonography, magnetic resonance imaging, and computed tomography have been used in diagnosing for PTC. The most effective test for distinguishing malignant from benign thyroid nodules is ultrasound-guided fine-needle aspiration biopsy which has approximately 93% sensitivity and 75% specificity. Advancing in the proteomics has introduced new techniques for screening the biomarkers and improving diagnosis of different kinds of cancers to a new horizon. Till now, some biomarkers for thyroid carcinoma diagnosis have been found including galectin-3 [6], fibronectin-1 [7], CITED-1 [8], HBME1, cytokeratin-19, TPO [9], etc., however, most of these biomarkers lacking specificity or having poor positive predictive value to some degree [6,7,8,9].
Protein acetylation in which the acetyl group transferring from acetyl-coenzyme A (Ac-CoA) to a specific site on a polypeptide chain is the major post-translational modifications (PTMs) in most eukaryotes. About 90% of proteins become co-translationally acetylated at their N-termini of the nascent polypeptide chains in humans. Till now, hundreds of different PTMs were identified including acetylated proteomics profiling with new techniques emerging and especially in the mass spectrometry field. Because of more consumption of Ac-CoA during acetylation and more consumption of NAD+ during deacetylation by specific KDACs (lysine deacetylase), acetylation process participates in the metabolic processes and energy homeostasis of body. Consequently, unbalanced acetylation machinery can lead to severe diseases, such as neurodegenerative diseases, cardiovascular disorders, cancers, etc. [10,11,12].
With the help of LC-MS/MS, we screened the global and the acetylated proteomics profiling of the tumor tissues from 10 PTC patients at TNM stage Ш, controlled with the normal tissues adjacent to the lesions from the same patients. Totally 147 out of 1923 identified proteins were considered as the differentially expressed protein (DEP), including 78 up-regulated and 69 down-regulated, respectively. Fifty-seven out of 311 identified acetylated proteins were differentially expressed acetylated proteins (DEAP), among them 32 were increased and 25 were decreased. Further, some bioinformatics analyses, such as hierarchical clustering, KEGG pathway, gene ontology (GO), based on DEPs and DEAPs were performed. We found that either involved GO processes or KEGG pathways were almost completely different between the global proteomics and acetylated proteomics.
Methods
Ethics statement
The research protocol was approved by the Ethic Committee of Peking University Cancer Hospital & Institute, and the subject signed the informed consent form approved by the Institutional Review Board at the Peking University Cancer Hospital & Institute.
Samples of thyroid cancer
To address the possible difference in the protein and the acetylated protein expressions between the carcinoma tissues and the normal tissues, we enrolled the specimens of the surgically removed tumor tissues and the normal tissues adjacent to the lesion sites without abnormal pathological signs from 10 patients of PTC. Approximately 700 to 800 mg specimen of each patient was obtained, 200 mg for global proteomics and 500 mg for acetylated proteomics profiling assay. All enrolled samples were pathologically confirmed by the pathologists in Peking University Cancer Hospital and Institute, excluding the evidence of other chronic thyroid diseases, such as chronic lymphocytic thyroiditis and autoimmune thyroid disease. As more numbers of female patients of PTC than male patients, all enrolled patients were female in this study, to eliminate the potential influence of gender. The ages of those 10 patients ranged from 56 to 83 years old, with the average of 65.6 year-old. All enrolled patients were at TNM stage III. The ages and TNM stages of those cases were summarized in Supplemental Table 1.
Protein extraction
Tissues were homogenized in the lysis buffer containing 5 mM C4H7NaO2, 2.5 mM NA3PO4, 1 mM Na3VO4, 8 M Urea and 30 mM HEPES. The lysates were mixed with 10% TCA-acetone and stored at -20℃ overnight, and then, collected by centrifuging at 4℃ for 30 min at 20,000 rpm and washed with acetone for three times. After adding lysis buffer, the preparations were exposed to ultrasonic under power 180W for 5 min. Next, after centrifugating for 30 min at 20,000 rpm, the supernatants were collected and incubated for 1 h at 56℃ with 10 mM DTT in the final concentration. The products were immediately mixed with IAM to the final concentration of 55 mM and stayed in dark for 1 h. The supernatants were collected after centrifugating at 20,000 rpm for 30 min. The protein concentrations in the final products were measured using Bradford protein assay (Sigma) and the qualities of the extracted proteins were analyzed by SDS-PAGE.
Protein digestion and peptide purification
Aliquots of 5 mg proteins each preparation were transferred into a 10 K ultrafilter (Sartorius Stedim Lab L Ltd) and centrifuged at 4℃,4,000 rpm for 40 min. 5 ml of 5 mM Triethylammonium bicarbonate (TEBA) was added into the filters and centrifuged at 4℃, 4,000 rpm for 40 min, and this procedure was repeated two times. Protein digestion was performed with 1 μg/μl trypsin at 37℃ for 24 h. The purification of peptides was conducted with C18 reversed-phase column (Agela Technologies) to remove salts and dried by vacuum. The lengths of the majority of the digested peptides were less than 30 amino acids.
Enrichment of the acetylated peptides
The dried products were solved in PTMScan® IAP (reagents supplied in the kit) buffer containing 50 mM NaCl, 10 mM Na3PO4 and 50 mM MOPS, pH7.2. The pellets were discarded by centrifuged at 10,000 rpm at 4℃ for 5 min. By using a commercial kit (ICP0388, ImmuneChem Pharmaceuticals), the acetylated peptides were enriched and the beads were incubated with 2% acetic acid and IAP buffer overnight rotatably after washing with PBS. Finally, the beads were eluted with 0.15% TFA (reagents supplied in the kit) and peptides in supernatants were collected after centrifugating.
Peptide TMT-labelling and identification of mass spectra
Prepared TMT-labeled reagents (Thermo Scientific) were mixed with 41 μl acetonitrile and vortexed for 1 min. Then the preparations of the tested peptide were mixed with different isotopes and incubated at RT for 1 h. The mixture was added with 8 μl 5% hydroxylamine for 15 min. Different labeled products were mixed 1:1:1:1 and dried by vacuum for further using.
Peptide mixtures were loaded onto an Acclaim PePmap C18-reversed phase column (75 μm × 2 cm, 3 μm, 100 Ǻ, Thermo Scientific) and separated with reversed phase C18 column (75 μm × 10 cm, 5 μm, 300 Ǻ, Agela Technologies) mounted on a Dionex ultimate 3 000 nano LC system. Peptides were eluted using a gradient of 5–80% (v/v) acetonitrile in 0.1% formic acid over 65 min at a flow rate of 300 nl min−1 combined with a Q Exactive mass spectrometer (Thermo Fisher Scientific, MA, USA).
The eluates were directly entered Q-Exactive MS (Thermo Fisher Scientific, Waltham, MA, USA), setting in positive ion mode and data-dependent manner with full MS scan from 350–2000 m/z, full scan resolution at 70,000, MS/MS scan resolution at 35,000. MS/MS scan with minimum signal threshold 1E+5, isolation width at 2 Da. To evaluate the performance of this mass spectrometry on the TMT labeled samples, two MS/MS acquisition modes, higher collision energy dissociation (HCD) was employed. To optimize the MS/MS acquisition efficiency of HCD, normalized collision energy (NCE) was systemically examined 30, stepped 20%.
Bioinformatics analysis
Spectrum screening, protein identification and protein quantitative parameters used in mass spectrometry data processing were showed as following. Spectrum screening parameters included that mass range of parent ions was 350–6,000 Da, minimum number of peaks in secondary mass spectrometry was 10, signal-to-noise ratio value was 1.5. Identification retrieval parameters included that Mascot version was 2.3.0, database was uniport_2019_human 9606 (Time files compressed: Wed Jun 17 13:20:26 2020 and Number of sequences: 24,873). The quantitative parameters included that protein ratio type was median, the minimum peptide was 1 and the normalization method was median.
In this study, the peptides or the acetylated peptides in the carcinoma tissues changing > 1.2 fold compared with the data of normal control were considered as the differentially expressed peptides (DEPs) or differentially expressed acetylated peptides (DEAPs). P < 0.05 was generally taken as the difference screening criteria to evaluate the significance of difference in protein quantification. Volcano Plot, Gene Ontology (GO functional annotation), KEGG pathway analysis (including top 20 pathway enrichment, KEGG classification), Heat map clustering and protein interaction network were included in the further bioinformatics analysis for proteomics profiling for the global and acetylated proteins of PTC.
Western blot
Different tumor tissues were homogenized in lysis buffer (100 mM NaCl, 10 mM EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 10 mM Tris, pH 7.5) on ice containing protease inhibitor cocktail set III (Merck Millipore, 535140-1SET). The supernatant fractions were collected after the homogenates being centrifuged at 2,000 g for 10 min, separated by 12% SDS-PAGE and electro-transferred onto nitrocellulose membranes. Membranes were incubated with various primary antibodies, including fibronectin 1 (Fib) antibody (Santa Cruz Biotechnology, sc8422), carbonic anhydrase 1 (CA1) antibody (R&D Systems, AF2180-SP), chitinase 3 like protein 1 (CHI3L1) antibody (R&D Systems, AF2599-SP) and metalloproteinase inhibitor 1 (TIMP1) antibody (Santa Cruz Biotechnology, sc365905), at 4 °C overnight and then incubated with HRP conjugated secondary antibodies (Jackson Immuno Research Labs, 115-035-003 and 111-035-003) at RT for 1 h after washing with PBS. For detecting total acetylated proteins, anti-acetylated-lysine antibody (ICP0380, Immunechem, Burnaby, BC, Canada) was used. The blots were developed using enhanced chemiluminescence (ECL) system. Images were captured by ChemiScope 6000 (CLiNX).
Statistical analysis
The results of immunoblot images were analyzed using software Image J and gray values of target blots were evaluated. Statistical analyses were carried using Student’s t test.
Results
Global protein and acetylated protein profiles of tumor and normal tissues of PTC
The tissue homogenates of the surgically removed PTC from 10 patients diagnosed were pooled as the carcinoma-tumor specimen (Ca-T) and the normal thyroid tissues adjacent to the lesion from the same patients were pooled as the carcinoma-normal samples (Ca-N). The extracted acetylated proteins from tumor and normal tissues were nominated as Ca-T-A and Ca-N-A. The quality of extracted proteins, quantitative accuracy of proteins, trypsin hydrolysis efficiency, mass spectral mass deviation, mass spectrometry acquisition intensity and data volume fulfilled the requirements of the quality control for proteomic assays (Suppl Fig. 1).
For global proteomics profiling, 33,726 matched spectra were achieved, and 3,214 proteins, elicited from 11,350 unique peptides, were identified with 95% confidence interval by the Peptide Prophet Algorithm. Totally 1,923 proteins were identified both in the samples of Ca-T and Ca-N. The general pattern of DEPs and non-differential expressed proteins were showed in the Volcano plot (Fig. 1A). One hundred forty-seven proteins in Ca-T were considered as DEPs in comparison with Ca-N when utilizing the standard of 1.2-fold changed and p < 0.05, among them 78 DEPs were up-regulated and 69 DEPs were down-regulated, respectively. Thirty-nine DEPs showed ≥ 1.5-fold changed, including 27 up- and 12 down-regulated ones. Nine DEPs were ≥ 2.0-fold changed, including 8 up- and 1 down-regulated ones (Fig. 1B).
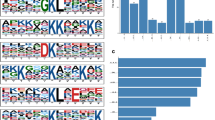
Volcano plots and numbers of DEPs identified in global proteomics and DEAPs in acetylated proteomics in comparison of Ca-T with Ca-N. Volcano plots of DEPs in global proteomics (A) and DEAPs in acetylated proteomics (C). Green dots represent the down-regulated proteins and red dots represent up-regulated ones. Gray dots are non-significantly changed proteins. The numbers of DEPs (B) and DEAPs (D). The changed folds of the up- and down-regulated DEPs and DEAPs are indicated on X-axis. The numbers of the changed proteins are indicated above the columns
For acetylated proteomics profiling, 5,628 matched spectra were achieved, and 636 proteins, elicited from 1,722 unique peptides, were identified with 95% confidence interval by the Peptide Prophet Algorithm. Totally 311 proteins were identified both in the samples of Ca-N-A and Ca-T-A. The general pattern of DEAPs and non-differential ones were illustrated in the Volcano plot (Fig. 1C). Total 57 DEAPs in the sample of Ca-T-A were obtained in comparison with Ca-N-A, showing ≥ 1.2-fold changed (32 up- and 25 down-regulated ones). 23 DEAPs were 1.5-fold changed (17 up- and 6 down-regulated ones) and 7 DEAPs were ≥ 2.0-fold changed (6 up- and 1 down-regulated ones) (Fig. 1D).
In the context of global proteomics, 7.6% (147) out of 1923 identified proteins showed differentially expressed between Ca-T and Ca-N, while in the context of acetylated proteomics, 18.3% (57) out of 311 identified proteins were differentially expressed. Among 147 DEPs, only 30 proteins (20.4%) were also identifiable in acetylated proteomics, whereas 43 out of 57 DEAPs (75.4%) were also detectable in global proteomics (Fig. 2A). Among 13 up-regulated and 17 down-regulated DEPs, the up-regulated (1.2-fold), unchanged (< 1.2- to > 0.83-fold) and down-regulated (< 0.83-fold) proteins in acetylated proteomics were 5, 6, 2 and 1, 5, 11 (Fig. 2A). Among 25 up-regulated and 18 down-regulated DEAPs, the up-regulated, unchanged and down-regulated proteins in global proteomics were 4, 20, 1 and 1, 11, 6, respectively (Fig. 2B).
The numbers and statuses of the DEPs identifiable in acetylated proteomics (A) and the DEAPs identifiable in global proteomics (B). Yellow graphic: unfound; green graphic: downregulated < 0.83 fold; grey graphic: insignificantly changed between < 1.2 and > 0.83 fold; red graphic: upregulated > 1.2 fold. The numbers of DEPs and DEAPs identifiable in the opposite proteomic assays are separately showed on the bottom tables
Top 20 up- and down-regulated DEPs and DEAPs
The top 20 up- and down-regulated DEPs and DEAPs were summarized in Tables 1 and 2. Three of the most increased DEPs were cDNA FLJ53365 highly similar to Homo sapiens fibronectin 1, fibronectin 1 (FN1), KRT1B protein (KRT1), while three most decreased DEPs were adhesion G-protein-coupled receptor G1 (ADGRG1), cDNA FLJ53570 highly similar to Keratin, type I cytoskeletal 16 (KRT16), A-gamma globin Osilo variant, respectively (Table 1). Three up-regulated DPEs (fibronectin 1, TNC protein and epididymis secretory sperm binding protein) and five down-regulated DEPs (trefoil factor 3, hemoglobin alpha-2 globin mutant, carbonic anhydrase 1, hemoglobin subunit alpha and creatine kinase B-type) were also identifiable in acetylated proteomics. Three of the most upregulated DEAPs were ribosomal protein L18a-like protein, alpha-1-acid glycoprotein 2, eukaryotic peptide chain release factor GTP-binding subunit ERF3A, while three most downregulated ones were trefoil factor 3, thyroglobulin, histone H2B, respectively (Table 2). Among the top 20 DEAPs, 14 up- and down-regulated proteins were also detectable in global proteomics.
Hierarchical clustering heat map
Totally 221 proteins from Ca-T and Ca-N samples were identified both in global and acetylated proteomics. To see the general profile of those proteins in global and acetylated proteomics between the groups of Ca-T and Ca-N, hierarchical clustering was performed. As shown in Fig. 3, the profile of acetylated proteomics between the groups of Ca-T and Ca-N revealed a more pronounced change than that of global proteomics. More proteins displayed increased acetylated profiling in Ca-T and decreased acetylated profiling in Ca-N.
Validation of the selected DEPs and DEAPs by Western blots
To validate the screening results for DEPs and DEAPs based on MS/MS, the expressions of four proteins, including fibronectin 1 (Fib), carbonic anhydrase 1 (CA1), chitinase 3 like protein 1 (CHI3L1) and metalloproteinase inhibitor 1 (TIMP1), in the samples of Ca-T and Ca-N were evaluated by the Western blots. The changed states of those four proteins of Ca-T vs Ca-N in global proteomics and acetylated proteomics were 3.181 and 1.853 (Fib), 0.67 and 0.65 (CA1), 2.666 and undetectable (CHI3L1), 2.23 and undetectable (TIMP1), respectively (Fig. 4A). Western blots revealed that the signals of Fib, CHI3L1 and TIMP1 in the samples of Ca-T were stronger than that of Ca-N, while the band of CA1 in Ca-T sample was much weaker (Fig. 4B). Further, the acetylated extracts from Ca-T and Ca-N were prepared and subjected into the Western blots with individual specific antibodies. As shown in Fig. 4B, the signal of acetylated Fib in Ca-T was stronger than that of Ca-N. Acetylated CA1 band was observed only in Ca-N. No specific acetylated CHI3L1 and TIMP1 signal was identified both in Ca-T and Ca-N tissues. Those data validate, at least partially, the results of the proteomic assays.
Western blot validation of four selected proteins (Fib, CA1, CHI3L1, TIMP1) identified in omic assays. A The changed folds or statuses of the four selected proteins in omics assays. B Western blots. Upper blots each panel: the extracts from Ca-T and Ca-N samples blotted with the specific antibodies against individual target proteins. Middle blots each panel: the extracts from Ca-T and Ca-N samples blotted with the antibody to β-actin. Lower blots each panel: the acetylated extracts from Ca-T and Ca-N samples blotted with the specific antibodies against individual target proteins
Involvement of the significant pathways
To analyze the involved metabolic and signaling pathways by DEPs or DEAPs in tumor tissues, Kyoto Encycloppedia of Genes and Genomes (KEGG) pathway analyses were conducted using software of KOBAS2.0. In global proteomics, 78 pathways were involved but only 4 pathways were significantly changed (P < 0.05), including mineral absorption, protein digestion and absorption, nicotinate and nicotinamide metabolism and relax in signaling pathway (Table 3). In acetylated proteomics, 37 pathways were identified, among them 3 pathways showed significance (P < 0.05), including proteoglycans in cancer, viral carcinogenesis and tight junction (Table 3).
GO ontology description
To characterize the distributions of DEPs and DEAPs in cellular processes, GO analysis was conducted and the significantly affected processes (P < 0.05) were counted. In the categories of cellular component, biological process and molecular function, the numbers of the affected processes based on global proteomics were 67, 400 and 73, while those based on acetylated proteomics were 28, 121 and 3, respectively.
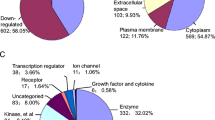
The top ten affected biological processes each category based on the P values and the involved protein numbers in global and acetylated proteomics were separately calculated. In the category of cellular component (Fig. 5A), they were extracellular region, extracellular matrix, extracellular region part, collagen-containing extracellular matrix, hemoglobin complex, intermediate filament, extracellular matrix component, supramolecular fiber, supramolecular complex and supramolecular polymer in global proteomics, while DNA packaging complex, protein-DNA complex, nucleosome, chromosomal part, chromatin, intracellular organelle, organelle, extracellular space, extracellular region and extracellular region part in acetylated proteomics. Two processes (extracellular region and extracellular region part) were identified in both global and acetylated proteomics.
In the category of biological process (Fig. 5B), the top ten affected functions were collagen fibril organization, biological adhesion, cell adhesion, skin development, peptide cross-linking, positive regulation of T cell activation, regulation of biological quality, positive regulation of leukocyte cell-cell adhesion, T cell co-stimulation and lymphocyte co-stimulation in global proteomics, while inflammatory response, negative regulation of phosphorus metabolic process, negative regulation of protein modification process, negative regulation of phosphorylation, negative regulation of protein phosphorylation, negative regulation of phosphate metabolic process, cytokine production, response to bacterium, positive regulation of cytokine production and defense response in acetylated proteomics. These top 10 processes were not overlap between two omics assays.
In the category of molecular function (Fig. 5C), the top ten affected functions were structural molecule activity, extracellular matrix structural constituent, oxygen binding, oxygen carrier activity, metal ion binding, calcium ion binding, cation binding, molecular carrier activity, heme binding, tetrapyrrole binding in global proteomics, while protein binding, extracellular matrix structural constituent conferring compression resistance, protein N-terminus binding, kinase binding, protein kinase binding, protein heterodimerization activity, ribonuclease activity, phosphoglycerate kinase activity, G-protein coupled receptor binding and Ran GTPase binding in acetylated proteomics. None of the top 10 processes overlapped in two omics assays.
Discussion
In this study the global expressing proteomic and acetylated proteomic profiles of the tumor tissues of PTC were analyzed compared to the normal tissues adjacent to the lesions. Although the data in this study are not obtained from the proteomic assay of individual cases, pooled samples from 10 cases of PTC with the same TNM stage (III) might, to some extents, balance the individual differences. More proteins and differentially expressed ones has been identified in the assay of global proteomics than that of acetylated proteomics. We believe that it is reasonable, as compared with thousands of proteins there are about 1,750 proteins acetylated nearly at 3,600 sites [13, 14]. Another possibility may drive from the preparing processes for mass spectrometry, as the enrichment process of the acetylated peptides from the tested samples is conducted after the extraction of the whole proteins, which may result in loss of some peptides. On the other hand, we have found that 75.4% of DEAPs are detectable in global proteomics, while only 20.4% DEPs are seen in acetylated proteomics. It highlights that the extraction of acetylated peptides may help to enrich those of relatively low abundance.
Obviously, there is a small portion of the differentially expressed proteins can be identified both in global and acetylated proteomics. 7 out of 40 mostly changed DEPs are detectable in acetylated proteomics and 6 of them are considered as DEAPs. 28 out of 40 mostly changed DEAPs are identifiable in global proteomics and only 6 are classified as DEPs. Only two DEAPs show different altering direction, i.e., reticulon 4 receptor-like 2 (1.579 in acetylated and 0.756 in global proteomics) and collagen alpha-2(I) chain (0.749 in acetylated and 1.263 in global proteomics). The rest of the mostly changed DEPs and DEAPs shows the same changing direction, or maintains unchanged, or is undetectable in the opposite technique.
We have also reviewed the top 10 up- and down-regulated DEPs and DEAPs in thyroid cancer and other cancers from literatures and summarized the results in Table 4. All 10 of the mostly up-regulated DEPs in this study have already addressed in other types of cancers showing overexpression, i.e., fibronectin 1 [7, 15], chitinase-3-like protein 1 [16, 17], metalloproteinase inhibitor 1 [18, 19], thrombospondin-2 [20, 21], thrombospondin type 1 [22, 23], TNC protein [24, 25], KRT1B protein, dermcidin [26], keratin, type II cytoskeletal 5 [27]. Among them, KRT1B protein, dermcidin and keratin, type II cytoskeletal 5 are not well addressed in thyroid cancer. Except UPF0764 protein and A-gamma globin Osilo variant, 8 of the top 10 down-regulated DEPs have also mentioned in other types of cancers, adhesion G-protein-coupled receptor G1 (decreased) [28], keratin-type I cytoskeletal 16 [27], Huntingtin interacting protein-1 (decreased) [29], ribosomal protein L19 (increased) [30], Beta-1,4-galactosyltransferase 1 (negatively regulates cell survival) [31], trefoil factor 3 (decreased) [32, 33], hemoglobin alpha-2 globin mutant [34], sulfotransferase [35, 36]. Contrarily, only 2 proteins, trefoil factor 3 and sulfotransferase are addressed in thyroid cancer.
Unlike the proteomic assays for cancers, the protein acetylation status in carcinomas is relatively poorly understood. Reviewing the top 10 increased and decreased DEAPs in this study from the literatures (Table 4), we find a few of proteins having their acetylated data, such as alpha-1-acid glycoprotein 2 in cystic ovarian tumors [37] and histone H2B in a bioluminescent orthotopic surgical xenograft model of ovarian cancer [38], although majority of them has been proposed to be associated with various malignant tumors. Notably, three acetylated forms of histone proteins, H2B, H2B type 1-O and liver H1e, are down-regulated in the tumor tissues. Acetylation and deacetylation of histone are the two important processes amongst the different modes of epigenetic modulation that are involved in regulating cancer initiation and development [39]. Abnormal expression of histone deacetylases (HDACs) is often reported in various types of cancers, e.g., overexpression of HDAC1-3 in ovarian cancer, overexpression of HDAC 1 and 3 in lung cancer and overexpression of HDAC2 in gastric cancer [39]. Overexpression of HDAC leads to enhanced deacetylation of histones, and subsequently increases the gravitational attraction between DNA and histones and compacts the loose nucleosome against the expression of certain genes, including some tumor suppressor genes, which makes HDAC inhibitors be potent drug molecules [40]. Low acetylated levels of different histones in tumor tissues in comparison with the adjacent normal tissues in this study may also reflect an active status of HDACs in papillary thyroid cancer, though the level of HDAC in tumor tissues seems not change significantly in comparison with that of normal tissues by global proteomics (Ca-T/ Ca-N, 0.937 in histone deacetylase and 1.065 in histone deacetylase complex subunit SAP18, respectively).
Functional GO annotation analysis and KEGG pathway analysis based on the DEPs and DEAPs show completely different pictures, in which only two biological processes are commonly seen among the top 10 involved processes in the context of three categories of GO analysis. We believe that it is because of the difference in the proteins differentially expressed in the assays of global and acetylated proteomics. Global proteomics evaluates the differences in the components and alterations of proteins among the tested samples, whereas acetylated proteomics reflects the difference in protein acetylation, one of the protein post-translational processes,. Taken the whole profiling of those two proteomics together will give us more broad view of protein alterations on the carcinogenesis of PTC.
Conclusions
In this study, we have, for the first time, screened the profiles of global and acetylated proteomics of PTC controlled with the normal tissues adjacent to the lesions in parallel. In total, 147 proteins in global proteomics and 57 proteins in acetylated proteomics in tumor tissues are considered as differentially expressed. Only small proportion of the significantly changed proteins are found both in global and acetylated proteomics, leading to only two out of the top 10 processes overlap in GO assay and no pathway overlap in KEGG pathway assay. As global proteomics evaluates the changes of protein levels and acetylated proteomics reflects the alterations of protein post-translation affecting protein activity, evaluation of the changes in those two proteomics techniques together will help to reveal the pathogenesis of PTC more comprehensively.
Availability of data and materials
The data and material are available.
Abbreviations
- Ac-CoA:
-
Acetyl-coenzyme A
- Ca-T:
-
Sample of the tumor tissue of PTC
- Ca-N:
-
Sample of the adjacent normal tissues of PTC
- DEAPs:
-
Differentially expressed acetylated peptides
- DEPs:
-
Differentially expressed proteins
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encycloppedia of Genes and Genomes
- PTC:
-
Papillary thyroid carcinoma
- PTMs:
-
Post-translational modifications
References
Dralle H, Machens A, Basa J, Fatourechi V, Franceschi S, Hay ID, et al. Follicular cell-derived thyroid cancer. Nat Rev Dis Primers. 2015;1:15077. https://doi.org/10.1038/nrdp.2015.77. Epub 2015/01/01. PubMed PMID: 27188261.
Lin JS, Bowles EJA, Williams SB, Morrison CC. Screening for thyroid cancer: updated evidence report and systematic review for the US Preventive Services Task Force. JAMA. 2017;317(18):1888–903. https://doi.org/10.1001/jama.2017.0562. Epub 2017/05/12. PubMed PMID: 28492904.
Mattiuzzi C, Lippi G. Current cancer epidemiology. J Epidemiol Glob Health. 2019;9(4):217–22. https://doi.org/10.2991/jegh.k.191008.001. Epub 2019/12/20. PubMed PMID: 31854162; PubMed Central PMCID: PMCPMC7310786.
Wiltshire JJ, Drake TM, Uttley L, Balasubramanian SP. Systematic review of trends in the incidence rates of thyroid cancer. Thyroid. 2016;26(11):1541–52. https://doi.org/10.1089/thy.2016.0100. Epub 2016/11/02. PubMed PMID: 27571228.
Kim J, Gosnell JE, Roman SA. Geographic influences in the global rise of thyroid cancer. Nat Rev Endocrinol. 2020;16(1):17–29. https://doi.org/10.1038/s41574-019-0263-x. Epub 2019/10/17. PubMed PMID: 31616074.
Kalfert D, Ludvikova M, Kholova I, Ludvik J, Topolocan O, Plzak J. Combined use of galectin-3 and thyroid peroxidase improves the differential diagnosis of thyroid tumors. Neoplasma. 2020;67(1):164–70. https://doi.org/10.4149/neo_2019_190128N86. Epub 2019/11/30. PubMed PMID: 31777257.
Sponziello M, Rosignolo F, Celano M, Maggisano V, Pecce V, De Rose RF, et al. Fibronectin-1 expression is increased in aggressive thyroid cancer and favors the migration and invasion of cancer cells. Mol Cell Endocrinol. 2016;431:123–32. https://doi.org/10.1016/j.mce.2016.05.007. Epub 2016/05/14. PubMed PMID: 27173027.
Liu YY, Morreau H, Kievit J, Romijn JA, Carrasco N, Smit JW. Combined immunostaining with galectin-3, fibronectin-1, CITED-1, Hector Battifora mesothelial-1, cytokeratin-19, peroxisome proliferator-activated receptor-{gamma}, and sodium/iodide symporter antibodies for the differential diagnosis of non-medullary thyroid carcinoma. Eur J Endocrinol. 2008;158(3):375–84. https://doi.org/10.1530/EJE-07-0492. Epub 2008/02/27. PubMed PMID: 18299472.
Prasad ML, Pellegata NS, Huang Y, Nagaraja HN, de la Chapelle A, Kloos RT. Galectin-3, fibronectin-1, CITED-1, HBME1 and cytokeratin-19 immunohistochemistry is useful for the differential diagnosis of thyroid tumors. Mod Pathol. 2005;18(1):48–57. https://doi.org/10.1038/modpathol.3800235. Epub 2004/07/24. PubMed PMID: 15272279.
Tardiolo G, Bramanti P, Mazzon E. Overview on the effects of N-acetylcysteine in neurodegenerative diseases. Molecules. 2018;23(12):3305. https://doi.org/10.3390/molecules23123305. Epub 2018/12/16. PubMed PMID: 30551603; PubMed Central PMCID: PMCPMC6320789.
Li D, Yang Y, Wang S, He X, Liu M, Bai B, et al. Role of acetylation in doxorubicin-induced cardiotoxicity. Redox Biol. 2021;46:102089. https://doi.org/10.1016/j.redox.2021.102089. Epub 2021/08/08. PubMed PMID: 34364220; PubMed Central PMCID: PMCPMC8350499.
Gil J, Ramirez-Torres A, Encarnacion-Guevara S. Lysine acetylation and cancer: a proteomics perspective. J Proteomics. 2017;150:297–309. https://doi.org/10.1016/j.jprot.2016.10.003. Epub 2016/10/25. PubMed PMID: 27746255.
Deng W, Wang C, Zhang Y, Xu Y, Zhang S, Liu Z, et al. GPS-PAIL: prediction of lysine acetyltransferase-specific modification sites from protein sequences. Sci Rep. 2016;6:39787. https://doi.org/10.1038/srep39787. Epub 2016/12/23. PubMed PMID: 28004786; PubMed Central PMCID: PMCPMC5177928.
Carrico C, Meyer JG, He W, Gibson BW, Verdin E. The mitochondrial acylome emerges: proteomics, regulation by sirtuins, and metabolic and disease implications. Cell Metab. 2018;27(3):497–512. https://doi.org/10.1016/j.cmet.2018.01.016. Epub 2018/03/08. PubMed PMID: 29514063; PubMed Central PMCID: PMCPMC5863732.
Zhou Y, Cao G, Cai H, Huang H, Zhu X. The effect and clinical significance of FN1 expression on biological functions of gastric cancer cells. Cell Mol Biol (Noisy-le-grand). 2020;66(5):191–8 Epub 2020/10/13. PubMed PMID: 33040835.
Cheng SP, Lee JJ, Chang YC, Lin CH, Li YS, Liu CL. Overexpression of chitinase-3-like protein 1 is associated with structural recurrence in patients with differentiated thyroid cancer. J Pathol. 2020;252(2):114–24. https://doi.org/10.1002/path.5503. Epub 2020/07/03. PubMed PMID: 32613636.
Salomon J, Piotrowska A, Matusiak L, Dziegiel P, Szepietowski JC. Chitinase-3-like protein 1 (YKL-40) expression in squamous cell skin cancer. Anticancer Res. 2018;38(8):4753–8. https://doi.org/10.21873/anticanres.12783. Epub 2018/08/01. PubMed PMID: 30061245.
Lorenc Z, Waniczek D, Lorenc-Podgorska K, Krawczyk W, Domagala M, Majewski M, et al. Profile of Expression of Genes Encoding Matrix Metallopeptidase 9 (MMP9), Matrix Metallopeptidase 28 (MMP28) and TIMP Metallopeptidase Inhibitor 1 (TIMP1) in Colorectal Cancer: Assessment of the Role in Diagnosis and Prognostication. Med Sci Monit. 2017;23:1305–11. https://doi.org/10.12659/msm.901593. Epub 2017/03/16. PubMed PMID: 28293015; PubMed Central PMCID: PMCPMC5363457.
Bumber B, Marjanovic Kavanagh M, Jakovcevic A, Sincic N, Prstacic R, Prgomet D. Role of matrix metalloproteinases and their inhibitors in the development of cervical metastases in papillary thyroid cancer. Clin Otolaryngol. 2020;45(1):55–62. https://doi.org/10.1111/coa.13466. Epub 2019/10/28. PubMed PMID: 31646745.
Byrling J, Kristl T, Hu D, Pla I, Sanchez A, Sasor A, et al. Mass spectrometry-based analysis of formalin-fixed, paraffin-embedded distal cholangiocarcinoma identifies stromal thrombospondin-2 as a potential prognostic marker. J Transl Med. 2020;18(1):343. https://doi.org/10.1186/s12967-020-02498-3. Epub 2020/09/06. PubMed PMID: 32887625; PubMed Central PMCID: PMCPMC7487897.
de Fraipont F, Nicholson AC, Feige JJ, Van Meir EG. Thrombospondins and tumor angiogenesis. Trends Mol Med. 2001;7(9):401–7. https://doi.org/10.1016/s1471-4914(01)02102-5. Epub 2001/09/01. PubMed PMID: 11530335.
Sid B, Langlois B, Sartelet H, Bellon G, Dedieu S, Martiny L. Thrombospondin-1 enhances human thyroid carcinoma cell invasion through urokinase activity. Int J Biochem Cell Biol. 2008;40(9):1890–900. https://doi.org/10.1016/j.biocel.2008.01.023. Epub 2008/03/07. PubMed PMID: 18321763.
Yee KO, Connolly CM, Duquette M, Kazerounian S, Washington R, Lawler J. The effect of thrombospondin-1 on breast cancer metastasis. Breast Cancer Res Treat. 2009;114(1):85–96. https://doi.org/10.1007/s10549-008-9992-6. Epub 2008/04/15. PubMed PMID: 18409060; PubMed Central PMCID: PMCPMC2631620.
Gocheva V, Naba A, Bhutkar A, Guardia T, Miller KM, Li CM, et al. Quantitative proteomics identify Tenascin-C as a promoter of lung cancer progression and contributor to a signature prognostic of patient survival. Proc Natl Acad Sci USA. 2017;114(28):E5625–34. https://doi.org/10.1073/pnas.1707054114. Epub 2017/06/28. PubMed PMID: 28652369; PubMed Central PMCID: PMCPMC5514763.
Tseleni-Balafouta S, Gakiopoulou H, Fanourakis G, Voutsinas G, Balafoutas D, Patsouris E. Tenascin-C protein expression and mRNA splice variants in thyroid carcinoma. Exp Mol Pathol. 2006;80(2):177–82. https://doi.org/10.1016/j.yexmp.2005.09.006. Epub 2005/11/02. PubMed PMID: 16259977.
Stewart GD, Skipworth RJ, Ross JA, Fearon K, Baracos VE. The dermcidin gene in cancer: role in cachexia, carcinogenesis and tumour cell survival. Curr Opin Clin Nutr Metab Care. 2008;11(3):208–13. https://doi.org/10.1097/MCO.0b013e3282fb7b8d. Epub 2008/04/12. PubMed PMID: 18403914.
Schumann H, Roth W, Has C, Volz A, Erfurt-Berge C, Magin TM, et al. Verrucous carcinoma in epidermolysis bullosa simplex is possibly associated with a novel mutation in the keratin 5 gene. Br J Dermatol. 2012;167(4):929–36. https://doi.org/10.1111/j.1365-2133.2012.11075.x. Epub 2012/05/30. PubMed PMID: 22639907.
Yang L, Xu L. GPR56 in cancer progression: current status and future perspective. Future Oncol. 2012;8(4):431–40. https://doi.org/10.2217/fon.12.27. Epub 2012/04/21. PubMed PMID: 22515446.
Hsu CY, Lin CH, Jan YH, Su CY, Yao YC, Cheng HC, et al. Huntingtin-interacting protein-1 is an early-stage prognostic biomarker of lung adenocarcinoma and suppresses metastasis via Akt-mediated epithelial-mesenchymal transition. Am J Respir Crit Care Med. 2016;193(8):869–80. https://doi.org/10.1164/rccm.201412-2226OC. Epub 2015/11/26. PubMed PMID: 26595459.
Bee A, Ke Y, Forootan S, Lin K, Beesley C, Forrest SE, et al. Ribosomal protein l19 is a prognostic marker for human prostate cancer. Clin Cancer Res. 2006;12(7 Pt 1):2061–5. https://doi.org/10.1158/1078-0432.CCR-05-2445. Epub 2006/04/13. PubMed PMID: 16609016.
Tang W, Weng S, Zhang S, Wu W, Dong L, Shen X, et al. Direct interaction between surface beta1,4-galactosyltransferase 1 and epidermal growth factor receptor (EGFR) inhibits EGFR activation in hepatocellular carcinoma. Biochem Biophys Res Commun. 2013;434(3):449–54. https://doi.org/10.1016/j.bbrc.2013.03.094. Epub 2013/04/16. PubMed PMID: 23583406.
Abols A, Ducena K, Andrejeva D, Sadovska L, Zandberga E, Vilmanis J, et al. Trefoil factor 3 is required for differentiation of thyroid follicular cells and acts as a context-dependent tumor suppressor. Neoplasma. 2015;62(6):914–24. https://doi.org/10.4149/neo_2015_111. Epub 2015/10/16. PubMed PMID: 26458316.
Espinoza I, Agarwal S, Reddy A, Shenoy V, Subramony C, Sakiyama M, et al. Expression of trefoil factor 3 is decreased in colorectal cancer. Oncol Rep. 2021;45(1):254–64. https://doi.org/10.3892/or.2020.7829. Epub 2020/11/20. PubMed PMID: 33210724; PubMed Central PMCID: PMCPMC7716703.
Li X, Wu Z, Wang Y, Mei Q, Fu X, Han W. Characterization of adult alpha- and beta-globin elevated by hydrogen peroxide in cervical cancer cells that play a cytoprotective role against oxidative insults. PLoS One. 2013;8(1):e54342. https://doi.org/10.1371/journal.pone.0054342. Epub 2013/01/26. PubMed PMID: 23349856; PubMed Central PMCID: PMCPMC3547883.
Kalathas D, Theocharis DA, Bounias D, Kyriakopoulou D, Papageorgakopoulou N, Stavropoulos MS, et al. Alterations of glycosaminoglycan disaccharide content and composition in colorectal cancer: structural and expressional studies. Oncol Rep. 2009;22(2):369–75. Epub 2009/07/07. PubMed PMID: 19578779.
Schulten HJ, Al-Mansouri Z, Baghallab I, Bagatian N, Subhi O, Karim S, et al. Comparison of microarray expression profiles between follicular variant of papillary thyroid carcinomas and follicular adenomas of the thyroid. BMC Genomics. 2015;16(Suppl 1):S7. https://doi.org/10.1186/1471-2164-16-S1-S7. Epub 2015/04/30. PubMed PMID: 25923053; PubMed Central PMCID: PMCPMC4315165.
Kolwijck E, Engelke UF, van der Graaf M, Heerschap A, Blom HJ, Hadfoune M, et al. N-acetyl resonances in in vivo and in vitro NMR spectroscopy of cystic ovarian tumors. NMR Biomed. 2009;22(10):1093–9. https://doi.org/10.1002/nbm.1417. Epub 2009/07/14. PubMed PMID: 19593761.
Helland O, Popa M, Bischof K, Gjertsen BT, McCormack E, Bjorge L. The HDACi Panobinostat shows growth inhibition both in vitro and in a bioluminescent orthotopic surgical xenograft model of ovarian cancer. PLoS One. 2016;11(6):e0158208. https://doi.org/10.1371/journal.pone.0158208. Epub 2016/06/29. PubMed PMID: 27352023; PubMed Central PMCID: PMCPMC4924861.
Shetty MG, Pai P, Deaver RE, Satyamoorthy K, Babitha KS. Histone deacetylase 2 selective inhibitors: a versatile therapeutic strategy as next generation drug target in cancer therapy. Pharmacol Res. 2021;170:105695. https://doi.org/10.1016/j.phrs.2021.105695. Epub 2021/06/04. PubMed PMID: 34082029.
Ramaiah MJ, Tangutur AD, Manyam RR. Epigenetic modulation and understanding of HDAC inhibitors in cancer therapy. Life Sci. 2021;277:119504. https://doi.org/10.1016/j.lfs.2021.119504. Epub 2021/04/20. PubMed PMID: 33872660.
Acknowledgements
Not applicable.
Funding
This work was supported by National Key R&D Program of China (2020YFE0205700), Grants (2021SKLID101, 2019SKLID501, 2019SKLID307) from the State Key Laboratory for Infectious Disease Prevention and Control, China CDC.
Author information
Authors and Affiliations
Contributions
WW contributed to cases diagnosis and sample preparations. CDD and SQ contributed to material preparations, data collections. WYZ, SYT and XGH contributed to sample preparations and data analysis. WW, SQ and DXP drafted the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Written consents for further investigation and publication were obtained from the patients or the patients’ relatives, respectively. Usage of the stored human samples in this study was approved by the Ethical Committees of Peking University Cancer Hospital & Institute and National Institute for Viral Disease Prevention and Control, China CDC.
Consent for publication
Written informed consent for publication of their clinical details and/or clinical images was obtained from the patient. A copy of the consent form is available form is available for review by the Editor of this journal.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplement Figure 1.
Protein quality control. A. Protein quantitative standard curve. B. Protein SDS-PAGE gel map. C. Peptide length distribution diagram.
Additional file 2: Supplemental Table 1.
The ages and TNM stages of 10 enrolled patients of PTC
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wei, W., Wu, Y., Chen, DD. et al. Proteomics profiling for the global and acetylated proteins of papillary thyroid cancers. Proteome Sci 21, 6 (2023). https://doi.org/10.1186/s12953-023-00207-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12953-023-00207-8