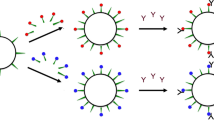
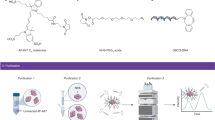
Abstract
Background
Various bacteria and archaea, including halophilic archaeon Halobacterium sp. NRC-1 produce gas vesicle nanoparticles (GVNPs), a unique class of stable, air-filled intracellular proteinaceous nanostructures. GVNPs are an attractive tool for biotechnological applications due to their readily production, purification, and unique physical properties. GVNPs are spindle- or cylinder-shaped, typically with a length of 100 nm to 1.5 μm and a width of 30–250 nm. Multiple monomeric subunits of GvpA and GvpC proteins form the GVNP shell, and several additional proteins are required as minor structural or assembly proteins. The haloarchaeal genetic system has been successfully used to produce and bioengineer GVNPs by fusing several foreign proteins with GvpC and has shown various applications, such as biocatalysis, diagnostics, bioimaging, drug delivery, and vaccine development.
Results
We demonstrated that native GvpC can be removed in a low salt buffer during the GVNP purification, leaving the GvpA-based GVNP's shell intact and stable under physiological conditions. Here, we report a genetic engineering and chemical modification approach for functionalizing the major GVNP protein, GvpA. This novel approach is based on combinatorial cysteine mutagenesis within GvpA and genetic expansion of the N-terminal and C-terminal regions. Consequently, we generated GvpA single, double, and triple cysteine variant libraries and investigated the impact of mutations on the structure and physical shape of the GVNPs formed. We used a thiol–maleimide chemistry strategy to introduce the biotechnological relevant activity by maleimide-activated streptavidin–biotin and maleimide-activated SpyTag003-SpyCatcher003 mediated functionalization of GVNPs.
Conclusion
The merger of these genetic and chemical functionalization approaches significantly extends these novel protein nanomaterials' bioengineering and functionalization potential to assemble catalytically active proteins, biomaterials, and vaccines onto one nanoparticle in a modular fashion.
Graphical Abstract

Similar content being viewed by others
Introduction
Self-assembling protein nanoparticles (NPs) like virus-like particles (VLPs) and gas vesicle nanoparticles (GVNPs) have gained extensive interest as next-generation functional bionanomaterials because of their readily production and unique properties, including modular nature, biocompatibility, and higher-order complexity [1,2,3]. VLPs are not contagious viral protein complexes with sizes ranging from 20–200 nm [4]. GVNPs are relatively large (widths up to 250 nm and lengths up to 1.5 µm), rigid, low-density hollow intracellular air-filled organelles solely made of protein (lipid or carbohydrate-free), exceptionally stable, inert, nontoxic, self-adjuvants, accessible to bioengineering, and can easily be purified by simple flotation due to their buoyancy [5,6,7]. The interior surface of the GVNP envelope is hydrophobic and, therefore, water-impermeable, while gases can be exchanged with the surrounding [8,9,10]. Various aquatic prokaryotes naturally produce GVNPs, enabling them to float in light- and oxygen-rich upper water layers by increasing the microorganism's surface area-to-volume ratio in response to environmental cues [2, 3, 6, 10, 11]. Furthermore, over the past decades, GVNPs have been used in various applications, such as biocatalysis [7], endotoxemia treatment [12], contrast-enhanced ultrasound [13], hyperpolarized xenon magnetic resonance imaging [14], 3D printing [15], and as biocompatible vaccine vectors in vaccine research [9, 12, 16,17,18].
In-depth studies on haloarchaeal GVNP gene cluster composition and GVNP expression were performed with GVNPs from Halobacterium sp. NRC-1 and Haloferax mediterranei [7, 9, 17,18,19,20]. Depending on the species, 8–14 proteins (Gvp proteins) are required for GVNP biogenesis [6]. In Halobacterium sp. NRC-1, 14 gvp genes contribute to the biosynthesis of GVNPs [9, 19]. The GVNPs from Halobacterium sp. NRC-1 is valuable for various processes, although one significant challenge is the slow growth rate, expensive antibiotics, and inconsistent induction systems of the Halobacterium expression host [6, 15, 21, 22]. Therefore, we recently developed a new, efficient haloarchaeal GVNP expression system using a Haloferax volcanii as a host, with significantly short growth times that enable the production of haloarchaeal GVNP fusions on a larger scale without the need for antibiotics [15]. Moreover, H. volcanii has well-developed microbiological and molecular genetics techniques [23, 24], including a facile DNA transformation system, shuttle plasmids, and selectable markers [25, 26]. The fragile nature of the S-layer cell walls of H. volcanii allows for efficient lysis in an aqueous solution, releasing cellular proteins and reducing the cost of protein purification [27,28,29,30].
Haloarchaeal GVNP biogenesis genes are organized into two directionally opposing clusters, gvpDEFGHIJKLM upstream and gvpACNO downstream [6, 7, 9]. GvpA and GvpC are the major components of the GVNP complex. The GvpA protein (76 aa) forms the main ribbed structure, and the GvpC protein (380 aa) forms an exterior mesh on the GVNPs surface [6, 9, 11]. The haloarchaeal GVNPs have been bioengineered by fusing several foreign proteins with or within the GvpC protein [7, 9, 12, 16,17,18, 31]. However, GVNPs from Halobacterium sp. NRC-1 tends to lose the GvpC protein during the purification process when performed at lower saline conditions, resulting in the exposure of the ribbed structure formed by GvpA [32]. Pfeifer and co-workers conducted a mutagenesis study on GvpA and showed GvpA viability for the genetic engineering [33, 34].
Inspired by the possibility of bioengineering GvpA, we choose a combinatorial approach to functionalize the GVNPs by the site-specific introduction of cysteines, due to the natural deficiency in GvpA, to facilitate thiol–maleimide reactions. We constructed single, double, and triple variant libraries by selective substitutions of alanine and serine residues to cysteine in exposed loops of GvpA. Further, we extended GvpA by introducing cysteine with linkers at the N- and C-terminus. We examined the impact of insertion and mutations on the structure and physical shape of the GVNPs formed. Additionally, we report the functionalization of the bioengineered GVNPs by the SpyTag003-SpyCatcher003 system [35] and streptavidin to enable the attachment of any recombinant protein conjugated with a biotin tag, supported by thiol–maleimide reactions. These SpyTag003-SpyCatcher003 and streptavidin–biotin GVNP conjugates are diverse, efficient, and powerful tools for GVNPs biotechnological application.
Materials and methods
Materials
Gibson Assembly Cloning Kit, restriction enzymes, and Q5® High-Fidelity DNA polymerase were purchased from New England Biolabs (Ipswich, MA, USA). All other chemicals and solvents were purchased from Sigma-Aldrich (St. Louis, MO, USA). Escherichia coli (E. coli) One Shot® TOP10 chemical competent cells and Measure-IT™ thiol assay kits were purchased from Invitrogen (Karlsbad, CA, USA). The E. coli BL21 (DE3) strain has been purchased from Agilent Technologies (Santa Clara, CA, USA), and electrocompetent cells have been prepared according to an in-house protocol. Halobacterium sp. NRC-1 was obtained from Carolina Biological Supply (Burlington, NC, USA). We used Haloferax volcanii H1895 and its corresponding vector pTA963 in the study [24, 25]. N-terminal maleimides modified SpyTag003 peptide (GGSGGGGRGVPHIVMVDAYKRYK with ≥ 98% purity) was synthesized by GenScript Biotech Corporation, HK.
GVNP plasmid generation and transformation in Haloferax volcanii
Halobacterium sp. NRC-1 was cultured in a CM+ medium containing 4.3 M NaCl and trace metals at 42 °C with shaking, as previously described [7, 15,16,17, 36]. The GVNP operon from Halobacterium sp. NRC-1 (ATCC 700,922 / JCM 11,081) [37] was PCR amplified from the genome and cloned using the Gibson Assembly Cloning Kit into pTA963 to generate pTA963.GVNP expression plasmid (Additional file 1: Table S1). For the mutagenesis study, the GvpA gene was replaced by a codon-optimized mutated GvpA synthetic gene (GenScript Biotech Corporation, HK or Twist Bioscience, USA). The construct was validated by PCR amplification and DNA sequencing. GVNP-containing vectors were transformed into the Haloferax volcanii H1895 using PEG/EDTA method [21].
Saturation mutagenesis for GvpA
Ab initio protein prediction for gvpA1 (gas vesicle structural protein 1) from Halobacterium salinarum (strain ATCC 700,922/JCM 11,081/NRC-1, Uniport: P08958) was performed by I-TASSER [38,39,40,41]. Cysteine substitutions were made based on Grantham's distance [42] and Sneath's index [43] to create a combinatorial cysteine mutant library. The initial substituted positions were further analyzed by predicting pairs of residues that will likely form a disulfide bond if mutated to cysteines [44], and dimerization analysis. Based on the GvpA of Bacillus megaterium (PDB: 7R1C), a new model was performed by I-TASSER, creating a more up-to-date model.
Expression of GVNPs in Haloferax volcanii
H. volcanii and derivatives were cultured in the Hv-YPC medium containing 4.0 M NaCl at 45 °C with shaking as previously described [25, 28,29,30, 45]. For solid media, 2% (w/v) agar was added. The processes for producing and culturing GVNPs were performed as previously described [7, 15, 17, 45]. H. volcanii lawns or floating cells were grown for 2 weeks and lysed osmotically with PBS solution (137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate dibasic, and 2 mM potassium phosphate monobasic, pH 7.4) containing 10 mM MgSO4 and 50 μg/mL of DNase I (Sigma-Aldrich, USA). The cell lysate suspension was incubated for 1 h at 37 °C before overnight centrifugation at 60 ×g in a swinging bucket rotor in a Sorvall XFR centrifuge (Thermo Scientific, Rockford, IL) to accelerate the floatation of the GVNPs. Intact GVNPs were collected and resuspended in PBS solution, then floated by overnight centrifugation and harvested. This floatation procedure was repeated until a white, milky suspension of GVNPs was obtained. GVNP concentration was quantified via Nanodrop by measuring 2 µl of sonicated and broken GVNPs.
Dynamic light scattering (DLS)
GVNPs were characterized by dynamic light scattering (DLS) in triplicate using a ZSPNano Zetasizer (Malvern Inc., Malvern, UK) for stability, temperature (20–60 °C), salt (0–4 M NaCl), pH (5.0–12.0) and organic solvents (10%, v/v of glycerol, ethanol, and DMSO). The particle number, diameter, and polydispersity of GVNP populations were determined, and the size distribution values were represented as mean diameter and polydispersity index (PDI). For thermal stability, GVNP suspensions (purified in PBS/2 M NaCl) were incubated in the water bath at different temperatures for up to 48 h. Samples were removed at fixed intervals, cooled at room temperature, and analyzed by dynamic light scattering. To determine the influence of pH and salt concentrations, GVNPs purified in PBS/2 M NaCl were diluted 100 times in different pH/salt concentration buffers. The results were transformed into residual GVNPs, with the control sample on each measurement being set to 100%.
Western blotting analysis
The methods used were those previously described [7, 17]. Briefly, the SDS-PAGE analysis was performed using the precast Novex® Tris-glycine gels (4–20%, Invitrogen, Carlsbad, CA, USA). Proteins were then transferred to 0.45 μm nitrocellulose membranes (Millipore Corp., Boston, MA). Membranes were blocked for 30 min in Pierce Fast Blocking Buffer (Thermo Fisher Scientific), incubated in blocking buffer supplemented with Anti-GFP Polyclonal Antibody (Thermo Fisher Scientific) or Anti-GvpC antibodies (GenScript, USA), followed by three washing steps with TBST and then incubation with Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (Invitrogen, Catalog # A-11008).
Tryptic digest and LC–MS/MS analysis
The identification of corresponding peptides was performed by LC-MS/MS analysis. The sample (10 μg) was digested with trypsin using the FASP protocol [46]. Peptides were measured using an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) and analyzed by MASCOT v2.3 (Matrix Sciences Ltd, UK).
Fluorescence microscopy imaging
Fluorescence imaging of GVNPs was performed in microfluidic chambers (Ibidi GmbH, sticky-slide VI 0.4) with a cleaned coverslip attached to the bottom. For imaging of GVNP-sfGFP, the coverslip was surface-coated with protein A by incubating with 10 μg ml−1 Protein A (Invitrogen) in Hank's Balanced Salt Solution (HBSS) and incubated with 0.1 μg ml−1 anti-GvpA antibody (GenScript Biotech Corporation, HK) at 4 °C for 1 h. Finally, the chamber was incubated with 1% (w/v) casein to minimize the nonspecific binding. GVNPs were taken from the concentrated floating portion of the purified GVNP solution and diluted 100 times by PBS or PBS containing 1 M or 2 M NaCl. Then the sample was injected into the chamber. The chamber was flipped with the coverslip side up to promote the interaction with the coated coverslip surface with floating GVNPs. The solution in the chamber was exchanged to remove the unbonded excess after 30 min of incubation.
For the imaging of GVNP-SpyCatcher003-mKate2 (SC.mKate2), the coverslip was incubated with 0.1 mg ml−1 poly-L-ornithine hydrobromide (Sigma-Aldrich) for two hours to coat the surface. The GVNPs were taken from the concentrated floating portion of the purified GVNP solution and diluted 100 times. Then the sample was injected into the chamber. The chamber was flipped with the coverslip side up to promote the GVNPs attaching to the coverslip. The solution in the chamber was exchanged to remove the unbonded excess after 5 min of incubation.
Fluorescence imaging experiments were conducted using a home-built wide-field fluorescence microscopy setup with an inverted microscope frame (Olympus, IX71) equipped with a high NA phase-contrast objective lens (Olympus, 100 × oil Ph3 NA = 1.30). A continuous-wave (CW) solid-state laser operating at 488 nm (Cobolt, MLD) was used to excite sfGFP, and 561 nm (Cobolt) was used for mKate2. Imaging was performed by a dichroic mirror (FF506-Di03-25 × 36 for 488 nm excitation and FF580-FDi01 for 561 nm excitation) and mission bandpass filter (FF01-550/88 for 488 nm excitation and FF01-630/92–25 for 561 nm). A fluorescence signal was detected by an EMCCD camera (Andor Technology, iXon3 897) with an exposure time of 300 ms (EM gain of 300). Phase contrast image was measured by the same configuration using a Ph3 condenser annulus plate.
Electron microscopy measurements
For cryogenic transmission electron microscopy (cryo-TEM), GVNP samples were vitrified by ultra-rapid freezing. Typically, 3 μl of the sample was put on a pre-glow-discharged copper grid coated with a holy carbon layer and blotted from both sides. The blotted grid was plunge-frozen in liquid ethane using Vitrobot Mark IV (ThermoFisher Scientific) and transferred into liquid nitrogen. Samples imaging was performed on Titan Krios G2 (Thermo Fisher Scientific) equipped with a K2 camera (Gatan) placed at the end of the energy filter (Gatan) and operated at 300 kV. Typically, electron doses of ∼50 electrons/ Å2 were used.
For room-temperature (RT-)TEM, 4 μl of the purified GVNPs sample was drop casted on pre-glow-discharged carbon film copper grids (Electron Microscopy Sciences) and incubated for 1 min for adsorption. After that, the sample was removed with filter paper. Negative staining was performed by adding 4 µl of 2% uranyl acetate for 1 min. Afterward, the uranyl acetate stain was removed with filter paper, and the grids were air-dried. Images were taken in different magnifications with a US4000 CCD camera (Gatan, Inc., USA) or OneView camera (Gatan, Inc., USA) on a Titan 80–300 (Thermo Fisher Scientific). The lengths and diameters of 25 GVNPs were measured manually in DigitalMicrograph GMS3 (Gatan, Inc., USA) with a selected library.
Thiol assay
GVNPs variants (20 µM each) were treated with a 50-fold molar excess (1 mM) of a reducing agent, Tris(2-cyanoethyl)phosphine (TCP), for two hours at room temperature with gentle mixing and then dialyzed overnight in 1000 kDa MWCO tubing against PBS. Following the established procedures, the free cysteine GVNPs variant is estimated in triplicate with the Measure-iT™ Thiol Assay kit (M30550, Invitrogen). Triplicate 10 μL GVNP variant samples were assayed; fluorescence was measured at 490/520 nm. The variation (CV) of replicate samples was < 7%.
Thiol-Maleimide binding
The maleimide-cysteine coupling reaction efficiency of each GVNP-producing variant in 50 mM phosphate buffer, pH 7.0, is tested with thiol-reactive Alexa Fluor 647 C2 maleimide (Thermo Scientific). In this reaction, we mixed GVNPs GvpA variants with the reagent (1:10 mol ratio of estimated cysteine). The reaction proceeded at 4 °C overnight with gentle mixing protected from light. Then the unreacted Alexa Fluor 647 C2 maleimide was removed by overnight dialysis in 1000 kDa MWCO tubing against PBS and three repeated centrifugally assisted flotations. GVNPs retained their milky white appearance during the functionalization process. Labeling efficiencies were assessed by measuring the fluorescence (Ex. 640/ Em. 671 nm) of the GVNP samples.
Homogeneous fluorescence assay for biotin binding on streptavidin-conjugated GVNPs
The determination of accessible and functional binding pockets of streptavidin (SAV) was done by a fluorescence titration assay using the biotin-4-fluorescein (B4F) dye (Sigma-Aldrich) [47]. The free SAV binding pockets were determined by quenching the fluorescence of the B4F dye upon binding to the SAV pocket. A standard curve was generated using the known amount of commercial SAV protein from Streptomyces avidinii (Sigma-Aldrich). The "reverse titration" assay in triplicate was set up in a 96-well plate to estimate the streptavidin binding with GVNPs (20 μM). 20 μl of diluted (60x) streptavidin-conjugated GVNPs were mixed with a fixed amount (8 nM) of B4F (10 μl of 160 nM) and filled up to 200 μl with assay buffer B. After mixing and incubating for 10 min at RT, the fluorescence was measured using Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek, Winooski, VT) according to standard procedure (490 nm/525 nm, gain 100).
Thiol-maleimide SpyCatcher003 binding
For thiol-maleimide SpyCatcher003 binding, we mixed GvpA.C variants with the maleimide-modified SpyTag003 peptide (1:10 mol ratio of estimated cysteine) for two hours with gentle mixing at room temperature.
SpyCatcher003 proteins expression
The SpyCatcher003-mKate2 and SpyCatcher003-Esterase amino acid sequences have been codon optimized for E. coli and synthesized by Twist Bioscience, USA, and afterward cloned into pET29b vector (Additional file 1: Figure S1). The respective pET29b_SpyCatcher003-mKate2 and pET29b_SpyCatcher003-Esterase plasmids were transformed into electrocompetent E. coli BL21 (DE3). A fresh colony was used to inoculate an overnight LB medium (15 mL) supplemented with kanamycin (50 μg mL−1) grown at 37 °C at 180 rpm. 10 mL of the overnight culture was used to inoculate 1 L of LB medium containing kanamycin (50 μg mL−1) in a 2 L Erlenmeyer flask at 37 °C at 180 rpm. The recombinant protein expression was induced by adding isopropyl-β-D-1-thiogalactopyranoside (IPTG) (0.5 mM, final) when OD600 reached 0.6–0.8. The temperature was lowered to 25 °C, and the culture was incubated for 16 h. The cells were harvested by centrifugation at 4 °C (4000 g, 30 min) and resuspended (1 g in 10 mL) in lysis buffer (100 mM Tris/HCl pH 7.4, 100 mM NaCl, 20 mM imidazole). Before lysis, Pierce™ Protease Inhibitor EDTA-free tablet (Thermo Scientific) was added together with DNase (Sigma Aldrich). Cell disruption occurred using a Cell Disruptor (Constant System Inc.). After 4 °C centrifugation (16,000 g, 20 min), the cleared lysate was used for protein purification by Ni–NTA agarose column (5 mL, GE Healthcare). The protein was eluted by gradient elution with elution buffer (100 mM Tris/HCl pH 7.4, 100 mM NaCl, 500 mM imidazole). The collected fractions were concentrated using an Amicon® Ultra Centrifugal filter (10,000 MWCO). The purified protein was overnight dialyzed against storage buffer (50 mM Tris/HCl pH 7.4, 50 mM NaCl, 5% glycerol) and aliquoted and stored at − 20 °C.
Biotinylation of horseradish peroxidase
Thermo Scientific Pierce Horseradish Peroxidase (HRP) (Cat. 31,490) was biotinylated with the EZ-Link™ Sulfo-NHS-LC-Biotinylation Kit (Cat. 21,435) according to the supplied procedure. In short, the Sulfo-NHS-LC-Biotin was equilibrated to room temperature, 5 mg of HRP protein was resuspended in 2 mL of PBS, and 30 µl of 10 mM Sulfo-NHS-LC-Biotin was added. The reaction was incubated on ice for two hours. Afterward, Thermo Scientific Zeba Spin Desalting Column was used for separating unreacted NHS-LC-Biotin from biotinylated-HRP.
Horseradish peroxidase assay
The activity of the horseradish peroxidase (HP) enzyme was determined calorimetrically using 0.3% (w/w) hydrogen peroxide solution (H2O2), 2.5 mM 4-aminoantipyrine (4-AAP), phenol [48, 49]. Measurements were carried out at room temperature (25 °C) in 200 mM potassium phosphate buffer, pH 7.0, using an ultraviolet (UV)-visible spectrophotometer (Cary 60, Agilent, Santa Clara, CA, USA). We calculated ∆A510/minute from the linear portion of the curve. The decomposition of H2O2 in μM per minute was referred to in Units. The specific horseradish peroxidase activity is expressed as a unit per milligram of GVNP.
Esterase activity assay
Esterase activity was measured at 400 nm by spectrophotometer (Cary 60, Agilent, Santa Clara, CA, USA) using 50 mM p-nitrophenyl butyrate in 100 mM sodium phosphate buffer pH 7.2 with 150 mM NaCl at 37 °C [50]. One unit of enzyme activity was defined as the amount of enzyme producing one nanomole of 4-nitrophenol per min. The specific esterase activity is expressed as a unit per milligram of GVNP.
Result and discussion
Gas vesicle nanoparticles (GVNPs) expression in Haloferax volcanii
In Halobacterium sp. NRC-1, GvpA, and GvpC are the major components of the GVNP membrane. The GvpC protein is located on the outer surface of GVNPs and functions as a "molecular glue" to enhance membrane stability [6, 7, 9]. These characteristics are desirable and potentially crucial for protein display on the surface. Previous studies have suggested that the GvpC protein is sufficiently flexible in tolerating exogenous sequence inserts [7, 16, 20]. We inserted the Haloarchaeal codon-optimized gene of Superfolder Green Fluorescent Protein (sfGFP) into a loop of the acidic tail of the gvpC gene (Additional file 1: Figure S2) and expressed it with the GVNP operon in Haloferax volcanii [15]. sfGFP_GVNPs expressing H. volcanii cells appear pink and opaque on the plate compared to wild-type red transparent colonies (Additional file 1: Figure S3). The fluorescent signal can be detected in H. volcanii cells (Fig. 1A). When cells were grown in liquid culture and left to stand for 1-week, buoyant cells were observed floating at the top (Fig. 1B). Cylinder- or spindle-shaped GVNPs with 30 to 250 nm widths and 40 nm to 1.5 μm lengths with a mean diameter of 255 nm were observed by Cryo-EM (Fig. 1C) and dynamic light scattering (Fig. 1D).
Establishing a purification scheme for GvpA-only GVNPs
GvpC is an exceptionally acidic (pI 3.57) protein and requires salt for stability. Western blot analysis and fluorescence imaging of GvpC.sfGFP fused GVNPs revealed that GvpC.sfGFP quickly washes off the GVNP surface when GVNPs were purified with a low salt buffer such as PBS. The addition of 1–2 M NaCl to PBS significantly stabilizes the GVNPs during purifications (Fig. 2).
Effect of salt concentrations on GVNP conjugates (GVNP-sfGFP) purification. A Schematic illustration describing the procedure, B Western blot analysis of GVNPs from each washing condition, C Schematic illustration describing the sample preparation procedure to immobilize only GVNPs on coverslip surfaces, D sf-GFP fluorescence image of immobilized GVNPs from each washing condition. A small sample was taken from each condition of buffer (PBS, 1 M salt in PBS, 2 M salt in PBS) and the number of washings (1 to 3) and immobilized on the slide chamber. All the fluorescent images were obtained with the same condition (e.g., laser excitation power, detection exposure, etc.). Image contrast is unified for comparison among conditions (image scale 42 \(\mu m\) × 42 \(\mu m\)). Phase-contrast imaging measurement was also performed on the same region of the sample to confirm the presence of GVNPs on the sample
Characterization of GvpA-only GVNPs
GvpC protein is reported to stabilize GVNP's exterior surface wall [6]. GvpC was stripped off from the GVNPs, without collapse, by washing with low-salt solutions (Fig. 2, Additional file 1: Figure S4A). Removal of GvpC in the case of cyanobacterial GVNPs reported a marked decrease in the critical collapse pressure of the GVNPs [5, 51, 52], suggesting GvpC provides structural support and removal leads to reducing surface tension pressures. We utilized dynamic light scattering (DLS) to characterize the GVNPs purified in 2 M NaCl (containing GvpC) and PBS (GvpA-only) for salt, thermal, pH, and organic solvent stability (Additional file 1: Figure S4, Fig. 3). GVNPs were serially diluted, straight-line calibrated by DLS, and characterized in the same dilution range (Additional file 1: Figure S4B). Harsh conditions impact the shape and size and break the GVNPs, calculated as kilo counts per second (kcps) (Additional file 1: Figure S4C, D).
GVNPs characterization (GVNPs with GvpC vs. GvpC stripped-off) A Effect of temperature, B Effect of salt, C Effect of pH, D Effect of organic solvents (10%, v/v). Left: GVNPs purified in PBS (GvpC stripped off), Right: 2 M NaCl (containing GvpC). The results were transformed into residual GVNPs (%), with the control sample on each measurement being set to 100%
GVNPs showed high thermal stability up to 50 °C and degraded fast at 60 °C (Fig. 3A). However, diluted GVNPs purified in PBS are more sensitive at high temperatures, and 80% of GVNPs are degraded within 48 h at 60 °C compared to only 50% degradation in the case of GVNPs purified in 2 M NaCl (Additional file 1: Figure S4E). GVNPs showed exceptional stability in the absence or presence of up to 3 M NaCl. However, 4 M NaCl showed adverse effects, and GVNPs purified in PBS were more sensitive compared to GVNPs purified in 2 M NaCl. Acidic (pH 5.0 and 6.0) and highly alkaline (pH 12.0) solutions harm GNVPs, especially GVNPs purified in PBS (Figs. 3B and C). GVNPs rapidly degrade in DMSO and are sensitive to ethanol. However, glycerol stabilizes GVNPs purified in PBS (Fig. 3D).
To summarize, we demonstrated that native GvpC strips off during the GVNP purification in PBS buffer, leaving the GvpA-based GVNP's shell intact and stable under normal physiological conditions (Fig. 2, Additional file 1: Figure S4A). Interestingly, studies revealed that A. flos-aquae GVNPs also become weaker after rinsing off the GvpC; however, GVNPs remained intact over 7 years [11, 51, 53].
Next, we aimed to bioengineer GvpA and selectively introduce cysteines into GVNPs, followed by a thiol-maleimide reaction for functionalization.
Bioengineering of GvpA
A single NRC-1 GVNP is predicted to contain around 55,000 GvpA proteins [32]. Moreover, the GvpA protein consists of 76 amino acids (molecular weight 8.0 kDa, theoretical pI: 4.2, Additional file 1: Table S2) [54] and does not require salt for its stabilization according to our measurements (Fig. 2). Previous mutagenesis studies on GvpA demonstrated that GvpA is sensitive to alterations; however, alanine substitutions with non-polar residue at specific regions do not affect the GVNP formation [34]. Subsequently, we performed ab initio protein prediction for GvpA that served as a basis for our cysteine substitutions. The initial library for cysteine introductions in GvpA was further analyzed by predicting possible disulfide bond formations [44]. Considering the antiparallel assembly of the GvpA protein in the GVNP rib structure, next, we analyzed cysteine insertion positions for redundancy due to being located in the possible dimer interface [33, 34]. Following this approach, we created a combinatorial cysteine mutant library of 13 single, 12 double, and 8 triple GvpA variants.
Furthermore, we investigated the N- or C-terminal viability for genetic engineering of GvpA and introduced cysteine on the N- or C-terminal of GvpA with a short linker (Additional file 1: Table S3). We reevaluated our initial GvpA structural analysis based on the recently published structure of GvpA from Bacillus megaterium (PDB: 7R1C) (Fig. 4, see Additional file 1: Figure S5 for sequence alignment) [55]. Our results did not change severely since the structural assumption of two β-sheets and two α-helices was confirmed. Nonetheless, the structural rearrangement of the angles between the two antiparallel β-sheets and the α-helices changed drastically.
Overview of GvpA cysteine variants. Ab initio protein prediction for gvpA1 (gas vesicle structural protein 1) from Halobacterium salinarum (Uniport: P08958) by I-TASSER (left side), and based on GvpA of Bacillus megaterium (PDB: 7R1C) (right side). Cysteines are marked in yellow. Surface colors indicate negative and positive electrostatic potentials contoured from 50 kT/e (blue) to –50 kT/e (red). Visualized by PyMOL Molecular Graphics System, Version 2.4.2, Schrödinger, LLC
The codon-optimized GvpA genes with respective mutations in GvpA were synthesized and replaced the wild-type (WT) GvpA in pTA963.GVNP expression plasmid (Fig. 5A). Each modification was confirmed by PCR and DNA sequence analysis. The resulting plasmids were transformed in H. volcanii H1895 and processed for GVNP production as previously described [7, 17, 45]. After 2 weeks, lawns or floating cells were lysed osmotically with PBS solution, and GVNPs were purified by repeated washing in PBS or high salt-containing buffers. The colony phenotype determined the phenotype of H. volcanii and derivative strains containing GVNP expression plasmids with mutated GvpA (Fig. 5B). Red or orange colonies on agar plates indicate the abolition of GVNP formation, whereas pink color colonies indicate higher levels of GVNP production in respective transformants. Every transformant was analyzed by tryptic digest, LC–MS/MS, and transmission electron microscopy (TEM) (Fig. 5C, Additional file 1: Figure S6). A summary of these results is presented in Fig. 5 and Table 1. All transformants, except C.GvpA (N-ter), expressed well, evidenced by matched peptides in tryptic digest and LC–MS/MS analysis, even in the case of GVNP negative phenotypes (Additional file 1: Figure S6). However, the addition of cysteine with a linker, GGSGGGG, on the N-terminal of GvpA abolishes GVNP formation. In tryptic digest analysis, we found most GVNPs forming proteins, including GvpC, but we could not recognize any peptide of C.GvpA (N-ter). Surprisingly, similar modifications on the C-terminal of GvpA (GvpA.C) form GVNPs similar to wild-type (Fig. 5A). Several studies revealed that GvpA forms the essential core of the GVNP structure and is necessarily required to produce GVNPs [3, 19, 34]. Therefore, (i) non-production of a GvpA e.g., C.GvpA (N-ter) variant, or (ii) mutations close to the N-terminal of GvpA e.g., A2C, abolishes the GVNP formation. Among 33 GvpA variants (13 single,12 double, and 8 triple variants), eight transformants (two single variants, A2C and A10C, two double variants, S7C.A64C, S7C.A76C, and five triple variants, S7C.A64C.A70C, S7C.A64C.A72C, S7C.A64C.A73C, S6C.A64C.A76C, S7C.A64C.A76C) resulted to GVNP negative cells (Vac-) —colonies were translucent and red —and the other 25 resulted to gas vesiculation (Vac +)—colonies appeared turbid and pink-white (Fig. 5B). The diameters and lengths of these purified GVNPs were also determined by TEM for 25 particles each (Table 1). Pfeifer and co-workers performed a mutagenesis study on GvpA and demonstrated that GvpA tolerates alanine-to-serine substitutions at position 10 (A10S), resulting in GVNPs similar to wild-type [34]. However, here A10C resulted in the lack of GVNP, suggesting that cysteine is involved in disulfide bond formation and negatively impacts the GVNP formation.
Cysteine mutagenesis within GvpA. A The fourteen proteins involved in GVNP formation in Halobacterium sp. NRC-1 and amino acid sequences of GvpA (helices α1, α2 highlighted in yellow, β-sheets β1, β2 are in gray) [6, 34], N-terminal and C-terminal cysteine with linker addition in GvpA. GvpA amino acid substitutions are underlined. B Phenotype of H. volcanii and derivative strains containing GVNP expression plasmids with mutated GvpA. An orange and pink color indicates low and higher levels of GVNPs produced in cells. The numbers indicated variants are detailed in Fig. 5C and Table 1. C TEM images of purified GVNPs. The substitutions are shown at the bottom of each panel
Single mutations in GvpA, S6C, S47C, A45C, A46C, A68C, A70C, and A72C resulted in the spindle and cylinder-shaped GVNPs, like wild types; however, they were comparatively bigger (except S6C). Interestingly, the A64C transformant's GVNPs were tiny and mini-spindle-shaped, suggesting that the alteration disturbed the enlargement of the GVNP structure. On the other hand, A73C and A76C were bigger and cylinder-shaped. Most GvpA variants' GVNP populations' showed a broad distribution in shape and size, especially A68C, A70C, A73C, and A76C. Strunk et al. (2011) reported that deletion of 5 or 7 amino acids at the C-terminus of GvpA did not affect the GVNP formation and resulted in wild-type GVNPs, suggesting that the C-terminus of GvpA can tolerate alteration [56].
GVNPs from double and triple variants were predominantly longer and contained mixtures of spindle and cylindrical-shaped. Interestingly, double or triple variants of S6C and A64C transformant's GVNPs were spindle and longer, with an average diameter of 1.85 μm. A few GVNPs were more than 2 μm in length. However, all the double or triple variants of S7C and A64C resulted in GVNP-negative cells (Vac-).
Only GVNPs that were formed successfully and stable were used as a basis for the functionalization studies.
Functionalization of cysteine-GvpA only GVNPs
We introduced cysteine into a specific site along the GvpA protein amino acid chain to functionalize the GVNPs by bioconjugation with maleimides. The native GvpC layer was almost entirely removed by extensive washing of GVNPs in PBS before GvpA-cysteine functionalization (Fig. 2). Next, we estimated free cysteine on the GVNP variant and examined the GvpA variant's GVNP maleimide-cysteine coupling efficiency.
Detection of free thiols in intact cysteine-GvpA only GVNPs
Detection of free thiols in intact GVNPs can indicate cysteine residues accessible for maleimide conjugation. The free cysteines in the GVNPs variant are estimated in triplicate measurements with the Measure-iT™ Thiol Assay kit (Invitrogen) by following the producer's recommendations (Fig. 6). The amount of cysteine among the single variants varies from 0.46 µM (A76C) to 1.18 µM (GvpA.C). Interestingly, compared to single variants, cysteine was estimated about 2–3 times in double (1.3–2.3 µM) and triple variants (3.0–3.5 µM).
Binding reactivity with a small fluorophore
The maleimide-cysteine coupling reaction efficiency of each GVNP is tested with thiol-reactive Alexa Fluor 647 C2 maleimide. In this reaction, we mixed GVNPs GvpA variants with the reagent (1:10 mol ratio of estimated cysteine). GVNPs retain their milky white appearance during the functionalization process. Labeling efficiencies were assessed by measuring the fluorescence intensity of the GVNP samples (Fig. 7). Wild-type GVNPs treated similarly were used as a control. The highest fluorescence was recorded for triple mutated GvpA variants, followed by double and single GvpA variants.
Interestingly, double and triple-mutated GvpA variants show almost double and triple fluorescence compared to the single variants. Among the single variants, A64C and GvpA.C (C-ter) shows the highest coupling reaction. Because of the flexible linker cysteine of GvpA.C (C-ter) GvpA variant is possibly more accessible for the thiol reaction.
GvpA.C variant produces GVNPs like the wild type; therefore, used for further coupling applications. After demonstrating the ability of GvpA variants to serve as a cysteine-mediated GVNP functionalization platform, we examined the GvpA.C variant's GVNPs' capacity for maleimide chemical conjugation. As the first proof of concept, we tested maleimide-activated streptavidin, biotinylated horseradish peroxidase conjugation, and maleimide-activated SpyTag003 and SpyCatcher003 system conjugation.
Maleimide-activated streptavidin and biotinylated horseradish peroxidase
The biotin-streptavidin binding is known for specificity and high affinity and is frequently used [57]. Here, we conjugated maleimide-activated streptavidin with the GvpA.C variant GVNPs (20:1 mol ratio of free cysteine in the GVNPs) at 4 °C overnight with gentle mixing. The free maleimide-activated streptavidin molecules were removed by repeated GVNPs floatation, and bound streptavidin quantity was measured by Biotin-4-Fluorescein (B4F) based fluorescence quenching assay (Additional file 1: Figure S7). In the reduced state, each free thiol group of GVNPs appears to react with a single molecule of maleimide-activated streptavidin (0.8 μM streptavidin in 20 μM of GVNPs, Fig. 8A).
Next, the streptavidin-conjugated GVNPs were incubated overnight at 4 °C in PBS buffer with biotinylated horseradish peroxidase (B-HRP) at an 80-molar excess. The B-HRP-streptavidin-GVNP complex was purified by three repeated centrifugally assisted floatation and overnight dialysis in 1000 kDa MWCO tubing against PBS before horseradish peroxidase assay. Streptavidin on GVNPs coupled well with B-HRP and detected by high horseradish peroxidase activity in the floating top fractions (TP) containing the GVNPs-B-HRP complex compared to centrifugation-separated subnatant fraction (SF) (Fig. 8B).
These results showed that we could successfully functionalize the streptavidin-conjugated GVNPs with biotinylated horseradish peroxidase.
Maleimide-activated SpyTag003- SpyCatcher003 functionalization of GVNPs
To simplify GVNP functionalization, the SpyTag003/SpyCatcher003 protein conjugation under physiological conditions system was employed. Noteworthy, the system is versatile, works over a broad range of reaction conditions, and is flexible in the fusion sites [35]. As a proof of concept, the GVNP-maleimide-activated SpyTag003 complex is conjugated with a recombinantly expressed red fluorescent protein, SpyCatcher003-mKate2 (SC.mKate2). The resulting floating fraction GVNPs-SC.mKate2 complex gives high fluorescence compared to centrifugation-separated subnatant fraction (SF) (Fig. 9A). mKate2 colocalizes with GVNPs seen as fluorescent dots and perfectly superimposed with GVNPs appear on the phase contrast image (Fig. 9B).
Maleimide-activated SpyTag003-SpyCatcher003 functionalization of GVNPs. A mKate2 fluorescence (588/633 nm) of maleimide-activated SpyTag003- mKate2.SpyCatcher003, B Phase contrast and fluorescence images of the maleimide-activated SpyTag003- mKate2.SpyCatcher003 (bottom) and the GVNPs (top) (Image scale 38 \(\mu m\) × 38 \(\mu m\)), C Estimation of GVNPs bound esterase activity. TF: Top fraction, SF: subnatant fraction.
The resulting GVNP conjugated complexes were purified by repeated buoyancy enrichment and overnight dialysis in 1000 kDa MWCO tubing against PBS before fluorescence measurement or esterase assay (Fig. 9C). The esterase activity shows a successful binding. This improved system allows for faster functionalization of GVNPs in a more biocompatible way.
Conclusion
Here we describe the development of a new genetic and chemical approach for efficient and selective modification of the GvpA-only GVNP surface by cysteine residues. Serine or alanine residues in GvpA are selected based on the GvpA model analysis and replaced with cysteine to create a variant library of 13 single, 12 double, eight triple, and two N- or C-terminal modified GvpA variants. We examined the resulting GvpA variants for their impact on the GVNP formation, structure, and physical shape. The site-directed mutated GvpA proteins were efficiently and selectively labeled via cysteine-maleimide mediated reaction at physiological conditions and tested for foreign protein display on the GVNP surface via streptavidin-biotin or SpyTag003-SpyCatcher003 conjugation. In summary, the merger of these genetic and chemical approaches enabled a marked increase in GVNP's functional diversity and applicability for various medical, environmental, and biotechnological applications.
Availability of data and materials
The datasets generated during the current study are available from the corresponding author upon reasonable request.
References
Katyal P, Meleties M, Montclare JK. Self-assembled protein-and peptide-based nanomaterials. ACS Biomater Sci Eng. 2019;5:4132–47.
Hill AM, Salmond GP. Microbial gas vesicles as nanotechnology tools: exploiting intracellular organelles for translational utility in biotechnology, medicine and the environment. Microbiology. 2020;166:501.
Pfeifer F. Recent advances in the study of gas vesicle proteins and application of gas vesicles in biomedical research. Life. 2022;12:1455.
Nooraei S, Bahrulolum H, Hoseini ZS, Katalani C, Hajizade A, Easton AJ, Ahmadian G. Virus-like particles: preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. Journal of nanobiotechnology. 2021;19:1–27.
Lakshmanan A, Farhadi A, Nety SP, Lee-Gosselin A, Bourdeau RW, Maresca D, Shapiro MG. Molecular engineering of acoustic protein nanostructures. ACS Nano. 2016;10:7314–22.
Pfeifer F. Haloarchaea and the formation of gas vesicles. Life. 2015;5:385–402.
DasSarma S, Karan R, DasSarma P, Barnes S, Ekulona F, Smith B. An improved genetic system for bioengineering buoyant gas vesicle nanoparticles from Haloarchaea. BMC Biotechnol. 2013;13:112.
Daviso E, Belenky M, Griffin RG, Herzfeld J. Gas vesicles across kingdoms: a comparative solid-state nuclear magnetic resonance study. Microbial Physiology. 2013;23:281–9.
DasSarma S, DasSarma P. Gas vesicle nanoparticles for antigen display. Vaccines. 2015;3:686–702.
Walsby AE. Gas vesicles. Microbiol Mol Biol Rev. 1994;58:94–144.
Walsby AE, Hayes PK. Gas vesicle proteins. Biochemical Journal. 1989;264:313.
Balakrishnan A, DasSarma P, Bhattacharjee O, Kim JM, DasSarma S, Chakravortty D. Halobacterial nano vesicles displaying murine bactericidal permeability-increasing protein rescue mice from lethal endotoxic shock. Sci Rep. 2016;6:1–11.
Schnell C. Gas vesicles enable ultrasound imaging. Nat Methods. 2018;15:159–159.
Shapiro MG, Ramirez RM, Sperling LJ, Sun G, Sun J, Pines A, Schaffer DV, Bajaj VS. Genetically encoded reporters for hyperpolarized xenon magnetic resonance imaging. Nat Chem. 2014;6:629.
Alshehri S, Karan R, Ghalayini S, Kahin K, Khan Z, Renn D, Mathew S, Rueping M, Hauser CA. Air-loaded gas vesicle nanoparticles promote cell growth in three-dimensional bioprinted tissue constructs. Int J Bioprint. 2022. https://doi.org/10.18063/ijb.v8i3.489.
DasSarma P, Negi VD, Balakrishnan A, Karan R, Barnes S, Ekulona F, Chakravortty D, DasSarma S. Haloarchaeal gas vesicle nanoparticles displaying Salmonella SopB antigen reduce bacterial burden when administered with live attenuated bacteria. Vaccine. 2014;32:4543–9.
Andar AU, Karan R, Pecher WT, DasSarma P, Hedrich WD, Stinchcomb AL, DasSarma S. Microneedle-assisted skin permeation by nontoxic bioengineerable gas vesicle nanoparticles. Mol Pharm. 2017;14:953–8.
Stuart ES, Morshed F, Sremac M, DasSarma S. Antigen presentation using novel particulate organelles from halophilic archaea. J Biotechnol. 2001;88:119–28.
DasSarma S, Arora P, Lin F, Molinari E, Yin L. Wild-type gas vesicle formation requires at least ten genes in the gvp gene cluster of Halobacterium halobium plasmid pNRC100. J Bacteriol. 1994;176:7646–52.
Pecher WT, Kim J-M, DasSarma P, Karan R, Sinnis P, DasSarma S. Halobacterium expression system for production of full-length Plasmodium falciparum circumsporozoite protein. In biotechnology of extremophiles. Springer International Publishing Ag, Cham; 2016. p. 699–709.
Dattani A, Harrison C, Allers T. Genetic Manipulation of Haloferax Species. Methods Mol Biol. 2022;2522:33–56. https://doi.org/10.1007/978-1-0716-2445-6_3. PMID: 36125742. https://pubmed.ncbi.nlm.nih.gov/36125742/
Wendoloski D, Ferrer C, Dyall-Smith M. A new simvastatin (mevinolin)-resistance marker from Haloarcula hispanica and a new Haloferax volcanii strain cured of plasmid pHV2The GenBank accession number for the sequence reported in this paper is AF123438. Microbiology. 2001;147:959–64.
Haque R, Paradisi F, Allers T. Haloferax volcanii as immobilised whole cell biocatalyst: new applications for halophilic systems. Appl Microbiol Biotechnol. 2019;103:3807–17.
Allers T. Overexpression and purification of halophilic proteins in Haloferax volcanii. Bioengineered bugs. 2010;1:290–2.
Strillinger E, Grötzinger SW, Allers T, Eppinger J, Weuster-Botz D. Production of halophilic proteins using Haloferax volcanii H1895 in a stirred-tank bioreactor. Appl Microbiol Biotechnol. 2016;100:1183–95.
Pérez-Arnaiz P, Dattani A, Smith V, Allers T. Haloferax volcanii—a model archaeon for studying DNA replication and repair. Open Biol. 2020;10: 200293.
Vogler M, Karan R, Renn D, Vancea A, Vielberg M-T, Grötzinger SW, DasSarma P, DasSarma S, Eppinger J, Groll M. Crystal structure and active site engineering of a halophilic γ-carbonic anhydrase. Front Microbiol. 2020;11:742.
Karan R, Mathew S, Muhammad R, Bautista DB, Vogler M, Eppinger J, Oliva R, Cavallo L, Arold ST, Rueping M. Understanding high-salt and cold adaptation of a polyextremophilic enzyme. Microorganisms. 2020;8:1594.
Akal AL, Karan R, Hohl A, Alam I, Vogler M, Grötzinger SW, Eppinger J, Rueping M. A polyextremophilic alcohol dehydrogenase from the Atlantis II Deep Red Sea brine pool. FEBS Open Bio. 2019;9:194–205.
Grotzinger SW, Karan R, Strillinger E, Bader S, Frank A, Al Rowaihi IS, Akal A, Wackerow W, Archer JA, Rueping M, et al. Identification and experimental characterization of an extremophilic brine pool alcohol dehydrogenase from single amplified genomes. ACS Chem Biol. 2018;13:161–70.
Kim J-M, Kim Y-S, Kim Y-R, Choi M-J, DasSarma P, DasSarma S. Bioengineering of Halobacterium sp. NRC-1 gas vesicle nanoparticles with GvpC fusion protein produced in E. coli. Applied Microbiology and Biotechnology 2022;106:2043–2052.
Fernando A, Gariépy J. Coupling Chlorin e6 to the surface of nanoscale gas vesicles strongly enhance their intracellular delivery and photodynamic killing of cancer cells. Sci Rep. 2020;10:2802.
Offner S, Ziese U, Wanner G, Typke D, Pfeifer F. Structural characteristics of halobacterial gas vesicles. Microbiology. 1998;144(Pt 5):1331–42.
Knitsch R, Schneefeld M, Weitzel K, Pfeifer F. Mutations in the major gas vesicle protein GvpA and impacts on gas vesicle formation in Haloferax volcanii. Mol Microbiol. 2017;106:530–42.
Keeble AH, Turkki P, Stokes S, Anuar INK, Rahikainen R, Hytönen VP, Howarth M. Approaching infinite affinity through engineering of peptide–protein interaction. Proc Natl Acad Sci. 2019;116:26523–33.
DasSarma S, Robb FT, Place AR, Sowers KR, Schreier HJ, Fleischmann EM. Archaea: a laboratory manual – halophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor 1995.
Grote A, Hiller K, Scheer M, Munch R, Nortemann B, Hempel DC, Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526-531.
Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 2007;5:17.
Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40.
Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–38.
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat Methods. 2015;12:7–8.
Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–4.
Sneath PHA. Relations between chemical structure and biological activity in peptides. J Theor Biol. 1966;12:157–95.
Craig DB, Dombkowski AA. Disulfide by design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinf. 2013;14:346.
Grötzinger SW, Karan R, Strillinger E, Bader S, Frank A, Al Rowaihi IS, Akal A, Wackerow W, Archer JA, Rueping M. Identification and experimental characterization of an extremophilic brine pool alcohol dehydrogenase from single amplified genomes. ACS Chem Biol. 2018;13:161–70.
Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359.
Ebner A, Marek M, Kaiser K, Kada G, Hahn CD, Lackner B, Gruber HJ. Application of biotin-4-fluorescein in homogeneous fluorescence assays for avidin, streptavidin, and biotin or biotin derivatives. Methods Mol Biol. 2008;418:73–88. https://doi.org/10.1007/978-1-59745-579-4_7. PMID: 18287651.
Mogharrab N, Ghourchian H, Amininasab M. Structural stabilization and functional improvement of horseradish peroxidase upon modification of accessible lysines: experiments and simulation. Biophys J. 2007;92:1192–203.
Morawski B, Quan S, Arnold FH. Functional expression and stabilization of horseradish peroxidase by directed evolution in Saccharomyces cerevisiae. Biotechnol Bioeng. 2001;76:99–107.
Quinn DM, Shirai K, Jackson RL, Harmony JA. Lipoprotein lipase-catalyzed hydrolysis of water-soluble p-nitrophenyl esters Inhibition by apolipoprotein C-II. Biochemistry. 1982;21:6872–9.
Walsby A, Hayes P. The minor cyanobacterial gas vesicle protein, GVPc, is attached to the outer surface of the gas vesicle. Microbiology. 1988;134:2647–57.
Sundararajan A, Ju L-K. Use of cyanobacterial gas vesicles as oxygen carriers in cell culture. Cytotechnology. 2006;52:139–49.
Hayes P, Lazarus C, Bees A, Walker J, Walsby A. The protein encoded by gvpC is a minor component of gas vesicles isolated from the cyanobacteria Anabaena flos-aquae and Microcyctis sp. Mol Microbiol. 1988;2:545–52.
Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A: Protein identification and analysis tools on the ExPASy server. The proteomics protocols handbook. Springer Protocols Handbooks. Humana Press. 2005: 571–607.
Huber ST, Terwiel D, Evers WH, Maresca D, Jakobi AJ. Cryo-EM structure of gas vesicles for buoyancy-controlled motility. Cell. 2023;186(5):975-986.e13. https://doi.org/10.1016/j.cell.2023.01.041. PMID: 36868215; PMCID: PMC9994262. https://pubmed.ncbi.nlm.nih.gov/36868215/
Strunk T, Hamacher K, Hoffgaard F, Engelhardt H, Zillig MD, Faist K, Wenzel W, Pfeifer F. Structural model of the gas vesicle protein GvpA and analysis of GvpA mutants in vivo. Mol Microbiol. 2011;81:56–68.
Dundas CM, Demonte D, Park S. Streptavidin–biotin technology: improvements and innovations in chemical and biological applications. Appl Microbiol Biotechnol. 2013;97:9343–53.
Acknowledgements
We thank the research intern, A. Huang, for protein nanoparticle production and purification assistance.
Funding
The research reported in this publication was supported by funding from the King Abdullah University of Science and Technology.
Author information
Authors and Affiliations
Contributions
MR conceived and supervised the study; RK and DR designed, and RK performed the experiments; DR performed computational analysis, expressed and purified proteins in E. coli, and conducted initial experiments on GVNP functionalization; SN performed Fluorescence microscopy imaging; LZ performed Electron microscopy measurements; RK, DR, SH, and TA analyzed the data. RK, DR, and SH wrote the manuscript. All the authors contributed to and commented on this paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors approved the manuscript and gave their consent for submission and publication.
Competing interests
The authors confirm that no known competing interest is associated with this publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Oligonucleotides used in this study. Table S2. Amino acid composition of GvpA. Table S3. Cysteine site-saturation mutagenesis within GvpA. Figure S1: Amino acid composition and plasmids of SpyCatcher003-mKate2 and SpyCatcher003-esterase. Figure S2. Arrangement of gvp genes in Halobacterium sp. NRC-1. Figure S3. Gas vesicle nanoparticles (GVNPs) engineering and expression in Haloferax volcanii. Figure S4. Gas vesicles characterization (GVNPs with GvpC vs. GvpC stripped-off). Figure S5. Sequence alignment gvpA1 (gas vesicle structural protein 1) from Halobacterium salinarum (Uniport: P08958) and GvpA of Bacillus megaterium (PDB: 7R1C). Figure S6. Tryptic digest and LC-MS/MS analysis of GvpA mutants and GVNP conjugates. Figure S7. Quenching of biotin-4-fluorescein (B4F) fluorescence by adding various amounts of pure streptavidin. Discussion 1 C.GvpA (N-ter). Discussion 2 GVNP morphology changes and cysteine influence. Figure S8. Pseudo-atomic model of an entire GVNP particle. Figure S9 GVNP subunit with GvpA models based on the pseudo-atomic model of an entire GVNP particle by Huber et al.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Karan, R., Renn, D., Nozue, S. et al. Bioengineering of air-filled protein nanoparticles by genetic and chemical functionalization. J Nanobiotechnol 21, 108 (2023). https://doi.org/10.1186/s12951-023-01866-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12951-023-01866-7