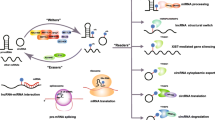
Abstract
RNA modifications have recently emerged as critical posttranscriptional regulators of gene expression programmes. Significant advances have been made in understanding the functional role of RNA modifications in regulating coding and non-coding RNA processing and function, which in turn thoroughly shape distinct gene expression programmes. They affect diverse biological processes, and the correct deposition of many of these modifications is required for normal development. Alterations of their deposition are implicated in several diseases, including cancer. In this Review, we focus on the occurrence of N6-methyladenosine (m6A), 5-methylcytosine (m5C) and pseudouridine (Ψ) in coding and non-coding RNAs and describe their physiopathological role in cancer. We will highlight the latest insights into the mechanisms of how these posttranscriptional modifications influence tumour development, maintenance, and progression. Finally, we will summarize the latest advances on the development of small molecule inhibitors that target specific writers or erasers to rewind the epitranscriptome of a cancer cell and their therapeutic potential.
Similar content being viewed by others
Introduction
The epitranscriptome landscape is very complex, with more than 170 different types of chemical modifications of RNA described to date to decorate coding and non-coding RNAs (ncRNAs) [1]. Their occurrence has been well documented for over 50 years, however their function remains still widely unknown [2]. Thus, while known since the emergence of molecular biology, RNA modifications were only coined as the “epitranscriptome” in 2015. The study of the function of these modifications is now emerging and has shown to have big implications in human pathologies [3, 4]. For example, the role of 6-methyladenosine (m6A), the most abundant and better characterized internal modification in messenger RNA (mRNA), is to regulate embryonic stem cells and cancer cells self-renewal and to favour survival upon heat shock or DNA damage [5,6,7]. In addition to the roles of m6A modification in mRNAs, adenosine methylation is also found in non-coding RNAs, such as microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) regulating their biogenesis and function [8,9,10,11,12,13,14,15]. We now begin to appreciate the plethora of molecular processes that are finely regulated by RNA modifications ranging from RNA metabolism, decay, splicing or translation, localization, stability, turnover, binding to RNA binding proteins (RBPs) or other RNAs, and thereby diversifying genetic information. Similar to epigenetics, groups of proteins have been identified that specifically “write” (catalyse the deposition of a specific modification), “erase” (catalyse the removal of a specific modification), and “read” (recognize and bind modified nucleotides) thereby affecting the fate of RNA. Other modifications have been recently documented in mRNA including N6,2′-O-dimethyladenosine (m6Am), 5-methylcytosine (m5C), 5-hydroxymethylcytosine (hm5C), pseudouridine (Ψ), 1-methyladenosine (m1A) or 2'-O-ribose methylation, although their molecular functions remain still widely unknown [5, 16].
RNA modifications are also present in other regulatory ncRNAs, in fact the most modified RNAs are transfer RNA (tRNAs) and ribosomal RNAs (rRNAs) and their modifications shape protein synthesis efficiency and fidelity. More than 100 modifications have been described for tRNA, being the anticodon loop one hotspot of modifications and play key roles in accurate and efficient decoding in translation [17]. In rRNA, most modifications cluster around functional sites including the decoding site and the peptidyl transfer centre (PTC), suggesting their functional relevance in regulating protein synthesis [18, 19]. Studies in humans, yeast, and bacteria have shown that dynamic deposition of these modifications in rRNA regulate cell growth, and drug and stress sensitivity by fine-tuning translation and is a very conserved mechanism [17, 20,21,22]. For instance, in yeast, flies, worms, and humans, alterations of m5C levels in rRNA favours the translation of stress response-decoding transcripts in order to increase survival [23, 24]. Occurrence of Ψ residues increases in mRNA in yeast under starvation and heat shock [25,26,27]. And lack of 2′-O-ribose methylations in rRNAs decreases efficient translation and affects growth and sensitivity to antibiotics [28]. Similarly, the overall levels of tRNA modifications change to reprogram protein translation by changing codon usage [29,30,31].
The deposition of RNA modifications is dynamic, and thereby allows rapid cellular responses to environmental signals [16, 25, 31,32,33,34]. The ability to adapt to changing microenvironments such as that of stress or chemotherapeutic drugs is crucial to ensure survival of tumour cells, indicating that RNA modifications could play important roles in cancer. Historically, cancer has been considered fundamentally as a disease characterized by stepwise accumulation of genetic or epigenetic alterations of different oncogenes and tumour suppressor genes. However, compelling evidence indicates that epitranscriptomics could also play a fundamental role in this pathology. Through its ability to modulate many processes of RNA metabolism, dynamic RNA modifications have been shown to be important emerging regulators in cancer [3, 33, 35,36,37,38]. Although RNA modifications are not generally considered cancer drivers, cumulative evidence shows that their aberrant expression is functionally related to survival, proliferation, self-renewal, differentiation, stress adaptation, invasion, and resistance to therapy, all of which are hallmarks of cancer [24, 33, 35, 37, 39,40,41,42,43]. For example, dynamic changes for multiple RNA modifications can be observed in the urine of cancer patients [44]. Most striking it has been the extraordinary enlargement of experimental evidence that implicates alterations in the expression of m6A writers, erasers or readers are associated with increased risk of obesity and diabetes, infertility and with tumour-suppressive or tumour-promoting scenarios [3, 45]. Other RNA modifying enzymes have been found to be altered in cancer. For example, in an aggressive breast cancer cell lines, 2′-O methylation appeared to be hypermodified in rRNA and correlated with altered translation [46]. Mutations in the rRNA pseudouridine synthase DKC1, cause X-linked dyskeratosis congenita (X-DC) characterized by impaired translation, hematopoietic stem cells differentiation failure and increased cancer susceptibility [47]. Alterations in tRNA modifications have been also reported in cancer including m5C or 5-methoxycarbonylmethyluridine (mcm5U) and correlate with altered protein translation [33, 48,49,50,51]. All these studies show that aberrant RNA modifications contribute to proliferation, self-renewal, migration, stress adaptation and survival of cancer cells and suggest that targeting aberrant posttranscriptional modifications in cancer cells may hold promise as an efficient therapy for tumours [52].
In this review we will discuss the molecular and cellular functions of RNA modifications in modulating gene expression programmes, with a focus on their roles in cancer. We further summarize here recent studies that elucidate the therapeutic potential of targeting their aberrant deposition in cancer. We will focus our review article on m6A, m5C and Ψ in coding and non-coding RNAs as notable examples due to the advances in our understanding of the role of these epitranscriptomic marks in cellular functions including proliferation, self-renewal, survival to stress or migration. In addition, expression alterations or mutations in m6A, m5C and Ψ depositing machineries have been documented in cancer.
6-methyladenosine
m6A deposition in coding and non-coding RNA
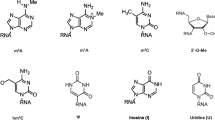
N6-methyladenosine (m6A), a well-known posttranscriptional modification first discovered in 1974 [53, 54], has been regarded as the most frequent internal modification found in mRNA from viruses to mammals, but also occurs in small ncRNA and lncRNA in many eukaryotic species [11, 55]. Around 0.1–0.4% of all mRNA adenines are methylated at position N6, representing approximately 3–5 modifications per mRNA (Fig. 1) [56, 57]. The recent advent of genome-wide m6A mapping of polyadenylated RNAs has yielded unprecedented insights into the m6A-methylome landscape. Most methods for global m6A detection rely on immunoprecipitation of methylated RNAs using specific antibodies that recognise m6A [32, 58]. Subsequent improvements using ultraviolet crosslinking steps to bind the methylated RNA to antibodies have allowed the identification of m6A sites at single-nucleotide resolution [59, 60]. These methods have revealed that this modification is enriched at 3′ untranslated regions (3′UTRs), near stop codons, within long internal exons, in intergenic regions, introns, and at 5’UTRs (Fig. 1) [32, 41, 59,60,61].
Schematic diagram of the location of m6A, m6Am, m5C and Ψ modifications on mRNA. Blue ribbon represents the mRNA with m7G-cap and a poly(A) tail. ATG and STOP codons are indicated. The writers, erasers, readers and function are listed in the text box attached to each modification. Modifications: m6A, 6-methyladenosine; m6Am, N6,2ʹ-O-dimethyladenosine; m5C, 5-methylcytosine; Ψ, pseudouridine. Proteins: METTL, methyltransferase-like; FTO, fat mass and obesity-associated protein; PUS, pseudouridine synthase; NSUN, NOL1/NOP2/SUN domain family member; ALyREF, Aly/REF Export Factor; ZCCHC4, zinc-finger CCHC domain-containing protein 4; ALKBH5, Alpha-Ketoglutarate-Dependent Dioxygenase AlkB Homolog 5; YTHDC, YTH domain-containing; YTHDF, YTH domain-containing family
Deposition of m6A has been also reported in ncRNAs such as miRNA, lncRNA and circRNA [9, 14, 62,63,64,65]. Deposition in miRNA is enriched in primary miRNAs (pri-miRNA), but not in precursor miRNAs (pre-miRNA) and m6A marks are usually located in both intergenic and intragenic pri-miRNAs that contain canonical METTL3 motifs [9]. As for circRNAs, despite being derived from mRNA exons, m6A-modified circRNAs are frequently derived from exons not methylated in mRNAs [62]. LncRNAs are generally defined as transcripts longer than 200 nucleotides, and yet despite sharing several features with coding mRNAs such as being 5′capped, spliced and, polyadenylated, m6A residues in lncRNAs are distributed along the whole body of the transcript and are more present in lncRNAs that undergo alternative splicing [65].
m6A writers
The deposition of m6A occurs into nascent pre-mRNAs during transcription and it is carried out in the nucleus by a multicomponent methyltransferase complex, that includes the S-adenosyl methionine (SAM) binding protein methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14) heterodimeric catalytic core (Fig. 2a) [66,67,68]. METTL3 catalyses the conversion of adenosine to m6A through its methyltransferase domain, while METTL14 is responsible for the recognition of RNA substrates, and therefore the whole METTL3-METTL14 heterodimer is required for the methylation process [69, 70]. In addition, the currently defined methyltransferase complex is also composed of adapters. The RNA-binding motif protein 15 (RBM15) is one of these adapters and is responsible for the initial recruitment of the complex to its target site in the mRNA. The regulatory proteins Wilms’ tumour 1-associating protein (WTAP) and KIAA1429 (also known as VIRMA) are responsible for the complex formation [71, 72]. The recently characterized zinc finger CCCH domain-containing protein 13 (ZC3H13) has been found to act as a bridge between the adaptor RBM15 and WTAP [73]. miRNAs are methylated by the METTL3-METTL14-WTAP-RBM15/15B-KIAA1429 complex [9]. More recently, a new study has identified a single enzyme, METTL16, as another active m6A methyltransferase in human cells [74]. METTL16 has been shown to methylate mostly small nuclear RNAs, a number of intronic sites in pre-mRNAs and in addition other ncRNAs (Fig. 2c) [15, 74, 75].
Molecular mechanism of m6A deposition in RNA, biological function and implications in human cancer. a-c m6A RNA methylation landscape in mRNA (a) and ncRNA (b & c) mediated by writers (blue balloons), including METTL3, METTL14, WTAP, RBM15B, KIAA1429, ZC3H13 and METTL16, erasers (pink balloons) FTO and ALKBH5 and reader proteins (yellow balloons) YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2 and HNRNPC, HNRNPG, HNRNPA2, HNRNPB1, IGF2BPs and eIF3, and their role in mRNA metabolism. d-g Aberrant m6A deposition in mRNA and ncRNAs promotes or suppresses tumour progression through METTL3/METTL14 upregulation (d) or downregulation (e) and FTO/ALKBH5 upregulation (f) or downregulation (g). Red arrows indicate induction. Blue arrows with flat end represent inhibition
m6A erasers
Deposition of N6-methyladenosine is reversible and relies on an orchestrated and dynamic network of specific methyltransferases but also demethylases or “erasers”. Two 6-methyladenosine demethylases have been identified, fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5), which are members of the nonheme Fe (II)/α-ketoglutarate-dependent dioxygenases (Figs. 1 and 2a) [76, 77]. FTO shows a preference for demethylating N6,2′O-dimethyladenosine (m6Am) but also demethylates m6A [78, 79]. Furthermore, the AlkB homolog 3 (ALKBH3), another member of this family which preferentially acts on m6A in tRNAs [80]. While we start to appreciate the complexity of the m6A methylation machinery, yet it remains to be fully understood how the methylation machinery selectively and dynamically targets specific regions of the transcriptome.
m6A readers
m6A methylation acts as a unique recognition element for reader binding proteins that drive the biochemical processes that occurred to marked RNAs [81]. Some of the m6A readers contain a common RNA binding domain, the YTH domain, which include the family of YTH domain-containing proteins 1 and 2 (YTHDC1 and YTHDC2, respectively) and the YTH domain family proteins 1, 2 and 3 (YTHDF1, YTHDF2 and YTHDF3, respectively) [82,83,, 83]. Subsequently, other readers have been discovered; eukaryotic translation initiation factor 3 (eIF3), heterogeneous nuclear ribonucleoprotein (HNRNPC and HNRNPA2/B1), insulin-like growth factor (IGF2BP1, IGF2BP2, and IGF2BP3), proline-rich and coiled-coil-containing protein 2A (PRRC2A), and the fragile X mental retardation protein FMRP, among other protein readers described to date (Figs. 1 and 2a) [8, 13, 61, 84,85,86,87].
Molecular function of m6A deposition in RNA
Role of m6A in mRNA
Regarding the biological function of m6A deposition, this dependents on the protein reader that identifies and binds the modified mRNA (Fig. 2a). For example, the YTHDC protein subtypes are found mainly in the nucleus, and specifically YTHDC1 has been described as an alternative splicing factor of pre-mRNA [88]. YTHDC1 facilitates mRNA export too by recruiting nuclear transport receptors or by binding to m6A methyltransferases complexed with TREX mRNA export complex and modified mRNAs [89, 90]. In contrast, YTHDF protein subtypes are predominantly found in the cytoplasm and regulate the fate of cytoplasmic modified mRNAs. For example, YTHDF1 and YTHDF3 improve the efficiency of mRNA translation [91, 92], and YTHDF2 facilitates mRNA decay through the CC4-NOT deadenylase complex [93, 94]. YTHDF3 may accelerate mRNA decay too through interacting with YTHDF2 [92]. YTHDC2 protein can exist both inside and outside the nucleus, being able to selectively bind to m6A mark of ncRNA, yet the biological consequence upon binding is unknown [11]. In the cytosol, YTHDC2 affects the translation efficiency and abundance of its target mRNAs [95]. While the evidence so far has shown that m6A deposition mediate diverse effects through a complex network of readers, a recent study has revealed evidence for a unifying functional model of m6A readers, where m6A predominantly influences mRNA degradation through the combined action of three YTHDF member readers [96].
Apart from the YTHDC and YTHDF members, other reader proteins can modulate translation or stability. For instance, eIF3 subunits have been reported to affect canonical and non-canonical cap-dependent translation. This protein binds preferentially to m6A within the 5′ UTR of mRNA leading to enhanced translation [61]. In addition, evidence shows that IGF2BP promotes the stability and storage of their targeted mRNAs [87].
It must be considered too that the methylation of adenines causes alterations in the secondary structure of RNAs, which in turn may alter their interaction with reader proteins [97]. m6A compared to A promotes the destabilization of A/U pairings and alters the thermostability of RNA duplexes [98]. This structural change has been already correlated with an alteration in the interaction between HNRNPC1 and HNRNPG with mRNA [13, 84]. Other members of the HNRNP family, HNRNPA2/B1 could bind directly to m6A to regulate alternative splicing events and processing of precursor miRNAs [8].
Role of m6A in non-coding RNAs
Similar to mRNA, the presence of m6A on miRNA, lncRNA, and circRNA can regulate their binding to m6A-readers which in turn regulate their processing and maturation, abundance, translation, stability, location, function, or degradation [8,9,10,11,12, 99,100,101], but also dynamically regulate many physiological and pathological processes including tumourigenesis [102,103,104] (Fig. 2b). For example, the methylation of primary miRNAs (pri-miRNAs) by METTL3 increases their binding to HNRNPA2/B1, which interacts and enables microprocessor complex unit DGCR8 to target pri-miRNAs, promoting the initiation of miRNA biogenesis and their maturation into miRNAs [8, 9]. METTL14 can directly recruit DCGR8 on the m6A modified pri-miRNA encoding for miR-126a and consequently affecting the levels of miR-126a [12]. In addition, METTL14 has been found to be associated with chromatin during transcriptional elongation [105], which may suggest that METTL14 may contribute to the co-transcriptional recruitment of the microprocessor complex on pri-miRNA transcripts. Therefore, alternations of m6A levels by methylases or erasers differential expression could result in significant changes in the mature pool of miRNAs.
As recently reported m6A deposition can alter lncRNA stability. GAS5-AS1 transcript improves its stability by m6A when modulated by ALKBH5, however m6A can promote GAS5 lncRNA degradation through YTHDF2 and YTHDF3 binding [99, 100]. Although it is unclear whether lncRNA localization is also regulated by m6A, it has been shown that overexpression of METTL3 can significantly increase the nuclear localization of RP11 lncRNA [101]. The modification of m6A in lncRNA could as well affect their structure and influence the interactions between RNAs and between RNAs and the proteins that regulate their specific biological functions [11, 13, 14]. For instance, HNRNPC was found to bind to the m6A-modified hairpin compared to the unmethylated hairpin of the lncRNA MALAT1 in an “m6A switch”-regulated manner, which indicated that m6A modification disrupts lncRNA hairpin-stem structure and thus promoting its binding to HNRNPC [13]. LncRNA X-inactive-specific transcript (XIST) is methylated by METTL3, and METTL3 knockdown was shown to impair XIST-mediated transcriptional silencing of genes on the X chromosome both in vitro and in vivo [11]. Cytoplasmic lncRNA linc1281 is methylated at its 3′-end region, and the methylation marks are required for the binding of let-7 through the interaction of yet unknown proteins [14].
Regarding METTL16-mediated deposition in snRNAs, initial studies have shown to methylate A43 position of U6 snRNA, which is found near the region that base pairs with 5′ splice sites of pre-mRNAs, suggesting that METTL16 plays an important role in mRNA splicing [15].
Interestingly, ncRNAs also play significant roles in regulating the methylation levels of adenosine-6 [106, 107]. For example, in differentiating stem cells, miRNAs would regulate the binding of METTL3 to mRNAs, by using a sequence pairing mechanism, and thus modulating the abundance of m6A [107]. Also in hepatocellular carcinoma (HCC), the inhibition of miR-145 causes an increase in YTHDF2 expression which in turn leads to a fall in the levels of m6A, probably due to increased mRNA degradation [64].
While the functions of m6A deposition are currently not fully understood, a picture begins to emerge and shows that m6A methylation takes part in the regulation of mRNA processing, decay, stability, splicing, polyadenylation, nuclear export and translation. In the case of ncRNAs, m6A has been shown to promote their processing or enhance their functions [8, 11].
The role of m6A in cancer
Numerous studies have shown that m6A deposition in RNA plays a critical role in many physiological processes, including circadian rhythms regulation, spermatogenesis, embryogenesis, DNA damage and stress response, pluripotency and cell reprogramming [7, 36, 41, 75, 108,109,110]. Furthermore, emerging evidence suggests that m6A regulators are also closely associated with oncogenic or tumour suppressive functions including proliferation [111], tumourigenesis [112], invasion [113], metastasis [12, 114, 115] or immune system evasion [116] in malignant tumours. Below, we summarize the main emerging roles of m6A writers, erasers and readers in cancer (Table 1).
Role of m6A writers in cancer
METTL3 and METTL14 expression have been reported to promote tumourigenesis in several cancer types (Fig. 2d). For example, METTL3 is overexpressed in acute myeloid leukaemia (AML) [117]. In this study, METTL3 was shown to methylate BCL-2, PTEN, and c-Myc mRNAs which resulted in their increased translation, inhibiting cell differentiation and apoptosis, and promotion leukaemia progression [117]. However, the mechanism of the m6A-dependent translation remains undetermined. Furthermore, another study identified that METTL3 expression in vivo was essential to maintain AML cells in an undifferentiated state, and thus maintaining myeloid leukaemia growth [118]. In comparison, METTL14 was shown to act as an oncogene in AML by increasing MYB and c-Myc mRNA stability and translation [40]. METTL3 acts as an oncogene that facilitates the growth, survival, and invasion of cells in lung and colon cancer by promoting the translation of EGFR and TAZ [113]. METTL3 is also upregulated in breast cancer, where it increases methylation and stability of HBXIP mRNA, which induces cell proliferation and survival of tumour cells via inhibiting the tumour suppressor let-7 g [119]. Recent studies have shown that aberrant overexpression of m6A modifiers is observed in colorectal cancer too. The aberrant deposition of m6A increased miRNA-1246 expression, resulting in the downregulation of the tumour suppressor of SPRED2 and metastasis induction [120]. Other recent studies have reported that METTL3 is overexpressed in prostate cancer cells, contributing to the growth and invasion of cancer cells through SHH-GLI1 signalling [121]. In addition, METTL3 also regulates the expression of ITGB1, thus affecting its binding to Collagen I, the mobility of tumour cells, and promoting prostate cancer bone metastasis [122].
Despite the unanimously oncogenic functions of METTL3 and METTL14 in all these cancer types, in glioblastoma and HCC several reports have demonstrated both, oncogenic and tumour suppressive roles. Initial studies showed that m6A methylation inhibited the growth, self-renewal, and tumourigenesis of glioblastoma stem cells by decreasing the stability and expression of key oncogenic transcripts such as ADAM19 [35]. In contrast, in a subsequent study, METTL3 was found to be upregulated, and to be a predictor of poor patient survival. In this study, METTL3 was found to increase the stability and expression of SOX2 mRNA, promoted the growth of glioblastoma stem cells (GSCs), and prolonged survival in mice [123]. Similarly, in HCC the expression of m6A writes can inversely promote or suppress tumourigenesis. For instance, METTL14 was initially identified as a tumour suppressor in HCC [12]. In this study, the authors showed that METTL14 expression induced an increase in pri-miR-126 expression, suppressing tumour metastases [12]. Conversely, in another study METTL3 was found upregulated and associated with poor prognosis in patients with HCC. In another study, METTL3 inhibited SOCS2 mRNA expression through a m6A-YTHDF2 dependent manner [56]. From the reported data one can conclude that the variable expression of m6A writers and their bound factors, erasers and readers may account for differences in m6A deposition levels, as well as differences in targeted RNAs, which in turn will lead to the observed discrepancy and controversy. Yet more studies will be necessary to precisely determine the nature of METTL3 and METTL14 in glioblastoma and HCC and to identify the factors, pathways or tumour cell states responsible for the controversial findings.
In other tumours, METTL3 and METTL14 play an tumour suppressive role (Fig. 2e). For example, in endometrial cancer, 70% of tumours exhibit a reduction in m6A levels, either through METTL14 mutations or downregulation of METTL3 expression. METTL3-METTL14 complex loss leads to increased cell proliferation by upregulating the expression of members of the AKT pathway, and thus, showing a tumour suppressor function in endometrial cancer [111]. Similarly, in renal cell carcinoma, depletion of METTL3 promotes cell proliferation, growth, and colony formation through the PI3K-AKT-mTOR pathway activation and enhances cell migration and invasion through the epithelial-mesenchymal transition (EMT) pathway [124].
The implication of METTL16 in cancer is poorly understood, in fact only few studies have linked METTL16 to cancer [15, 125]. In recent studies, loss-of-function mutations and expression alterations were found in colon cancer, suggesting a role for METTL16 in the tumourigenesis of colorectal cancer [126, 127]. Other studies have associated METTL16 with the maturation of MALAT1 mRNA which can act as an oncogene and a tumour suppressor in different types of cancer [125]. In addition, given the role of METTL16 in regulating snRNA methylation and hence their processing, METTL16 dysregulation could favour tumour development by inducing changes in alternative splicing. Studies in the last decade have demonstrated the potential role of alternative splicing in the aetiology of cancer [128]. Indeed, the change in the expression of key enzyme isoforms in apoptosis, metabolism, cell signalling and resistance to therapy has been attributed to the acquisition of the tumour phenotype too [128].
Role of m6A erasers in cancer
Dysregulation of m6A erasers have been found too associated to cancer risk, in fact FTO polymorphisms have been known to be associated to several human disorders including increased risk of cancer for decades [129]. After the discovery of FTO catalytic activity, we begin to understand the molecular mechanisms underlying FTO oncogenic activity. Initial studies showed that FTO is highly expressed in AML and exerts its oncogenic function by reducing m6A levels in the mRNA of the tumour suppressors RARA and ASB2, leading to inhibition of their expression [36]. Similarly, FTO was shown to induce GSCs growth, self-renewal, tumour progression, and was associated with poor survival through regulation of ADAM19 (Fig. 2f) [35]. More recent studies have linked increased expression of FTO to other tumours. For example in melanoma, FTO expression promotes tumourigenesis and resistance to immunotherapy (anti-PD-1) by directly removing m6A from PDCD1, CXCR4, and SOX10 mRNAs, thereby increasing their stability (Fig. 2f) [38, 130]. FTO is also upregulated in cervical cancer where it induces resistance to chemo-radiotherapy and enhances the response to DNA damage [131, 132]. In cervical cancer, FTO can activate the β-catenin pathway and increase ERCC1 expression that is associated with worse prognosis. Furthermore, FTO promotes cell migration and proliferation by positive regulation of E2F1 and MYC [131, 132]. In lung cancer FTO is found overexpressed and is associated with poorer prognosis, facilitating cell proliferation and invasion, and inhibiting apoptosis by regulating MZF1 expression (Fig. 2f) [133]. In breast cancer too, high levels of FTO promotes cell proliferation, colony formation and metastasis in vitro and in vivo through downregulation of BNIP3 expression [134]. The compelling evidence clearly indicates a role for FTO in cancer, yet although FTO has been described to remove m6A in the mRNA of tumour suppressors or genes that could confer resistance to immunotherapy, currently there is insufficient evidence to confirm that the effects detected in cancer are due exclusively to its demethylase activity.
Aberrant overexpression of ALKBH5, the second m6A demethylase to be identified, has been also associated to several cancer types (Fig. 2f) [135,136,137,138,139,140,141,142]. ALKBH5 has been found overexpressed in glioblastoma and its expression is associated with poorer prognosis, and it promotes GSCs proliferation and tumour progression by enhancing FOXM1 expression [136]. In breast cancer, ALKBH5 has been shown to promote tumourigenesis by decreasing adenosine methylation in KLF4 and NANOG mRNAs, enhancing their stability in breast cancer stem cells [135, 137]. In gastric cancer, it promotes invasion and metastasis by demethylating the lncRNA NEAT1 [138]. ALKBH5 expression is found increased in ovarian cancer too and its expression correlates with poorer survival. The upregulated expression promotes proliferation, invasion and autophagy through the mTOR and BLC2-Beclin1 pathways [139, 142]. Similar to other m6A regulators, ALKBH5 has been demonstrated to have a tumour suppressive role in other tumours (Fig. 2g). For example, in pancreatic cancer cells, loss of ALKBH5 induces increased methylation of the lncRNA KCNK15-AS1, leading to its downregulation and increased cell migration. ALKBH5 in pancreatic cancer also has been shown to inhibit tumourigenesis by reducing WAF-1 levels and hindering Wnt signalling activation [140, 141].
Role of m6A readers in cancer
m6A readers are also aberrantly expressed in cancer, and their defective function has been linked to oncogenic or tumour suppressive roles, however we begin to appreciate only now the interplay of these RNA binding proteins, m6A and cancer. YTHDC2 and YTHDF1 are overexpressed in colorectal cancer, both are associated with poor prognosis in patients and high cell proliferation and metastatic potential. In these tumours, YTHDC2 regulates the expression of tumour promoter genes such as HIF1A [143]. Silencing of YTHDF1 inhibits tumourigenicity in vitro and tumour growth in vivo by inhibiting the Wnt-β-catenin pathway and its expression induces resistance to chemotherapy [144]. YTHDF1 is also upregulated in cancer. In particular, in ovarian cancer YTHDF1 increases EIF3C translation, facilitating tumourigenesis and metastasis [145]. In contrast, YTHDF1 acts as a tumour suppressor in melanoma where it promotes the translation of the tumour suppressor HINT2, thus inhibiting tumour development [146]. In the case of YTHDF2, its overexpression in AML has been shown to reduce the half-life of various m6A-containing transcripts which are involved in TNF signalling and whose upregulation promotes cell apoptosis [147]. Furthermore, YTHDF2 was found to be upregulated in lung cancer, and to aberrantly promote the translation of 6PGD mRNA which is critical for the promotion of tumour growth [148]. Nevertheless, YTHDF2 is also capable to suppress cell proliferation, tumour growth and activation of MEK and ERK signalling via promoting the degradation of EGFR mRNA in HCC cells [149].
Targeting m6A machinery in cancer
Considering the critical roles of the m6A regulatory proteins in several cancer hallmarks, they are promising therapeutic targets, specially the writers and erasers, since their activity can be modulated by small molecules. Although there are currently no small molecule inhibitors of RNA methyltransferases, several demethylase inhibitors have been discovered or developed by biochemical- or cell-based small-molecule compound library screening or chemical synthesis. Most commonly, those developed or discovered inhibitors target FTO. In a seminal study, Su R et al. FTO was found to be inhibited by the oncometabolite R-2-hydroxyglutarate (R-2HG) [37]. In this study, R-2HG was used to directly inhibiting FTO in AML and glioma cells, which resulted in increased methylation and decreased expression of c-MYC and CEBPA mRNAs, blocking proliferation, cell cycle, and inducing apoptosis in AML and glioma cells [37]. Since then, other studies have attempt to develop small-molecule inhibitors to target FTO or AKLBH5 RNA demethylases, given promising results at the pre-clinical level. For example, the ethyl ester form of Meclofenamic acid (MA) MA2, a US Food and Drug Administration (FDA)-approved nonsteroidal anti-inflammatory drug, was found to be a FTO inhibitor which led to elevated levels of m6A modification in mRNAs in glioblastoma cells, suppressing tumour progression and prolonging the lifespan of GSC-grafted mice [35, 150]. In addition, based on the structures of FTO and ALKBH5 domains other groups have designed 2-oxoglutarate and iron-dependent oxygenases (2OGX) inhibitors to target m6A erasers [151], such as for example the IOX3 inhibitor [152]. However the promising inhibitors have some limitations and need to be considred before their use in the clinic. All 2-oxoglutarate derivates are not selective and may also suppress the activity of other Fe (II) and 2OG dependent oxygenases. More recent studies have developed two FTO inhibitors, namely FB23 and FB23–2, which have been shown to suppress proliferation and promote AML cell differentiation/apoptosis in vitro and significantly inhibit the progression of human AML in xenotransplanted mice [153].
Despite the contradictory results for some types of tumours, m6A regulators have shown to have wide implications in several cancer hallmarks. The controversial results reflect however that the consequences of aberrant m6A deposition may dependent on the cancer or cell type context, dysregulation of other signalling pathways and distinct set of substrates. Thus, future studies will accurately determine the biological function of each individual m6A regulator in different types of cancer, and they will identify each critical target transcript revealing the exact underlying mechanism. In addition, development of selective and clinically effective inhibitors for m6A regulatory enzymes may provide effective therapeutic strategies, alone or in combination with other therapeutic agents. For example, neoantigen-specific immunity was shown to be regulated through YTHDF1 in a m6A-dependet manner. Mechanistically, transcripts encoding lysosomal proteases were shown to be marked by m6A and bound by YTHDF1, leading to increased translation of lysosomal cathepsins and decreased cross-presentation of dendritic cells, implicating YTHDF1 and m6A as potential therapeutic targets in anticancer immunotherapy [116].
5-methylcytosine
m5C deposition in RNA
m5C is a conserved and prevalent mark in RNA in all life domains. m5C is found in a wide range of RNAs but it is most abundant in eukaryotic tRNAs and rRNAs (extensively reviewed in [16]). Current global m5C detection methods rely on the chemical reactivity of cytosines to be deaminated in the presence of sodium bisulphite, or immunoprecipitation methods that use either antibodies against m5C or RNA methyltransferases previously crosslinked to the RNA target (extensively reviewed in [16]). RNA bisulphite sequencing is the most common used technique to map m5C and while it has resulted effective in detecting m5C in abundant RNAs such as tRNA and rRNA [24, 42]. In mRNA different studies obtained very different results. From studies identifying m5C sites in 8000 RNAs [154], to other studies finding only a few methylated mRNAs [155]. These controversial findings have raised the need to develop more robust methods for truly identifying m5C depositions in mRNA [156]. More recent studies have reported only few hundred m5C sites in human and mouse transcriptomes using an improved bisulphite sequencing method and a novel computational approach (Fig. 1) [156, 157]. What is still undetermined is whether this low prevalence has biological value and it needs to be further investigated.
m5C writers
In humans, cytosine-5 methylation is catalysed by the NOL1/NOP2/sun (Nsun) family and the DNA methyltransferase member 2 (DNMT2, TRNA Aspartic Acid Methyltransferase 1 or TRDMT1) [158, 159]. DNMT2, NSUN2, NSUN3 and NSUN6 all methylate cytoplasmic tRNAs, yet with different specificity and at different residues (Figs. 3b, 4) [42, 160,161,162,163,164,165,166,167]. For example, NSUN2 methylates the vast majority of tRNAs at the variable loop, and in leucine at the wobble position [42, 168, 169], DNMT2 methylates three tRNAs at the anti-codon loop [161, 162, 170], and NSUN6 targets the acceptor stem of few tRNAs [164]. NSUN2 methylates also mRNA, ncRNAs and lncRNAS (Fig. 3a, c) [157, 168, 171,172,173]. NSUN3 targets tRNAs in the mitochondria, and its deposition is required for the formation of 5-formylcytosine (f5m) [160, 163, 166] (Figs. 3b, 4). NOP2 (NSUN1) and NSUN5 are nucleolar and methylate very conserved residues in 28S rRNA, close to the peptidyl-transferase centre [165, 174, 175] and at the interface between the large and small subunit respectively (Fig. 3d) [165, 174, 175]. Finally, NSUN4 targets the small subunit of mitochondrial rRNA (Fig. 3d) [176].
Molecular mechanism of m5C deposition in RNA, biological function implication in human cancer. a-d m5C RNA deposition landscape in mRNA (a), tRNA (b), rRNA (d), and ncRNAs (c) mediated by writers (blue balloons) NSUN2–6, DNMT2 and NOP2, erasers (pink balloons) TET and ALKBH1, and reader proteins (yellow balloons) YBX1 and AlyREF, and their role in mRNA metabolism. e-h Aberrant m5C deposition in RNAs promotes or suppresses tumour progression through NSUN2 upregulation in mRNA, miRNA and lncRNA (e), NSUN2 downregulation in tRNA (f), NOP upregulation in rRNA (g) or NSUN5 downregulation in rRNA (h). Red arrows indicate induction. Blue arrows with flat end represent inhibition
Simplified structure of a tRNA and all known Ψ and m5C events in humans. Ψ modification is shaded in blue and m5C in purple. The table on the right indicates the position and modification in tRNA, the human enzyme that has been identified to catalyse this modification, the sub-cellular localization of tRNAs containing the modification. m5C, 5-methylcytosine; f5C, 5-formylcytosine Ψ, pseudouridine; Ψ derivates: Ψm, 2ʹ-O-methylpseudouridine and m1Ψ, 1-methylpseudouridine. Proteins: PUS, pseudouridine synthase; RPUSD, RNA pseudouridine synthase domain-containing protein; NSUN, NOL1/NOP2/SUN domain family member; DNMT2, DNA methyltransferase 2. f5C* is formed from m5C by ALKBH1 (Alpha-Ketoglutarate-Dependent Dioxygenase AlkB Homolog 1) in mitochondrial tRNAs. Ψ derivates are formed from Ψ by methyltransferases
m5C erasers
While writers for m5C are now well documented, the existence of m5C erasers are still under debate. Some reports have however indicated that m5C can be oxidised to generate 5-hydroxymethylcytosine (hm5C) by enzymes of the ten-eleven translocator family (TET) in mRNA (Fig. 3a) [177, 178] and the formation of f5C by Alpha-Ketoglutarate-Dependent Dioxygenase AlkB Homolog 1 (ALKBH1) at the wobble position of mitochondrial tRNAs (Fig. 3b) [160, 163, 166]. While the biological relevance of f5C deposition in mitochondrial tRNAs has been well established [160, 163, 166], the biological relevance of the low abundant hm5C deposition in mRNAs remains yet to be determined.
Molecular function of m5C deposition in RNA
Role of m5C in non-coding RNAs
The functional consequences of m5C loss in tRNAs have been well documented. Deposition of m5C by DNMT2 and NSUN2 protects tRNAs from endonucleolytic cleavage by angiogenin [42, 162, 167]. NSUN2-mediated m5C deposition in tRNAs regulates differentiation and stress responses in tissue and in cancer stem cells [33, 42, 179,180,181]. Molecularly, 5-cytosine methylation of tRNA protects them from processing into tRNA-derived small RNA fragments (tRFs) by angiogenin [42, 162, 167, 170, 182]. The formation of tRFs is usually induced under stress and can repress canonical translation and favour ribosome assembly in unconventional 5’ start sites found in 5’ UTR of stress response transcripts [183]. In contrast, loss of DNMT2-mediated methylation of tRNA at C38 causes tRNA cleavage which leads to specific codon mistranslation [167]. Similarly, deletion of rRNA cytosine-5 methyltransferases in yeast, flies, worms, and mice are not lethal, however, in all cases, the level of m5C deposition plays a significant role in regulating the cellular response to stress including drugs, DNA damage, oxidative stress, or environmental cues [23, 175, 184]. Mechanistically, rRNA modifications such as NSUN5-mediated methylation fine-tune the translation capacity of the ribosome by adapting it to specifically translate mRNAs relevant in stress [24]. m5C has also been detected in small ncRNAs and lncRNA [154, 168, 169, 185,186,187]. m5C deposition on ncRNA such as vault RNA also affects its processing into miRNA-like regulatory small RNA [168, 171].
Role of m5C in mRNAs
Only two recent studies have reported finding readers that bind to m5C-modified mRNAs, revealing the biological relevance and the role of m5C deposition in mRNAs (Fig. 3a). In one report, nuclear mature methylated mRNAs at any nucleotide position interacted with the nuclear export factor AlyREF and were more likely to be exported to the cytoplasm [173]. More recently, another report showed that in cancer cells NSUN2 aberrant methylation of oncogenic mRNAs at the 3′ UTR increased their interaction with the reader protein Y-box-binding protein 1 (YBX1), which maintained the stability of its targeted mRNAs by recruiting ELAVL1 [157]. Despite these studies, the advances in finding the prevalence, deposition site preference, molecular function, writers and readers of m5C on mRNA still remain elusive.
The role of m5C in cancer
Role of m5C writers in cancer
Over the past decade, a number of cytosine-5 methyltransferases have been found to be associated with various human diseases including several cancer types (Table 2). NOP2 overexpression was long shown to increase proliferation of mouse fibroblasts (Fig. 3g) [188], and it was found to be a valuable proliferation marker [189]. NOP2 expression has been found to be upregulated in breast, lung, prostate cancer, and gallbladder carcinoma, and its expression correlates with poor prognosis [190,191,192]. Mechanistically, NOP2 has been shown to bind to the T-cell factor (TCF)-binding element of the cyclin D1 promoter, recruiting TERC elements (telomerase RNA component) and activating cyclin D1 transcription. Whether NOP2’s methylating activity of rRNA is involved remains unexplored [193]. NSUN2 expression alterations have been linked to several cancer types including breast, skin, colon, ovarian, oesophageal, bladder, gallbladder, gastric cancer, and head and neck squamous carcinoma [157, 172, 194,195,196,197,198,199,200]. DNMT2 is upregulated in hundreds of tumour samples and several somatic mutations in DNMT2 have been identified in different tumours types [201]. NSUN4 loci is associated to increase risk of breast, ovarian or prostate cancer and its high expression is associated with HCC [202, 203]. Lastly, NSUN5 expression has been found to correlate with poor survival in glioblastoma [24] and NSUN7 high expression is associated to shorter survival in low-grade gliomas [204].
Regarding the associated mechanisms, the methylases’ molecular role seems to be tissue and context dependent, showing different unique substrates preferences for each cancer type, but with a unifying oncogenic result. For example for NSUN2, in most reports its overexpression is shown to regulate the fate of one single transcript of tumour suppressor genes or oncogenes and thus promoting proliferation [205]. Very recently NSUN2 was also shown to promote tumour progression by methylation of NMR ncRNA in oesophageal cancer [172]. In another recent study, it was reported that aberrant NSUN2-mediated methylation at the 3′ UTR of oncogenic mRNAs such as heparin-binding growth factor (HDGF) mRNA can increase their stability by interacting with YBX1 (Fig. 3e) [157]. While all these reports focused on the fate of mRNAs or other transcripts whose m5C prevalence is low, and considering that tRNAs are the main NSUN2 targets and are highly methylated at C-5, it is not yet clear whether the potential role of NSUN2 in cancer might actually be mediated by modifications of mRNA.
Regarding tRNA methylation functions, NSUN2 has been reported to be upregulated in a population of proliferative progenitor cells in skin tumours that rely on the correct deposition of m5C onto tRNAs [33]. Unexpectedly, deletion of NSun2 in mouse cells and mouse cancer cells led to hypomethylated tRNAs which showed to be more vulnerable to the ribonuclease angiogenin, and consequently to the accumulation of 5′-tRNA fragments [33, 42, 182]. Notably, the increase cleavage of tRNAs did not resulted in depletion of mature tRNAs, supporting that cleaved tRNAs mediated the phenotypic consequences. In fact, there is a growing appreciation that biogenesis of tRNA fragments is largely induced under stress conditions and their aberrant expression, commonly found in cancer, may indicate important regulatory functions in tumourigenesis (extensively reviewed in [183]). The understanding of their function is still in its infancy, but the reported data indicate that 5′-tRNA fragments can reprogram translation to favour stress responses by regulating the binding of translation initiation factors to the translation initiation complex [206,207,208]. Indeed, in Nsun2 deficient mouse cancer cells, ribosome profiling data showed an increased translation of genes associated to stress response pathways and decreased translation of genes associated to differentiation [33]. Importantly, the data showed that Nsun2 deficiency in cells blocked them in an undifferentiated and more proliferative state necessary for the self-renewal of tissue or tumour stem cells [33, 179, 181, 182]. However, prolonged deficiency showed to increase the sensitivity to stress of the undifferentiated cells [33, 42]. The finding was further supported by showing that Nsun2 deficient skin tumour initiating cells were more efficiently killed by using chemotherapeutic agents such as 5′ FU or cisplatin, which could be further rescued upon treatment with angiogenin inhibitors [33]. Although the exact mechanism by which 5′-tRNA fragments directly favoured translation of specific set of transcripts is still unknown, the study demonstrated that the combinatory use of tRNA fragment biogenesis enhancers such as NSUN2 inhibitors, with convectional chemotherapeutic agents could result in an efficient strategy to specifically eliminate tumour initiating cells. Thus, these and other studies show that tumour initiating cells and cancer cells require tight control of tRNA methylation, tRNA fragment biogenesis and protein synthesis [209] for accurate cell responses and to maintain the bulk tumour, and suggest the use of tRNA methylation inhibitors as potent cancer initiating cells sensitizers to cytotoxic stress (Fig. 3f).
The contribution of tRNA modifications in survival and differentiation is further supported by findings that Dnmt2 deletion in mice and flies was characterized by defects in stress responses and differentiation [162, 167]. In line with Nsun2 −/− mice phenotype, Dnmt2-depletion in mice did not globally perturb protein synthesis rates but rather affected specific mRNAs through reduced translation fidelity caused by loss of tRNA methylation [167]. Yet the potential biological impact of elevated tRNA cleavage due to Dnmt2 loss or cytosine-5 methylation inhibition remains completely unexplored. Additional work will be necessary to unveil the contribution of tRNA fragment abundance to the complex phenotypes observed.
Alterations in rRNA methylation have been also linked to cancer. NSUN5 mRNA expression is strongly associated with poor survival in glioblastoma patients. Epigenetic loss of NSUN5 expression in gliomas leads to rRNA cytosine hypomethylation and to increased translation of survival factors, rendering glioma cells sensitive to substrates of the stress-related enzyme NAD(P)H quinone dehydrogenase 1 (NQO1) (Fig. 3h) [24].
Thus, these findings highlight the importance of the tight control of tRNA and rRNA methylation and specialised protein synthesis programmes and set the basis to search for tRNA methylase inhibitors as a potent new approach to treat cancer.
Role of m5C erasers in cancer
5-hydroxymethylation writers have been found altered in cancer too. TET and ALKBH1 mutations or expression alterations are highly associated to some malignancies. For example, TET2 and ALKBH1 mutations have been associated to lymphoblastic and myeloid leukaemia [210,211,212]. TET1 expression is upregulated in glioblastomas [213], TET2 is downregulated in gliomas [214], and TET3 is epigenetically repressed in gliomas [215]. While mechanistically the cause has been always associated to DNA demethylation or hydroxymethylation defects, TET and ALKBH1 have been shown to oxidise m5C in RNA too, raising the question as to whether defects of RNA hydroxymethylation could be also linked to cancer.
Targeting m5C machinery in cancer
To date no specific cytosine-5 RNA methyltransferase inhibitor has been developed. Yet, given that several drugs designed to interfere with cytosine-5 methylation of DNA rely on the use of chemical analogues of cytosine, they may well interfere with RNA methylation [50, 216]. In fact, in a study by Lyko and co-workers, complete inhibition of Dmnt2-mediated 5-cytosine tRNA methylation with azacytidine in cancer cells reduced their proliferative capacity, supporting the notion that reduced cytosine-5 methylation of tRNAs may be an efficient cancer therapeutic strategy [50]. Despite the promising results and potential clinical implications, the fact that those analogues can both inhibit RNA and DNA methyltransferases raise awareness of the use of this unselective drugs, which may affect the methylation of multiple targets (DNA, tRNA, rRNA, mRNA, ncRNA) and may have devastating consequences.
In line with the notion that inhibition of tRNA methylases may contribute to chemotherapy resistance, silencing of other tRNA methyltransferases such as the 7-guanosine methylase METTL1 which also methylates tRNAs at the variable loop of several tRNAs has shown to increase sensitivity of cancer cells to 5’FU [217].
Pseudouridine
Pseudouridine deposition in RNA
Pseudouridine (Ψ), the C5-glycoside isomer of uridine, was the first posttranscriptional modification discovered and is one of the most abundant modifications of RNA [218, 219]. Despite its discovery over seventy years ago, we are only now beginning to uncover its biological function [25,26,27, 220,221,222,223].
Pseudouridine was initially detected on yeast tRNAs and rRNA [224, 225], and now we know that different types of RNAs including tRNA, rRNA, small nuclear RNAs (snRNAs), small Cajal Body-specific RNAs (scaRNAs), miRNAs, lncRNAs are also Ψ-modified [226]. Most importantly, with the current technological advents which rely on chemical treatment of RNA using soluble carbodiimide(N-Cyclohexyl-N′-(2-morpholinoethyl) carbodiimide metho-ρ-toluenesulfonate (or CMCT) for the generation of reverse transcription-stops, several groups have successfully performed genome-wide mapping experiments validating established targets and revealing novel substrates like mRNAs (Fig. 1) [25, 26, 221]. Since then, several seminal studies have followed discovering novel substrates and molecular roles of Ψ [25,26,27, 221, 222, 227,228,229,230].
Pseudouridine synthetases
Pseudouridylation can be achieved through two distinct mechanisms, namely RNA-independent and RNA-dependent pseudouridylation. The RNA-dependent mechanism relies on RNA–protein complexes known as box H/ACA small ribonucleoproteins (snoRNPs), which consist of a box H/ACA snoRNA and four core proteins; dyskerin (also known as NAP57 or DKC1), non-histone protein 2 (Nhp2), nucleolar protein 10 (Nop10) and glycine-arginine-rich protein 1 (Gar1). In H/ACA ncRNA is responsible for substrate recognition through complementary base-pairing interactions with the RNA substrate, and the catalytic activity is provided by DKC1 (Fig. 5a) (reviewed in [227]). The RNA-independent pseudouridylation is catalysed by a single enzyme, pseudouridine synthases (PUS), which carry both substrate recognition and catalysis without an RNA template strand (Fig. 5b). The rules governing RNA substrate recognition by RNA-independent Ψ synthases have only been elucidated in a few cases (for a review, see [231]). Some of the characterized Ψ synthases have a rather strict substrate specificity and are only able to modify one position in only one type of cellular RNA, such as, for instance, tRNAs. The strict substrate specificity depends on the universally conserved G53UUCNANNC60 sequence in most tRNAs, and on the three-dimensional structure of the TΨC loop [232]. Similar results were found by analysing the co-crystal structures of the prokaryotic enzyme TruB with isolated tRNA stem-loops [233]. The crystals illustrated how the enzyme core domain makes extensive interactions with the RNA, and how TruB requires the consensus sequence around the TΨC loop [233]. The mechanism for enzymes with a broader susbtrate spectrum is different. For example, PUS1, which methylates three positions in tRNAs, relies on two positively charged RNA-binding clefts along the surface of the protein which uniquely interacts with the tRNA [234]. In the case of PUS7, the sequence and two-dimensional structure are required for substrate recognition [235]. How other Ψ synthases with a broader substrate repertoire can recognize all the different substrates with a high degree of specificity is still under study.
Ψ deposition mechanisms and pathological implications in cancer. a Schematic illustration of RNA-dependent pseudouridylation mechanisms. Substrate recognition is achieved by sequence and structure homology of the substrate with the structural stems and loops formed by box H/ACA small nucleolar RNAs (snoRNA). Dyskerin (DKC1) is the catalytic unit and non-histone protein 2 (Nhp2), nucleolar protein 10 (Nop10) and glycine-arginine-rich protein 1 (Gar1) are regulatory units. The RNA substrate is represented in green. b Illustration of RNA-independent pseudouridylation mechanisms. Substrate recognition and catalysis are performed by one single pseudouridine synthases (PUS) (blue). c Representative scheme illustrating the target specificity for each pseudouridine synthases. The fate of each modified RNA is also illustrated with orange arrows. Pseudouridylated RNAs may be also recognised by still unknown readers (?). d Altered expression of pseudouridine synthases can lead to cancer. For example, reduced expression of DKC1 induces a reduction of pseudouridylation in TERC and rRNA, leading to dysfunctional TERC and rRNA and increasing tumourigenesis. e In glioma, An increased expression of DKC1 can lead to an increased Ψ deposition on rRNA, snRNA and TERC, and thus promoting cancer cell growth and migration. f PUS7 decreased expression leads to hypomodified tRNA-derived fragments, leading to increased self-renewal and survival in bone marrow mononuclear cells, promoting tumourigenesis. Red arrows indicate an increase and blue arrows a decrease of processes or enzymes
In eukaryotes, there are over fourteen different PUS enzymes, ten of these enzymes belong to the PUS family, including PUS1-10 [227, 236]. Each of them has specific substrates, but also share some of the substrates (Fig. 5C). For example, PUS1 modifies tRNAs at positions 38, 39 and/or 40 [234]. PUS4 [237] and PUS10 modify too tRNAs, but at position 55 [238]. Regarding erasers, an important difference between pseudouridylation and base methylations is that pseudouridylation is an irreversible modification in mammals, and instead mammals excrete the intact nucleoside [239]. No readers have been described to date.
Molecular function of Ψ deposition in RNA
Pseudouridylation plays different physiological roles depending on the RNA that is modified, but most commonly experimental data confirmes an important role in different aspects of gene expression regulation. The presence of Ψ is capable of increasing the rigidity of the phosphodiester backbone of the RNA and affecting its thermodynamic stability and spatial conformation, making short RNAs more stable [240, 241]. In snRNAs, in vitro pseudouridylation generates RNA conformational changes that influences the snRNA-mRNA interactions [242, 243]. In vivo studies have shown that these conformational changes are important for the snRNA activity. For example, the yeast U2 snRNA is pseudouridylated during stress at positions U56 and U93 by the RNA-independent enzyme PUS7 and a box H/ACA RNP complex (snR81), having functional implications in the efficiency of pre-mRNA splicing [229]. Ψ’s role in small nuclear RNAs was thoroughly reviewed recently in [220].
Pseudouridylation also generates structural stability to different types of RNA, such as rRNAs, which is necessary for their function [244, 245]. In rRNAs, Ψs are found at the decoding site, mRNA channel, peptidyl transferase centre, tRNA binding site and ribosomal subunit interface, thus showing an important role for the correct assembly and function of the ribosome and for protein synthesis [246]. For example in yeast, alterations or substitutions of amino acids in the Ψ synthase domain of Cbf5p abolish pseudouridylation of rRNA, resulting in growth defects and reduced levels of cytoplasmic 40S and 60S subunits of rRNA [244]. In mouse embryonic fibroblast cells, expression of dyskerin mutants leads to altered rRNA processing, unstable secondary structure of rRNA and cell growth defects [245]. Another similar example occurs upon the loss or disruption of H/ACA snoRNAs that guide rRNA modification. Pseudouridylation at position U2919 in yeast rRNA is guided by five H/ACA snoRNAs, whose loss leads to reduced rRNA pseudouridylation at U2919, and impaired 18S rRNA biogenesis and growth defects in yeast [247].
In mRNAs, the incorporation of Ψ can mediate nonsense-to-sense codon conversion facilitating the base pairing in the ribosome decoding centre, and thus resulting in protein diversity [248]. Other in vitro studies have also reported that translation of mRNAs containing pseudouridines is slowed down and mRNA decoding affected compared to non-modified mRNAs [249]. In addition, Ψ-containing mRNAs have shown higher stability in cells undergoing stress, suggesting that increased pseudouridylation increases their stability [25,26,27]. In fact, in vitro-transcribed mRNAs containing pseudouridines that were introduced in mammalian cells or into mouse tissues displayed enhanced stability relative to uridine-containing in vitro-transcribed mRNA [250]. Notably, the majority of pseudouridines in mRNA are regulated in response to environmental signals, but its functional role is still poorly understood [25,26,27, 220,221,222,223].
In tRNAs, pseudouridylation is found at the anticodon stem and loop, at the D stem and in a conserved at the Ψ loop, position 55, all of which stabilize its tertiary structure and are important for codon–anticodon base-pairing [227] (Fig. 4). Novel insights into the role of tRNA pseudouridylation were recently obtained using human embryonic stem cells (hESCs) and haematopoietic stem and progenitor cells (HSPCs) [230]. This work unveiled that similar to m5C deposition at the anti-codon and variable loops, the deposition of Ψ at the eighth position of tRNAs by PUS7 regulated the bioproduction of a specific class of 18-nt 5′ tRNA fragments containing a 5′ terminal oligoguanine motif (mTOGs). In addition, the study showed that the presence of Ψ at position 8 of this novel class of small ncRNAs was required for efficient biding to polyadenylate-binding protein 1 (PABPC1), an integral component of the 5′ cap translation initiation complex, resulting in displacement of PABPC1 from capped mRNAS and reprograming translation [230]. Thus, the study showed that RNA modifications, by regulating the biogenesis of a novel class of ncRNAs, they can specialized the translation machinery.
The role of Ψ on other RNAs remains still poorly understood. For instance, TERC is also pseudouridylated [26], where two specific sites of pseudouridylation have been detected, although there is currently no evidence that this modification may be involved in telomerase activity [251]. In miRNA, depletion of PUS10 results in a marked reduction of the expression levels of mature miRNAs and concomitant accumulation of unprocessed pri-miRNAs, but unexpectedly, this process results independent of the catalytic activity of PUS10 [228].
In sum, Ψ deposition may confer different molecular properties to the modified RNAs, which changes their fate or activity. While most recent studies are focused on Ψ modifications in mRNA, yet its role on mRNA is still unclear. Thus, further studies will be required to fully understand the molecular role of Ψ on mRNAs. Ψ is highly abundant on other RNA types such as snRNA, rRNA or tRNA, and whose role has been studied for many years. Yet, recent findings are still revealing novel and unexpected functions, such as the role of Ψ on tRNA fragments, and thus indicating that other functions are still to be discovered.
Role of pseudouridylated RNAs in cancer
Role of DKC1 in cancer
One of the most studied disease linked to defects in pseudouridylation is the X-linked Dyskeratosis Congenita (X-DC), associated to DKC1 inactivating mutations [252]. Dyskerin modifies mainly rRNAs and snRNAs and it also participates in the active telomerase complex [219]. X-DC is characterized by defects in reticulate skin pigmentation, nail dystrophy, and mucosal leukoplakia, but bone marrow failure is the principal cause of early mortality in X-DC patients [253]. Patients with X-DC are also characterized by having higher risk for cancer development, and expression alterations are associated to cancer too [254]. DKC1 alterations have been found associated to skin cancer [255], breast [256, 257], colon [254, 258, 259], lung [254, 260], prostate [261], head and neck [262, 263], glioma [264], HCCs [265], and specially bone marrow failure syndromes and hematologic malignancies including chronic lymphocytic leukaemia [266, 267] or multiple myeloma [268,269,270] (Table 3). From the molecular point of view, DKC1 is a nucleolar protein that participates in the stabilization of the telomerase RNA component, necessary for telomerase activity [271, 272], and pseudouridylation of diverse rRNA residues at important ribosome domains for tRNA and mRNA binding, all of which are important functions in highly proliferating cells [273]. Thus, lack of dyskerin activity causes a reduced replicative potential and premature ageing by an impairment of telomerase activity [251] and impediment of ribosome translation of specific mRNAs [274, 275], primarily affecting tissues with rapid cell turnover (Fig. 5d). Mechanistically, it has been shown that low levels of pseudouridylated rRNA downregulates the internal ribosome entry (IRES)-dependent translation of tumour suppressors such as p53 [276], p27 and inhibitors of apoptosis (Bcl-xL, XIAP) [274, 277]. Increased expression of vascular endothelial growth factor has been associated to loss of DKC1 [278], resulting their depletion in high incidence of cancer development. Other recent studies investigating the role of Ψ in rRNA have also supported the emergent hypothesis of the existence of specialized ribosomes in cancer. In liver cancer cells, aberrant expression of snoRNA24, which mediates the pseudouridylation at U609 and U863 positions in 18S rRNA, leads to changes on tRNA selection efficiency, ribosome elongation rate and translation efficiency influencing cancer cell survival [279]. Other example of pseudouridine modifications in rRNA is the reduction of m1acp3Ψ modification of rRNA found in colorectal carcinoma, which affects the direct interaction of tRNA with ribosomal P site, altering and deregulating translation in cancer cells [280]
Although there is more evidence that Dyskerin acts as a tumour suppressor [274, 277, 278, 281], in contrast, other studies have indicated an oncogenic role. For example, in breast cancer, decreased levels of DKC1 expression, rRNA pseudouridylation and telomere length correlate with better prognosis [256]. In glioma, increased expression of DKC1 and pseudouridylation promote glioma cell growth and migration by inducing the upregulated expression of gliomagenesis regulators, although the direct role of increased pseudouridylation of RNAs and gliomagenesis remains unexplored (Fig. 5e) [264].
The data so far show that alterations in DKC1 expression or activity are significantly associated to cancer, however the exact mechanism may be cell, tissue or DKC1 substrate dependent. Further studies will be required to fully explore whether DKC1 has a pro-oncogenic or tumour suppressive role.
Role in cancer of RNA-independent pseudouridine synthetases
Despite the significant association of DKC1 activity or expression alterations to cancer, little is known on the role of other pseudouridylases and only few studies have associated alterations on their expression or activity to cancer (Table 3). For example, PUS1 mediates the interaction of steroid receptor RNA activator 1 (SRA1) with retinoic acid receptor-γ (RARγ) in melanoma cells, and with oestrogen receptor in breast cancer cell lines [282]. PUS10 is a mediator of TNF-related apoptosis inducing ligand (TRAIL)-induced apoptosis in prostate cancer cells [283], and genomic alterations of PUS10 locus are significantly associated with lung cancer risk [284], although it is not clear whether this effects are dependent on pseudouridylase activity. Loss of PUS7 occurs frequently in myelodysplastic syndromes, haematological clonal disorders characterised by haematopoietic stem cells dysfunction and high risk of transformation to AML [285]. Mechanistically, Guzzi et al. demonstrated that PUS7 depletion in hESC and HSPCs reduced PUS7-mediated pseudouridylation in a special class of tRNA-derived RNA fragments, inducing significantly higher protein synthesis rates leading to dramatic growth and differentiation defects (Fig. 5f) [230]. Thus this work supports the emerging view that ESCs and cancer cells are highly sensitive to perturbations of protein synthesis, and highlights an important role for tRNA fragment in reprograming translation [286]. Additional work using in vivo models and patient-derived cancer cells will be necessary to explore the clinical implications of targeting PUS7 loss or aberrant tRNA fragment bioproduction in cancer.
Targeting pseudouridine synthetases in cancer
From the clinical point of view, PUSs or Ψ may serve as potential anti-cancer targets and biomarkers. For instance, high amounts of Ψ have been detected in urine of colon, prostate or ovarian cancer patients, plasma of ovarian patients or in salivary metabolites of oral squamous cell carcinoma patients, suggesting to be a potential biomarker in liquid non-invasive biopsies for early cancer diagnoses [261, 287,288,289,290]. Yet, while mutations and expression alterations in DKC1 have been significantly reported in cancer, little has been reported on the status of other Ψ synthetases. With the advent of cancer genome- and transcriptome-wide studies, future computational analyses will reveal the mutational and expression status of all known human Ψ synthetases.
Regarding the design of drugs or screening for small molecules that inhibit PUSs activity, while several studies have attempted to generate or find compounds to diminish DKC1 activity as potential anti-cancer treatments, very little progress has followed [291,292,293]. Clinical trials were performed in ovarian carcinoma [294], sarcoma [295], colorectal carcinoma [296], acute myelogenous leukaemia [297], breast cancer [298], lung cancer [299], melanoma [300] and other cancers [301] to examine the effectivity as anti-cancer therapy of pyrazofurin, a small molecule inhibitor of the orotodine-5′-monophosphate-decarboxylase (ODCase) which also inhibits DKC1. In all cases pyrazofurin failed to demonstrate efficient anti-cancer activity. However, whether pyrazofurin could be an effective treatment for patients with overexpressed DKC1 was not concluded from those studies, since DKC1 expression levels were not taken into account.
Another potential small molecule inhibitor that is already used as effective anti-cancer agent in the clinic is 5’FU. Treatment with 5’FU is known to improve survival in various cancers [302]. While the mechanism of action for active metabolites of 5’FU has been always attributed to disruption of both DNA and RNA syntheses and DNA damage induction [302], 5’FU was shown to inhibit pseudouridine synthases, due to the substitution of uracil by the analogue 5’FU in RNAs [303, 304]. In those studies, Samuelsson et al. demonstrated that the use of fluorinated tRNAs generated specific and stable non-covalent complexes with yeast pseudouridine synthases, thus acting as potent and irreversible inhibitor [304]. Nonetheless, 5’FU is not an universal inhibitor for all pseudouridine synthases, in fact TruB E. Coli, and most likely its eukaryotic homologues, cannot form covalent adducts with fluorinated RNAs [303]. Thus, it would be interesting to test the specificity and efficacy of 5’FU in inhibiting mammalian pseudouridine synthases. In addition, it would be important to determine whether part of the associated cytotoxicity of 5’FU is due to an overall decrease of RNA pseudouridylation or to the loss of a particular modified RNA (e.g. rRNA).
More recent studies have used in silico approaches which can predict possible new inhibitors. One study predicted the use of a small molecule inhibitor based on the disruption of the interaction of DKC1 with TERC [293]. The virtual docking-based screen found ten molecules with high affinity values, of which three resulted as potent telomerase inhibitors in a breast cancer cell line. In another study, Floresta et al. hypothesized that nucleoside analogues such as the isoxazolidinyl derivative 5′-monophosphate could act as an inhibitor of pseudouridine 5′-monophosphate glycosidases competing with the natural substrate and hampering the glycosidic C–C bond cleavage [292]. They indeed found that the isoxazolidinyl derivative accommodated within the active site of the enzyme with higher ligand efficiency than the natural substrate, leading to the enzyme inhibition in vitro. While we still ignore the tumour growth inhibitory potential and the therapeutic benefits of using those first inhibitors, these studies set the basis to continue with the search for Ψ synthetase inhibitors for cancer treatment.
Conclusion
RNA modifications have emerged as critical posttranscriptional regulators of gene expression programmes. We start to appreciate the functional networks that the epitranscriptome interacts with, ranging from metabolisms [31] to epigenetics and chromatin remodelling [24, 216] or the immune system [116]. Despite the progress, most studies have focused on the molecular and physiological functions of only one mark, 6-methyladenosine on mRNA, however the epitranscriptome embraces over 170 RNA chemical modifications that decorate coding and ncRNAs, several other posttranscriptional RNA processing events, and RNA binding proteins that may be as well modified as histones in DNA [1]. Thus, association studies of hundreds of other RNA marks on coding but also ncRNAs such as tRNA or rRNA remain to be explored.
To achieve this, we first need to develop system-wide methods and tools for rapid and quantitative detection of RNA modifications. Most of the stablished methods rely on next-generation sequencing and, as such, they are typically blind to nucleotide modifications. Consequently, indirect methods are required that are based on immunoprecipitation techniques using specific antibodies, or enzymatic methods and chemical labelling and unique base-modification properties of RNA pairing [305]. These methodologies have allowed us to catalogue and identify endless modifications with high precision and at nucleotide resolution, yet these strategies have some limitations, reproducibility rate is low due to technical limitations and poor computational methods. For example, antibodies used to recognize modifications such as m6A still exhibit non-specific binding and can bind to N6,2′-O-dimethyladenosine (m6Am) too [59]. In fact, all antibody-related approaches suffer from poorly characterized, and thus unpredictable specificity of the antibody used for enrichment [306]. To overcome these limitations, novel detection methods have been introduced such as DART-seq (deamination adjacent to RNA modification targets), an antibody-free method for detecting m6A sites, where the cytidine deaminase APOBEC1 is fused to the m6A-binding YTH domain [307]. Yet, this methodology is also limited since it allows to simultaneously map only RNAs bound to YTH domain containing readers. For m5C detection, bisulphite-RNA sequencing is the gold standard, yet the reproducibility is low, especially in low abundant and unstable RNA such as mRNAs [16]. The large initial amount of RNA required to compensate for the high losses caused by the treatment, the resistance to C-U conversion from neighbouring modifications or double-stranded sequences, the inability to differentiate from other modifications protecting the C from the conversation such as 5-hydroxymethylation or 3-methylcytosine, and poor computational analysis are the most common found difficulties [16]. Yet, careful assessment of C-U conversion together with development of statistically robust bioinformatic tools that highly refined for data analysis are still generating contradictory results [155, 156]. Regarding the detection of pseudouridine, several labs have developed methodologies that rely only the chemical treatment of RNAs with CMCT, resulting in little overlap of pseudouridine sites on mRNA from the different studies [25, 26, 221]. Thus, despite to the variety of the established techniques, the reality is that there is currently no generic and precise method for mapping and quantifying modifications in RNA. In addition, current methods are complex and lack of single molecule resolution. In this regard, the emerging third-generation sequencing technologies, such as the platforms provided by Oxford Nanopore Technologies (ONT) and Pacific Biosciences (PacBio) have been proposed as a new means to directly detect RNA modifications [308]. RNA modifications can be detected by kinetic changes of reverse transcriptases when encountering a modified nucleotide (PacBio) [309]. Or by current changes as the native RNA molecule is pulled through a membrane pore (ONT) [310]. Although the detection of modifications using ONT direct RNA sequencing is already a reality [311], yet current efforts have not yielded an efficient and accurate RNA modification detection algorithm, largely due to the challenges in the alignment and re-squiggling of RNA current intensities. But emerging alternative base-calling strategies such as EpiNano algorithms which identifies m6A from RNA reads with an overall accuracy of ~90%, open new avenues to explore additional RNA modifications in the future [312].
The dynamic expression patterns of writer, reader and eraser proteins complicate the identification of the precise functional consequences of aberrant deposition of modifications on RNA metabolism. Thus uncovering the complete repertoire of cellular RNA substrates and the writers, readers, and erasers will unveil how the intricate network of epitranscriptomic events can converge into similar cellular processes and showing how their unbalanced deposition may lead to pathologies. For example, HIF1A mRNA is stabilized by m6A deposition, while in melanoma cells high levels of HIF1α protein are maintained by modifications at the U34 wobble position of tRNAs [51, 143]. Furthermore, it will be essential to understand the factors or signals that determine the specificity of the RNA modification writers, readers, and erasers and how these proteins are regulated in different cell types. For instance, METTL16 is sensitive to SAM levels and can regulate its synthesis by modifying the SAM synthase gene MAT2A as a feedback loop mechanism [313]. In addition, we need to develop innovative technologies for precise manipulation of the epitranscriptome and functional assays that enables to understand their dynamic mechanisms of action of each modified RNA, since depletion of individual RNA modifier may not be sufficient to comprehensively understand their roles. For example, while the main target for NSUN2 are tRNAs, it still remains unclear whether the phenotypic changes seeing upon Nsun2 deletion are caused by decrease methylation of tRNAs, or other RNA substrates may as well contribute to the observed phenotype [42, 168, 179, 181, 182].
We start to appreciate the wide range of functional consequences of the aberrant deposition of RNA modifications in human diseases including cancer. For example, aberrant deposition of tRNA modifications, including Ψ and m5C, leads to perturbed accumulation of tRNA fragments, a novel class of functional ncRNAs whose role is associated with aberrant protein synthesis rates and reprograming of the translational machinery in tissue and cancer stem cell populations [33, 230]. These findings support the current view that balanced protein synthesis and tight control of mRNA translation is central to cellular processes involved in tumorigenesis [314] and highlights a key role for tRNAs and tRNAs fragments in tumourigenic processes [315]. Yet, our understanding on how specific species of tRNA fragments govern these processes and the clinical implications of their aberrant expression are still very limited. Future studies will be necessary to decipher the molecular bases of the translation reprograming driven by tRNA fragments. Additional work will be necessary to differentiate the contribution of global changes in tRNA pools versus specific population tRNA-derived fragments in the tumour promoting effect of aberrant protein synthesis [315]. More importantly, given their association to cancer progression, their therapeutic potential must be explored exploiting the current advent in miRNA-based therapeutics [316].
Recent advancements in the rapidly evolving field of epitranscriptomics have linked the reprogramming of components of the epitranscriptomic machinery including writers, erasers or readers of the m6A, m5C or Ψ to cancer. The extensive number of RNA modifications that constitute the epitranscriptome and the reversible nature of epitranscriptomic aberrations hold promise to the emergence of a promising field of epitranscriptomic therapy, which is already making progress with the recent development of effective inhibitors against m6A modifying proteins. In the last years, several seminal studies have shed light onto the diagnostic and therapeutic potential of targeting the cancer epitranscriptomic code [3, 33, 317], yet we are far from understanding whether spatio-temporal modifications of the epitranscriptome can drive tumour initiation [318]. In addition, their use as therapeutic agents remains a great challenge due to the lack of consistent and consolidated evidence on the oncogenic or tumour suppressive nature of for example the aberrant deposition of m6A. The inconsistent evidence may reflect the precise functional outcome of the RNA modification on each modified RNA type, their crosstalk with other active signalling processes and the dynamic nature of the epitranscriptome. Identifying accurate epitranscriptome biomarkers and defining the oncogenic or tumour suppressive effect of a given aberrant modification within a specific cellular context, cell type, cellular proliferative capacity or tumour microenvironment will guide finding the exact molecular targets to develop selective and effective therapies for a given tumour type.
Though aberrant expression of several methylases and Ψ synthetases has now been described in cancer, it remains unclear whether they could be efficient targets for cancer therapy. Thus, the precise contribution of methylases and Ψ synthetases to tumour initiation, growth, metastasis and resistance need to be further investigated. It remains unclear the RNA target specificity of each enzyme and how specific modified targets can contribute to the malignant phenotype. In addition, little is known on the dynamics of the deposition of these modifications, their erasers and how the binding to their readers influence the RNA metabolism and tumour cell fate. Yet, the availability of the 3D structure of most of these enzymes and the fact that potent and selective inhibitors have been found [52], it is reasonable to expect that inhibition of these enzymes is achievable. These structures can provide the basis for structure-guided drug design which in combination with computational tools can be powerful resources for the development of RNA modifying enzymes inhibitors. Few small molecule inhibitors have been described that can target specifically m6A erasers, yet none of them have reached clinical stages [52]. For m5C methylases, azacytidine and decitabine (5-aza2′-deoxycytidine), which are cytidine analogues and can inhibit any cytosine-5 methylase, have been approved for clinical use in haematological malignancies [319]. However, their use should be taken carefully due to the lack of specificity of these inhibitors that can inhibit both RNA or DNA cytosine methylases. The validation of these enzymes and their modifications as good pharmacological targets will require the discovery of potent, selective, cell-permeable inhibitors to determine the therapeutic benefit and potential risks associated with inhibiting these enzymes.
Treatment failure in certain settings has been attributed to the presence of sub-populations of cancer stem cells or persister cells which are intrinsically resistant to many therapeutic approaches [320]. Given that m6A, m5C or Ψ regulate cell survival to stress in many settings and stem cell functions, targeting them represent a very promising opportunity to specifically target these cells populations and reduce chemoresistance and recurrence. Whether other epitranscriptomics changes can lead to drug resistance is not yet understood. This clearly opens the opportunity to explore novel avenues to develop diagnostic tools based on epitranscriptomic signatures that allow for better patient stratification. In addition, combinatorial therapeutic strategies with the potential to re-establish the normal epitranscriptomic landscape or inhibit survival signalling pathways are promising strategies to specifically eliminate cancer stem cells. Combinatorial therapies that target independent pathways may be a better option as the possibilities for the development of tumour drug resistance may be more limited. Understanding the machineries and factors that introduce, remove, or read RNA modifications will allow the development of novel drugs with pharmaceutical value, not only for cancer but other complex human pathologies that have been linked to aberrant deposition of RNA modifications such as diabetes, neurological, immune and mitochondrial-linked disorders [47].
Availability of data and materials
Supporting data will be available upon request to corresponding author.
Abbreviations
- 6PGD:
-
6-Phosphogluconate dehydrogenase
- ADAM19:
-
ADAM metallopeptidase domain 19
- AFF4:
-
AF4/FMR2 family member 4
- AKT:
-
AKT serine/threonine kinase
- ASB2:
-
Ankyrin repeat and SOCS box containing 2
- AXL:
-
AXL receptor tyrosine kinase
- BCL-2:
-
B cell leukaemia 2
- BNIP3:
-
BCL2 interacting protein 3
- MET:
-
MET proto-oncogene, receptor tyrosine kinase
- CDK2:
-
Cyclin dependent kinase 2
- CEBPA:
-
CCAAT enhancer binding protein alpha
- CMCT:
-
Carbodiimide(N-Cyclohexyl-N′-(2-morpholinoethyl) carbodiimide metho-ρ-toluenesulfonate
- CXCR4:
-
C-X-C motif chemokine receptor 4
- E2F1:
-
E2F transcription factor 1
- EGFR:
-
Epidermal growth factor receptor
- EIF3C:
-
Eukaryotic translation initiation factor 3 subunit C
- ELAVL1:
-
ELAV like RNA binding protein 1
- ERCC1:
-
ERCC excision repair 1
- ERK:
-
Mitogen-activated protein kinase 1
- ESR1:
-
Estrogen receptor 1
- FOXM1:
-
Forkhead box M1
- GLI1:
-
GLI family zinc finger 1
- HBXIP:
-
Hepatitis B virus X-interacting protein
- HIF1A:
-
Hypoxia inducible factor 1 subunit alpha
- HINT2:
-
Histidine triad nucleotide binding protein 2
- ITGA6:
-
Integrin subunit alpha 6
- ITGB1:
-
Integrin subunit beta 1
- JUNB:
-
JunB proto-oncogene, AP-1 transcription factor subunit
- KCNK15-AS1:
-
KCNK15 and WISP2 antisense RNA 1
- KLF4:
-
Kruppel like factor 4
- LATS-2:
-
Arge tumor suppressor kinase 2
- LEF1:
-
Lymphoid enhancer binding factor 1
- Let-7 g:
-
MicroRNA let-7 g
- MALAT1:
-
Metastasis-associated lung adenocarcinoma transcript 1
- MEK:
-
Mitogen-activated protein kinase kinase 7
- MMP2:
-
Matrix metallopeptidase 2
- mTOR:
-
Mechanistic target of rapamycin kinase
- MYB:
-
Myeloblastosis oncogene
- MYC:
-
Myelocytomatosis oncogene
- MZF1:
-
Myeloid zinc finger 1
- NANOG:
-
Nanog homeobox
- NEAT1:
-
Nuclear paraspeckle assembly transcript 1
- NF-κB:
-
Nuclear factor kappa B subunit 1
- NMR:
-
Long intergenic non-protein coding RNA 1672
- NOP2:
-
NOP2 nucleolar protein
- NSUN:
-
NOP2/Sun RNA methyltransferase
- P2RX6:
-
Purinergic receptor P2X 6
- P53:
-
Tumor protein p53
- PDCD1:
-
Programmed cell death 1
- PI3K:
-
Phosphatidylinositol 3-kinase,
- Pri-miRNA:
-
Primary microRNA
- Pre-miRNA:
-
Precursor microRNA
- PRMT1:
-
Protein arginine methyltransferase 1
- PTEN:
-
Phosphatase and tensin homolog
- RARA:
-
Retinoic acid receptor alpha
- SETD7:
-
SET domain containing 7, histone lysine methyltransferase
- SHH:
-
Sonic hedgehog signaling molecule
- SOCS2:
-
Suppressor of cytokine signaling 2
- SOX10:
-
SRY-box transcription factor 10
- SOX2:
-
Sex determining region Y box 2
- SPRED2:
-
Sprouty related EVH1 domain containing 2
- TAZ:
-
Tafazzin
- TERC:
-
Telomerase RNA component
- TET:
-
TET methylcytosine dioxygenase
- TNF:
-
Tumor necrosis factor
- TNFRSF2:
-
TNF receptor superfamily member
- VASH:
-
Vasohibin 1
- WAF-1:
-
Cyclin dependent kinase inhibitor 1A
- XIAP:
-
X-linked inhibitor of apoptosis
- XIST:
-
X inactive specific transcript
- YAP:
-
Yes1 associated transcriptional regulator
References
Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–7.
Cohn WE. Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: isolation, structure, and chemical characteristics. J Biol Chem. 1960;235:1488–98.
Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20:303–22.
Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. The RNA modification landscape in human disease. RNA. 2017;23:1754–69.
Davalos V, Blanco S, Esteller M. SnapShot: Messenger RNA Modifications. Cell. 2018;174:498–498 e491.
Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346–9.
Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573–6.
Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162:1299–308.
Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–5.
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–41.
Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–73.
Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, Wang TT, Xu QG, Zhou WP, Sun SH. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–43.
Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–4.
Yang D, Qiao J, Wang G, Lan Y, Li G, Guo X, Xi J, Ye D, Zhu S, Chen W, et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018;46:3906–20.
Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Hobartner C, Sloan KE, Bohnsack MT. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004–14.
Garcia-Vilchez R, Sevilla A, Blanco S. Post-transcriptional regulation by cytosine-5 methylation of RNA. Biochim Biophys Acta Gene Regul Mech. 1862;2019:240–52.
Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008;9:629–35.
Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–51.
Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I, Klaholz BP. Visualization of chemical modifications in the human 80S ribosome structure. Nature. 2017;551:472–7.
Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2011;2:611–31.
Schwartz MH, Waldbauer JR, Zhang L, Pan T. Global tRNA misacylation induced by anaerobiosis and antibiotic exposure broadly increases stress resistance in Escherichia coli. Nucleic Acids Res. 2016;44:10292–303.
Sloan KE, Warda AS, Sharma S, Entian KD, Lafontaine DLJ, Bohnsack MT. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017;14:1138–52.
Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E, Calle-Perez A, Pircher A, Gerstl MP, Pfeifenberger S, et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat Commun. 2015;6:6158.
Janin M, Ortiz-Barahona V, de Moura MC, Martinez-Cardus A, Llinas-Arias P, Soler M, Nachmani D, Pelletier J, Schumann U, Calleja-Cervantes ME, et al. Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 2019;138:1053–74.
Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–6.
Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–62.
Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol. 2015;11:592–7.
Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009;15:1716–28.
Chan CT, Deng W, Li F, DeMott MS, Babu IR, Begley TJ, Dedon PC. Highly Predictive Reprogramming of tRNA Modifications Is Linked to Selective Expression of Codon-Biased Genes. Chem Res Toxicol. 2015;28:978–88.
Chionh YH, McBee M, Babu IR, Hia F, Lin W, Zhao W, Cao J, Dziergowska A, Malkiewicz A, Begley TJ, et al. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat Commun. 2016;7:13302.
Gkatza NA, Castro C, Harvey RF, Heiss M, Popis MC, Blanco S, Bornelov S, Sajini AA, Gleeson JG, Griffin JL, et al. Cytosine-5 RNA methylation links protein synthesis to cell metabolism. PLoS Biol. 2019;17:e3000297.
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6.
Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortes-Garrido R, Gkatza N, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335–40.
Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010;6:e1001247.
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017;18:2622–34.
Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell. 2017;31:127–41.
Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell. 2018;172:90–105 e123.
Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, Aplin AE, Lu Z, Hwang S, He C, He YY. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10:2782.
Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–19.
Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell Stem Cell. 2018;22:191–205 e199.
Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–6.
Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–39.
Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, Di W, Hu B, An J, Kong L, et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12:135.
Seidel A, Brunner S, Seidel P, Fritz GI, Herbarth O. Modified nucleosides: an accurate tumour marker for clinical diagnosis of cancer, early detection and therapy control. Br J Cancer. 2006;94:1726–33.
Blanco S, Frye M. Role of RNA methyltransferases in tissue renewal and pathology. Curr Opin Cell Biol. 2014;31C:1–7.
Belin S, Beghin A, Solano-Gonzalez E, Bezin L, Brunet-Manquat S, Textoris J, Prats AC, Mertani HC, Dumontet C, Diaz JJ. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS One. 2009;4:e7147.
Frye M, Blanco S. Post-transcriptional modifications in development and stem cells. Development. 2016;143:3871–81.
Begley U, Sosa MS, Avivar-Valderas A, Patil A, Endres L, Estrada Y, Chan CT, Su D, Dedon PC, Aguirre-Ghiso JA, Begley T. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-alpha. EMBO Mol Med. 2013;5:366–83.
Delaunay S, Rapino F, Tharun L, Zhou Z, Heukamp L, Termathe M, Shostak K, Klevernic I, Florin A, Desmecht H, et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J Exp Med. 2016;213:2503–23.
Schaefer M, Hagemann S, Hanna K, Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 2009;69:8127–32.
Rapino F, Delaunay S, Rambow F, Zhou Z, Tharun L, De Tullio P, Sin O, Shostak K, Schmitz S, Piepers J, et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature. 2018;558:605–9.
Boriack-Sjodin PA, Ribich S, Copeland RA. RNA-modifying proteins as anticancer drug targets. Nat Rev Drug Discov. 2018;17:435–53.
Desrosiers RC, Friderici KH, Rottman FM. Characterization of Novikoff hepatoma mRNA methylation and heterogeneity in the methylated 5' terminus. Biochemistry. 1975;14:4367–74.
Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71:3971–5.
Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343–55.
Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–70.
Schibler U, Kelley DE, Perry RP. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J Mol Biol. 1977;115:695–714.
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635–46.
Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–72.
Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev. 2015;29:2037–53.
Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5' UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015;163:999–1010.
Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC, Mullen AC. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017;20:2262–76.
Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, Ma J. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun. 2018;9:420.
Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S, Liu Y, Ye L, Li Y, Zhang X. MicroRNA-145 Modulates N(6)-Methyladenosine Levels by Targeting the 3'-Untranslated mRNA Region of the N(6)-Methyladenosine Binding YTH Domain Family 2 Protein. J Biol Chem. 2017;292:3614–23.
Xiao S, Cao S, Huang Q, Xia L, Deng M, Yang M, Jia G, Liu X, Shi J, Wang W, et al. The RNA N(6)-methyladenosine modification landscape of human fetal tissues. Nat Cell Biol. 2019;21:651–61.
Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269:17697–704.
Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89.
Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vagbo CB, Geula S, Hanna JH, Black DL, Darnell JE Jr, Darnell RB. m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017;31:990–1006.
Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016;63:306–17.
Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–8.
Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, Hamakubo T. Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. 2013;288:33292–302.
Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014;8:284–96.
Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, Masiello I, Hares T, Villaseñor R, Hess D, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m. Genes Dev. 2018;32:415–29.
Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, Conrad NK. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017;169:824–835 e814.
Mendel M, Chen KM, Homolka D, Gos P, Pandey RR, McCarthy AA, Pillai RS. Methylation of Structured RNA by the m(6)A Writer METTL16 Is Essential for Mouse Embryonic Development. Mol Cell. 2018;71:986–1000 e1011.
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7.
Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29.
Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, et al. Differential m(6)A, m(6)Am, and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol Cell. 2018;71:973–985 e975.
Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, et al. Reversible methylation of m(6)Am in the 5' cap controls mRNA stability. Nature. 2017;541:371–5.
Ueda Y, Ooshio I, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K, Hase H, Harada K, Hirata K, Tsujikawa K. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep. 2017;7:42271.
Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500.
Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10:927–9.
Li F, Zhao D, Wu J, Shi Y. Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 2014;24:1490–2.
Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45:6051–63.
Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T, Chen Y, Xiao Y, Gao Y, Zhang Q, et al. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29:23–41.
Zhang F, Kang Y, Wang M, Li Y, Xu T, Yang W, Song H, Wu H, Shu Q, Jin P. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum Mol Genet. 2018;27:3936–50.
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95.
Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61:507–19.
Lesbirel S, Viphakone N, Parker M, Parker J, Heath C, Sudbery I, Wilson SA. The m(6)A-methylase complex recruits TREX and regulates mRNA export. Sci Rep. 2018;8:13827.
Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017;6:e31311.
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64. https://doi.org/10.7554/eLife.31311. PMID: 28984244; PMCID: PMC5648532.
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–28.
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20.
Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626.
Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J, et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–27.
Zaccara S, Jaffrey SR. A Unified Model for the Function of YTHDF Proteins in Regulating m(6)A-Modified mRNA. Cell. 2020;181:1582–1595 e1518.
Jacobs E, Mills JD, Janitz M. The role of RNA structure in posttranscriptional regulation of gene expression. J Genet Genomics. 2012;39:535–43.
Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc. 2015;137:2107–15.
Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. 2019;11:4909–21.
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, Liu J, Che L, Li J. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer. 2019;18:143.
Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, Li J, An P, Lu L, Luo N, et al. m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019;18:87.
Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18:103.
Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, Li S, Tan L, Mai D, Li G, et al. Excessive miR-25-3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10:1858.
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110.
Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, Chen Z, Deng X, Xiao G, Auer F, et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature. 2019;567:414–9.
Du M, Zhang Y, Mao Y, Mou J, Zhao J, Xue Q, Wang D, Huang J, Gao S, Gao Y. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem Biophys Res Commun. 2017;482:582–9.
Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, Wu Y, Lv Y, Hao J, Wang L, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301.
Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–4.
Aguilo F, Zhang F, Sancho A, Fidalgo M, Di Cecilia S, Vashisht A, Lee DF, Chen CH, Rengasamy M, Andino B, et al. Coordination of m(6)A mRNA Methylation and Gene Transcription by ZFP217 Regulates Pluripotency and Reprogramming. Cell Stem Cell. 2015;17:689–704.
Zhong X, Yu J, Frazier K, Weng X, Li Y, Cham CM, Dolan K, Zhu X, Hubert N, Tao Y, et al. Circadian Clock Regulation of Hepatic Lipid Metabolism by Modulation of m. Cell Rep. 2018;25:1816–1828.e1814.
Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, Tienda SM, Chryplewicz A, Zhu AC, Yang Y, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20:1074–83.
Liu S, Li Q, Chen K, Zhang Q, Li G, Zhuo L, Zhai B, Sui X, Hu X, Xie T. The emerging molecular mechanism of m(6)A modulators in tumorigenesis and cancer progression. Biomed Pharmacother. 2020;127:110098.
Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell. 2016;62:335–45.
Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, Luo G, Tauler J, Du J, Lin S, et al. RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat Commun. 2019;10:2065.
Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, Zhao G. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18:142.
Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, Huang X, Liu Y, Wang J, Dougherty U, et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019;566:270–4.
Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–76.
Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature. 2017;552:126–31.
Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, Liu Y, Zhang X, Zhang W, Ye L. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–9.
Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, Ji D, Wang Q, Zhang Z, Tang J, Sun Y. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38:393.
Cai J, Yang F, Zhan H, Situ J, Li W, Mao Y, Luo Y. RNA m(6)A Methyltransferase METTL3 Promotes The Growth Of Prostate Cancer By Regulating Hedgehog Pathway. Onco Targets Ther. 2019;12:9143–52.
Li E, Wei B, Wang X, Kang R. METTL3 enhances cell adhesion through stabilizing integrin β1 mRNA via an m6A-HuR-dependent mechanism in prostatic carcinoma. Am J Cancer Res. 2020;10:1012–25.
Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–33.
Li X, Tang J, Huang W, Wang F, Li P, Qin C, Qin Z, Zou Q, Wei J, Hua L, et al. The M6A methyltransferase METTL3: acting as a tumor suppressor in renal cell carcinoma. Oncotarget. 2017;8:96103–16.
Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA. Methyltransferase-like protein 16 binds the 3'-terminal triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci U S A. 2016;113:14013–8.
Yeon SY, Jo YS, Choi EJ, Kim MS, Yoo NJ, Lee SH. Frameshift Mutations in Repeat Sequences of ANK3, HACD4, TCP10L, TP53BP1, MFN1, LCMT2, RNMT, TRMT6, METTL8 and METTL16 Genes in Colon Cancers. Pathol Oncol Res. 2018;24:617–22.
Liu X, Liu L, Dong Z, Li J, Yu Y, Chen X, Ren F, Cui G, Sun R. Expression patterns and prognostic value of m(6)A-related genes in colorectal cancer. Am J Transl Res. 2019;11:3972–91.
Cherry S, Lynch KW. Alternative splicing and cancer: insights, opportunities, and challenges from an expanding view of the transcriptome. Genes Dev. 2020;34:1005–16.
Hernandez-Caballero ME, Sierra-Ramirez JA. Single nucleotide polymorphisms of the FTO gene and cancer risk: an overview. Mol Biol Rep. 2015;42:699–704.
Iles MM, Law MH, Stacey SN, Han J, Fang S, Pfeiffer R, Harland M, Macgregor S, Taylor JC, Aben KK, et al. A variant in FTO shows association with melanoma risk not due to BMI. Nat Genet. 2013;45:428–432, 432e421.
Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R, Wang YY, Zhe H. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog. 2018;57:590–7.
Zou D, Dong L, Li C, Yin Z, Rao S, Zhou Q. The m(6)A eraser FTO facilitates proliferation and migration of human cervical cancer cells. Cancer Cell Int. 2019;19:321.
Liu J, Ren D, Du Z, Wang H, Zhang H, Jin Y. m(6)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun. 2018;502:456–64.
Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, Wang Y, Li X, Xiong XF, Wei B, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18:46.
Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, Semenza GL. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113:E2047–56.
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bogler O, et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31:591–606 e596.
Zhang C, Zhi WI, Lu H, Samanta D, Chen I, Gabrielson E, Semenza GL. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7:64527–42.
Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY, Yang D, Zheng ZC, Zhao Y. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75:379–89.
Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J Exp Clin Cancer Res. 2019;38:163.
He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, Liu D, Tian L, Yin J, Jiang K, Miao Y. ALKBH5 Inhibits Pancreatic Cancer Motility by Decreasing Long Non-Coding RNA KCNK15-AS1 Methylation. Cell Physiol Biochem. 2018;48:838–46.
Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi Y, He S, Shimamoto F. m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol Cancer. 2020;19:3.
Jiang Y, Wan Y, Gong M, Zhou S, Qiu J, Cheng W. RNA demethylase ALKBH5 promotes ovarian carcinogenesis in a simulated tumour microenvironment through stimulating NF-kappaB pathway. J Cell Mol Med. 2020;24:6137–48.
Tanabe A, Tanikawa K, Tsunetomi M, Takai K, Ikeda H, Konno J, Torigoe T, Maeda H, Kutomi G, Okita K, et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1alpha mRNA is translated. Cancer Lett. 2016;376:34–42.
Bai Y, Yang C, Wu R, Huang L, Song S, Li W, Yan P, Lin C, Li D, Zhang Y. YTHDF1 Regulates Tumorigenicity and Cancer Stem Cell-Like Activity in Human Colorectal Carcinoma. Front Oncol. 2019;9:332.
Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, Cheng C, Li L, Pi J, Si Y, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48:3816–31.
Jia R, Chai P, Wang S, Sun B, Xu Y, Yang Y, Ge S, Jia R, Yang YG. Fan X: m(6)A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Mol Cancer. 2019;18:161.
Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, Mapperley C, Lawson H, Wotherspoon DA, Sepulveda C, et al. Targeting the RNA m(6)A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell. 2019;25:137–148 e136.
Sheng H, Li Z, Su S, Sun W, Zhang X, Li L, Li J, Liu S, Lu B, Zhang S, Shan C. YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation. Carcinogenesis. 2020;41:541–50.
Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, Ma H, Kang T. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–61.
Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, Gan J, Jiang H, Jia GF, Luo C, Yang CG. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373–84.
Islam MS, Leissing TM, Chowdhury R, Hopkinson RJ, Schofield CJ. 2-Oxoglutarate-Dependent Oxygenases. Annu Rev Biochem. 2018;87:585–620.
Rose NR, McDonough MA, King ON, Kawamura A, Schofield CJ. Inhibition of 2-oxoglutarate dependent oxygenases. Chem Soc Rev. 2011;40:4364–97.
Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, Ni T, Zhang ZS, Zhang T, Li C, et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell. 2019;35:677–691.e610.
Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–33.
Legrand C, Tuorto F, Hartmann M, Liebers R, Jacob D, Helm M, Lyko F. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 2017;27:1589–96.
Huang T, Chen W, Liu J, Gu N, Zhang R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat Struct Mol Biol. 2019;26:380–8.
Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, Chen RX, Wei WS, Liu Y, Gao CC, et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21:978–90.
Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–63.
Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38:1415–30.
Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA, Rorbach J, Lantaff R, Blanco S, Sauer S, et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun. 2016;7:12039.
Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–8.
Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–5.
Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C, Hubner B, Seikowski J, Dennerlein S, Rehling P, et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016;35:2104–19.
Haag S, Warda AS, Kretschmer J, Gunnigmann MA, Hobartner C, Bohnsack MT. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA. 2015;21:1532–43.
Burgess AL, David R, Searle IR. Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biol. 2015;15:199.
Nakano S, Suzuki T, Kawarada L, Iwata H, Asano K, Suzuki T. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat Chem Biol. 2016;12:546–51.
Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, Reitter S, Liebers R, Stoecklin G, Grone HJ, et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015;34:2350–62.
Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, Frye M. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255–61.
Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. 2013;31:458–64.
Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–5.
Sajini AA, Choudhury NR, Wagner RE, Bornelov S, Selmi T, Spanos C, Dietmann S, Rappsilber J, Michlewski G, Frye M. Loss of 5-methylcytosine alters the biogenesis of vault-derived small RNAs to coordinate epidermal differentiation. Nat Commun. 2019;10:2550.
Li Y, Li J, Luo M, Zhou C, Shi X, Yang W, Lu Z, Chen Z, Sun N, He J. Novel long noncoding RNA NMR promotes tumor progression via NSUN2 and BPTF in esophageal squamous cell carcinoma. Cancer Lett. 2018;430:57–66.
Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606–25.
Bourgeois G, Ney M, Gaspar I, Aigueperse C, Schaefer M, Kellner S, Helm M, Motorin Y. Eukaryotic rRNA Modification by Yeast 5-Methylcytosine-Methyltransferases and Human Proliferation-Associated Antigen p120. PLoS One. 2015;10:e0133321.
Sharma S, Yang J, Watzinger P, Kotter P, Entian KD. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013;41:9062–76.
Metodiev MD, Spahr H, Loguercio Polosa P, Meharg C, Becker C, Altmueller J, Habermann B, Larsson NG, Ruzzenente B. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014;10:e1004110.
Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, Cai Q, Ji D, Jin SG, Niedernhofer LJ, et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc. 2014;136:11582–5.
Shen Q, Zhang Q, Shi Y, Shi Q, Jiang Y, Gu Y, Li Z, Li X, Zhao K, Wang C, et al. Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature. 2018;554:123–7.
Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 2011;7:e1002403.
Hussain S, Benavente SB, Nascimento E, Dragoni I, Kurowski A, Gillich A, Humphreys P, Frye M. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J Cell Biol. 2009;186:27–40.
Hussain S, Tuorto F, Menon S, Blanco S, Cox C, Flores JV, Watt S, Kudo NR, Lyko F, Frye M. The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol Cell Biol. 2013;33:1561–70.
Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, Andersson-Rolf A, Selmi T, Blanco S, Tailor J, Dietmann S, Frye M. Cytosine-5 RNA Methylation Regulates Neural Stem Cell Differentiation and Motility. Stem Cell Reports. 2017;8:112–24.
Rosace D, Lopez J, Blanco S. Emerging roles of novel small non-coding regulatory RNAs in immunity and cancer. RNA Biol. 2020;17:1196–213.
Wu P, Brockenbrough JS, Paddy MR, Aris JP. NCL1, a novel gene for a non-essential nuclear protein in Saccharomyces cerevisiae. Gene. 1998;220:109–17.
Cui X, Liang Z, Shen L, Zhang Q, Bao S, Geng Y, Zhang B, Leo V, Vardy LA, Lu T, et al. 5-Methylcytosine RNA Methylation in Arabidopsis Thaliana. Mol Plant. 2017;10:1387–99.
Amort T, Souliere MF, Wille A, Jia XY, Fiegl H, Worle H, Micura R, Lusser A. Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 2013;10:1003–8.
David R, Burgess A, Parker B, Li J, Pulsford K, Sibbritt T, Preiss T, Searle IR. Transcriptome-Wide Mapping of RNA 5-Methylcytosine in Arabidopsis mRNAs and Noncoding RNAs. Plant Cell. 2017;29:445–60.
Perlaky L, Valdez BC, Busch RK, Larson RG, Jhiang SM, Zhang WW, Brattain M, Busch H. Increased growth of NIH/3T3 cells by transfection with human p120 complementary DNA and inhibition by a p120 antisense construct. Cancer Res. 1992;52:428–36.
Freeman JW, Busch RK, Gyorkey F, Gyorkey P, Ross BE, Busch H. Identification and characterization of a human proliferation-associated nucleolar antigen with a molecular weight of 120,000 expressed in early G1 phase. Cancer Res. 1988;48:1244–51.
Freeman JW, McGrath P, Bondada V, Selliah N, Ownby H, Maloney T, Busch RK, Busch H. Prognostic significance of proliferation associated nucleolar antigen P120 in human breast carcinoma. Cancer Res. 1991;51:1973–8.
Bantis A, Giannopoulos A, Gonidi M, Liossi A, Aggelonidou E, Petrakakou E, Athanassiades P, Athanassiadou P. Expression of p120, Ki-67 and PCNA as proliferation biomarkers in imprint smears of prostate carcinoma and their prognostic value. Cytopathology. 2004;15:25–31.
Saijo Y, Sato G, Usui K, Sato M, Sagawa M, Kondo T, Minami Y, Nukiwa T. Expression of nucleolar protein p120 predicts poor prognosis in patients with stage I lung adenocarcinoma. Ann Oncol. 2001;12:1121–5.
Hong J, Lee JH, Chung IK. Telomerase activates transcription of cyclin D1 gene through an interaction with NOL1. J Cell Sci. 2016;129:1566–79.
Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16:971–81.
Yang JC, Risch E, Zhang M, Huang C, Huang H, Lu L. Association of tRNA methyltransferase NSUN2/IGF-II molecular signature with ovarian cancer survival. Future Oncol. 2017;13:1981–90.
Frye M, Dragoni I, Chin SF, Spiteri I, Kurowski A, Provenzano E, Green A, Ellis IO, Grimmer D, Teschendorff A, et al. Genomic gain of 5p15 leads to over-expression of Misu (NSUN2) in breast cancer. Cancer Lett. 2010;289:71–80.
Gao Y, Wang Z, Zhu Y, Zhu Q, Yang Y, Jin Y, Zhang F, Jiang L, Ye Y, Li H, et al. NOP2/Sun RNA methyltransferase 2 promotes tumor progression via its interacting partner RPL6 in gallbladder carcinoma. Cancer Sci. 2019;110:3510–9.
Okamoto M, Hirata S, Sato S, Koga S, Fujii M, Qi G, Ogawa I, Takata T, Shimamoto F, Tatsuka M. Frequent increased gene copy number and high protein expression of tRNA (cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 2012;31:660–71.
Lu L, Zhu G, Zeng H, Xu Q, Holzmann K. High tRNA Transferase NSUN2 Gene Expression is Associated with Poor Prognosis in Head and Neck Squamous Carcinoma. Cancer Invest. 2018;36:246–53.
Mei L, Shen C, Miao R, Wang JZ, Cao MD, Zhang YS, Shi LH, Zhao GH, Wang MH, Wu LS, Wei JF. RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57(Kip2) by an m(5)C-dependent manner. Cell Death Dis. 2020;11:270.
Elhardt W, Shanmugam R, Jurkowski TP, Jeltsch A. Somatic cancer mutations in the DNMT2 tRNA methyltransferase alter its catalytic properties. Biochimie. 2015;112:66–72.
Kar SP, Beesley J, Amin Al Olama A, Michailidou K, Tyrer J, Kote-Jarai Z, Lawrenson K, Lindstrom S, Ramus SJ, Thompson DJ, et al. Genome-Wide Meta-Analyses of Breast, Ovarian, and Prostate Cancer Association Studies Identify Multiple New Susceptibility Loci Shared by at Least Two Cancer Types. Cancer Discov. 2016;6:1052–67.
He Y, Yu X, Li J, Zhang Q, Zheng Q, Guo W. Role of m(5)C-related regulatory genes in the diagnosis and prognosis of hepatocellular carcinoma. Am J Transl Res. 2020;12:912–22.
Sato K, Tahata K, Akimoto K. Five Genes Associated With Survival in Patients With Lower-grade Gliomas Were Identified by Information-theoretical Analysis. Anticancer Res. 2020;40:2777–85.
Wang W. mRNA methylation by NSUN2 in cell proliferation. Wiley Interdiscip Rev RNA. 2016;7:838–42.
Lyons SM, Gudanis D, Coyne SM, Gdaniec Z, Ivanov P. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat Commun. 2017;8:1127.
Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–23.
Ivanov P, O'Day E, Emara MM, Wagner G, Lieberman J, Anderson P. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci U S A. 2014;111:18201–6.
Ramon YCS, Castellvi J, Hummer S, Peg V, Pelletier J, Sonenberg N. Beyond molecular tumor heterogeneity: protein synthesis takes control. Oncogene. 2018;37:2490–501.
Weissmann S, Alpermann T, Grossmann V, Kowarsch A, Nadarajah N, Eder C, Dicker F, Fasan A, Haferlach C, Haferlach T, et al. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia. 2012;26:934–42.
Archer NP, Perez-Andreu V, Scheurer ME, Rabin KR, Peckham-Gregory EC, Plon SE, Zabriskie RC, De Alarcon PA, Fernandez KS, Najera CR, et al. Family-based exome-wide assessment of maternal genetic effects on susceptibility to childhood B-cell acute lymphoblastic leukemia in hispanics. Cancer. 2016;122:3697–704.
Li W, Xu L. Epigenetic Function of TET Family, 5-Methylcytosine, and 5-Hydroxymethylcytosine in Hematologic Malignancies. Oncol Res Treat. 2019;42:309–18.
Takai H, Masuda K, Sato T, Sakaguchi Y, Suzuki T, Suzuki T, Koyama-Nasu R, Nasu-Nishimura Y, Katou Y, Ogawa H, et al. 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 2014;9:48–60.
Garcia MG, Carella A, Urdinguio RG, Bayon GF, Lopez V, Tejedor JR, Sierra MI, Garcia-Torano E, Santamarina P, Perez RF, et al. Epigenetic dysregulation of TET2 in human glioblastoma. Oncotarget. 2018;9:25922–34.
Carella A, Tejedor JR, Garcia MG, Urdinguio RG, Bayon GF, Sierra M, Lopez V, Garcia-Torano E, Santamarina-Ojeda P, Perez RF, et al. Epigenetic downregulation of TET3 reduces genome-wide 5hmC levels and promotes glioblastoma tumorigenesis. Int J Cancer. 2020;146:373–87.
Cheng JX, Chen L, Li Y, Cloe A, Yue M, Wei J, Watanabe KA, Shammo JM, Anastasi J, Shen QJ, et al. RNA cytosine methylation and methyltransferases mediate chromatin organization and 5-azacytidine response and resistance in leukaemia. Nat Commun. 2018;9:1163.
Okamoto M, Fujiwara M, Hori M, Okada K, Yazama F, Konishi H, Xiao Y, Qi G, Shimamoto F, Ota T, et al. tRNA modifying enzymes, NSUN2 and METTL1, determine sensitivity to 5-fluorouracil in HeLa cells. PLoS Genet. 2014;10:e1004639.
Cohn WE, Volkin E. Nucleoside-5′-phosphates from ribonucleic acid. Nature. 1951;167:483–4.
Hamma T, Ferre-D'Amare AR. Pseudouridine synthases. Chem Biol. 2006;13:1125–35.
Zhao Y, Dunker W, Yu YT, Karijolich J. The Role of Noncoding RNA Pseudouridylation in Nuclear Gene Expression Events. Front Bioeng Biotechnol. 2018;6:8.
Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One. 2014;9:e110799.
Carlile TM, Martinez NM, Schaening C, Su A, Bell TA, Zinshteyn B, Gilbert WV. mRNA structure determines modification by pseudouridine synthase 1. Nat Chem Biol. 2019;15:966–74.
Safra M, Nir R, Farouq D, Vainberg Slutskin I, Schwartz S. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res. 2017;27:393–406.
Cohn WE. 5-Ribosyl uracil, a carbon-carbon ribofuranosyl nucleoside in ribonucleic acids. Biochim Biophys Acta. 1959;32:569–71.
Davis FF, Allen FW. Ribonucleic acids from yeast which contain a fifth nucleotide. J Biol Chem. 1957;227:907–15.
Zhao BS, He C. Pseudouridine in a new era of RNA modifications. Cell Res. 2015;25:153–4.
Spenkuch F, Motorin Y, Helm M. Pseudouridine: still mysterious, but never a fake (uridine)! RNA Biol. 2014;11:1540–54.
Song J, Zhuang Y, Zhu C, Meng H, Lu B, Xie B, Peng J, Li M, Yi C. Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat Chem Biol. 2020;16:160–9.
Wu G, Xiao M, Yang C, Yu YT. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J. 2011;30:79–89.
Guzzi N, Ciesla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, Pimkova K, Sommarin MNE, Munita R, Lubas M, et al. Pseudouridylation of tRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell. 2018;173:1204–1216 e1226.
Hur S, Stroud RM, Finer-Moore J. Substrate recognition by RNA 5-methyluridine methyltransferases and pseudouridine synthases: a structural perspective. J Biol Chem. 2006;281:38969–73.
Becker HF, Motorin Y, Sissler M, Florentz C, Grosjean H. Major identity determinants for enzymatic formation of ribothymidine and pseudouridine in the T psi-loop of yeast tRNAs. J Mol Biol. 1997;274:505–18.
Gu X, Yu M, Ivanetich KM, Santi DV. Molecular recognition of tRNA by tRNA pseudouridine 55 synthase. Biochemistry. 1998;37:339–43.
Foster PG, Huang L, Santi DV, Stroud RM. The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I. Nat Struct Biol. 2000;7:23–7.
Urban A, Behm-Ansmant I, Branlant C, Motorin Y. RNA sequence and two-dimensional structure features required for efficient substrate modification by the Saccharomyces cerevisiae RNA:{Psi}-synthase Pus7p. J Biol Chem. 2009;284:5845–58.
Rintala-Dempsey AC, Kothe U. Eukaryotic stand-alone pseudouridine synthases - RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 2017;14:1185–96.
Becker HF, Motorin Y, Planta RJ, Grosjean H. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of psi55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 1997;25:4493–9.
Roovers M, Hale C, Tricot C, Terns MP, Terns RM, Grosjean H, Droogmans L. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res. 2006;34:4293–301.
Rasmuson T, Bjork GR. Urinary excretion of pseudouridine and prognosis of patients with malignant lymphoma. Acta Oncol. 1995;34:61–7.
Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–6.
Arnez JG, Steitz TA. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry. 1994;33:7560–7.
Newby MI, Greenbaum NL. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA. 2001;7:833–45.
Newby MI, Greenbaum NL. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat Struct Biol. 2002;9:958–65.
Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol. 1999;19:7461–72.
Gu BW, Ge J, Fan JM, Bessler M, Mason PJ. Slow growth and unstable ribosomal RNA lacking pseudouridine in mouse embryonic fibroblast cells expressing catalytically inactive dyskerin. FEBS Lett. 2013;587:2112–7.
Penzo M, Montanaro L. Turning Uridines around: Role of rRNA Pseudouridylation in Ribosome Biogenesis and Ribosomal Function. Biomolecules. 2018;8(2):38. https://doi.org/10.3390/biom8020038.
King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell. 2003;11:425–35.
Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474:395–8.
Eyler DE, Franco MK, Batool Z, Wu MZ, Dubuke ML, Dobosz-Bartoszek M, Jones JD, Polikanov YS, Roy B, Koutmou KS. Pseudouridinylation of mRNA coding sequences alters translation. Proc Natl Acad Sci U S A. 2019;116:23068–74.
Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16:1833–40.
Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–5.
Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–8.
Kirwan M, Dokal I. Dyskeratosis congenita: a genetic disorder of many faces. Clin Genet. 2008;73:103–12.
Penzo M, Casoli L, Ceccarelli C, Trere D, Ludovini V, Crino L, Montanaro L. DKC1 gene mutations in human sporadic cancer. Histol Histopathol. 2013;28:365–72.
Dokal I. Dyskeratosis congenita in all its forms. Br J Haematol. 2000;110:768–79.
Montanaro L, Brigotti M, Clohessy J, Barbieri S, Ceccarelli C, Santini D, Taffurelli M, Calienni M, Teruya-Feldstein J, Trere D, et al. Dyskerin expression influences the level of ribosomal RNA pseudo-uridylation and telomerase RNA component in human breast cancer. J Pathol. 2006;210:10–8.
Rostami P, Zendehdel K, Shirkoohi R, Ebrahimi E, Ataei M, Imanian H, Najmabadi H, Akbari MR, Sanati MH. Gene Panel Testing in Hereditary Breast Cancer. Arch Iran Med. 2020;23:155–62.
Turano M, Angrisani A, De Rosa M, Izzo P, Furia M. Real-time PCR quantification of human DKC1 expression in colorectal cancer. Acta Oncol. 2008;47:1598–9.
Nersisyan L, Hopp L, Loeffler-Wirth H, Galle J, Loeffler M, Arakelyan A, Binder H. Telomere Length Maintenance and Its Transcriptional Regulation in Lynch Syndrome and Sporadic Colorectal Carcinoma. Front Oncol. 2019;9:1172.
Penzo M, Ludovini V, Trere D, Siggillino A, Vannucci J, Bellezza G, Crino L, Montanaro L. Dyskerin and TERC expression may condition survival in lung cancer patients. Oncotarget. 2015;6:21755–60.
Stockert JA, Gupta A, Herzog B, Yadav SS, Tewari AK, Yadav KK. Predictive value of pseudouridine in prostate cancer. Am J Clin Exp Urol. 2019;7:262–72.
Alawi F, Lin P, Ziober B, Patel R. Correlation of dyskerin expression with active proliferation independent of telomerase. Head Neck. 2011;33:1041–51.
Patel MM, Parekh LJ, Jha FP, Sainger RN, Patel JB, Patel DD, Shah PM, Patel PS. Clinical usefulness of telomerase activation and telomere length in head and neck cancer. Head Neck. 2002;24:1060–7.
Miao FA, Chu K, Chen HR, Zhang M, Shi PC, Bai J, You YP. Increased DKC1 expression in glioma and its significance in tumor cell proliferation, migration and invasion. Invest New Drugs. 2019;37:1177–86.
Liu B, Zhang J, Huang C, Liu H. Dyskerin overexpression in human hepatocellular carcinoma is associated with advanced clinical stage and poor patient prognosis. PLoS One. 2012;7:e43147.
Poncet D, Belleville A, t’kint de Roodenbeke C, Roborel de Climens A, Ben Simon E, Merle-Beral H, Callet-Bauchu E, Salles G, Sabatier L, Delic J, Gilson E. Changes in the expression of telomere maintenance genes suggest global telomere dysfunction in B-chronic lymphocytic leukemia. Blood. 2008;111:2388–91.
Perdigones N, Perin JC, Schiano I, Nicholas P, Biegel JA, Mason PJ, Babushok DV, Bessler M. Clonal hematopoiesis in patients with dyskeratosis congenita. Am J Hematol. 2016;91:1227–33.
Alter BP, Rosenberg PS, Giri N, Baerlocher GM, Lansdorp PM, Savage SA. Telomere length is associated with disease severity and declines with age in dyskeratosis congenita. Haematologica. 2012;97:353–9.
Zhang MY, Keel SB, Walsh T, Lee MK, Gulsuner S, Watts AC, Pritchard CC, Salipante SJ, Jeng MR, Hofmann I, et al. Genomic analysis of bone marrow failure and myelodysplastic syndromes reveals phenotypic and diagnostic complexity. Haematologica. 2015;100:42–8.
Ronchetti D, Todoerti K, Tuana G, Agnelli L, Mosca L, Lionetti M, Fabris S, Colapietro P, Miozzo M, Ferrarini M, et al. The expression pattern of small nucleolar and small Cajal body-specific RNAs characterizes distinct molecular subtypes of multiple myeloma. Blood Cancer J. 2012;2:e96.
Machado-Pinilla R, Sanchez-Perez I, Murguia JR, Sastre L, Perona R. A dyskerin motif reactivates telomerase activity in X-linked dyskeratosis congenita and in telomerase-deficient human cells. Blood. 2008;111:2606–14.
Trahan C, Dragon F. Dyskeratosis congenita mutations in the H/ACA domain of human telomerase RNA affect its assembly into a pre-RNP. RNA. 2009;15:235–43.
Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, Kopmar N, Krasnykh O, Dean AM, Thompson SR, et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell. 2011;44:660–6.
Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–6.
Penzo M, Rocchi L, Brugiere S, Carnicelli D, Onofrillo C, Coute Y, Brigotti M, Montanaro L. Human ribosomes from cells with reduced dyskerin levels are intrinsically altered in translation. FASEB J. 2015;29:3472–82.
Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 2010;29:1865–76.
Bellodi C, Krasnykh O, Haynes N, Theodoropoulou M, Peng G, Montanaro L, Ruggero D. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010;70:6026–35.
Rocchi L, Pacilli A, Sethi R, Penzo M, Schneider RJ, Trere D, Brigotti M, Montanaro L. Dyskerin depletion increases VEGF mRNA internal ribosome entry site-mediated translation. Nucleic Acids Res. 2013;41:8308–18.
McMahon M, Contreras A, Holm M, Uechi T, Forester CM, Pang X, Jackson C, Calvert ME, Chen B, Quigley DA, et al. A single H/ACA small nucleolar RNA mediates tumor suppression downstream of oncogenic RAS. Elife. 2019;8:e48847. https://doi.org/10.7554/eLife.48847.
Babaian A, Rothe K, Girodat D, Minia I, Djondovic S, Milek M, Spencer Miko SE, Wieden HJ, Landthaler M, Morin GB, Mager DL. Loss of m(1)acp(3)Psi Ribosomal RNA Modification Is a Major Feature of Cancer. Cell Rep. 2020;31:107611.
Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–62.
Zhao X, Patton JR, Davis SL, Florence B, Ames SJ, Spanjaard RA. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol Cell. 2004;15:549–58.
Jana S, Hsieh AC, Gupta R. Reciprocal amplification of caspase-3 activity by nuclear export of a putative human RNA-modifying protein, PUS10 during TRAIL-induced apoptosis. Cell Death Dis. 2017;8:e3093.
Ji P, Ding D, Qin N, Wang C, Zhu M, Li Y, Dai J, Jin G, Hu Z, Shen H, et al. Systematic analyses of genetic variants in chromatin interaction regions identified four novel lung cancer susceptibility loci. J Cancer. 2020;11:1075–81.
Sperling AS, Gibson CJ, Ebert BL. The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia. Nat Rev Cancer. 2017;17:5–19.
Robichaud N, Sonenberg N, Ruggero D, Schneider RJ. Translational Control in Cancer. Cold Spring Harb Perspect Biol. 2019;11(7):a032896. https://doi.org/10.1101/cshperspect.a032896.
Jiang T, Lin Y, Yin H, Wang S, Sun Q, Zhang P, Bi W. Correlation analysis of urine metabolites and clinical staging in patients with ovarian cancer. Int J Clin Exp Med. 2015;8:18165–71.
Krstulja A, Lettieri S, Hall AJ, Roy V, Favetta P, Agrofoglio LA. Tailor-Made Molecularly Imprinted Polymer for Selective Recognition of the Urinary Tumor Marker Pseudouridine. Macromol Biosci. 2017;17.
Zeleznik OA, Eliassen AH, Kraft P, Poole EM, Rosner BA, Jeanfavre S, Deik AA, Bullock K, Hitchcock DS, Avila-Pacheco J, et al. A Prospective Analysis of Circulating Plasma Metabolites Associated with Ovarian Cancer Risk. Cancer Res. 2020;80:1357–67.
Sridharan G, Ramani P, Patankar S, Vijayaraghavan R. Evaluation of salivary metabolomics in oral leukoplakia and oral squamous cell carcinoma. J Oral Pathol Med. 2019;48:299–306.
Rocchi L, Barbosa AJ, Onofrillo C, Del Rio A, Montanaro L. Inhibition of human dyskerin as a new approach to target ribosome biogenesis. PLoS One. 2014;9:e101971.
Floresta G, Pistara V, Amata E, Dichiara M, Damigella A, Marrazzo A, Prezzavento O, Punzo F, Rescifina A. Molecular modeling studies of pseudouridine isoxazolidinyl nucleoside analogues as potential inhibitors of the pseudouridine 5'-monophosphate glycosidase. Chem Biol Drug Des. 2018;91:519–25.
Armando RG, Mengual Gomez DL, Juritz EI, Lorenzano Menna P, Gomez DE. Homology Model and Docking-Based Virtual Screening for Ligands of Human Dyskerin as New Inhibitors of Telomerase for Cancer Treatment. Int J Mol Sci. 2018;19(10):3216. https://doi.org/10.3390/ijms19103216.
Edmonson JH, Decker DG, Malkasian GD, Webb MJ. Concomitant phase II studies of pyrazofurin and razoxane in alkylating agent-resistant cases of epithelial ovarian carcinoma. Cancer Treat Rep. 1981;65:1127–9.
Gralla RJ, Sordillo PP, Magill GB. Phase II evaluation of pyrazofurin in patients with metastatic sarcoma. Cancer Treat Rep. 1978;62:1573–4.
Carroll DS, Kemeny NE, Gralla RJ. Phase II evaluation of pyrazofurin in patients with advanced colorectal carcinoma. Cancer Treat Rep. 1979;63:139–40.
Vogler WR, Trulock PD. Phase I study of pyrazofurin in refractory acute myelogenous leukemia. Cancer Treat Rep. 1978;62:1569–71.
Nichols WC, Kvols LK, Ingle JN, Edmonson JH, Ahmann DL, Rubin J, O'Connell MJ. Phase II study of triazinate and pyrazofurin in patients with advanced breast cancer previously exposed to cytotoxic chemotherapy. Cancer Treat Rep. 1978;62:837–9.
Gralla RJ, Currie VE, Wittes RE, Golbey RB, Young CW. Phase II evaluation of pyrazofurin in patients with carcinoma of the lung. Cancer Treat Rep. 1978;62:451–2.
Budman D, Currie V, Wittes R. Phase II trial of pyrazofurin in malignant melanoma. Cancer Treat Rep. 1977;61:1733–4.
Cummings FJ, Stoller RG, Calabresi P. Clinical trial of weekly pyrazofurin. Cancer Treat Rep. 1979;63:1363–5.
Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–8.
Spedaliere CJ, Mueller EG. Not all pseudouridine synthases are potently inhibited by RNA containing 5-fluorouridine. RNA. 2004;10:192–9.
Samuelsson T. Interactions of transfer RNA pseudouridine synthases with RNAs substituted with fluorouracil. Nucleic Acids Res. 1991;19:6139–44.
Motorin Y, Helm M. Methods for RNA Modification Mapping Using Deep Sequencing: Established and New Emerging Technologies. Genes (Basel). 2019;10(1):35. https://doi.org/10.3390/genes10010035.
Mishima E, Jinno D, Akiyama Y, Itoh K, Nankumo S, Shima H, Kikuchi K, Takeuchi Y, Elkordy A, Suzuki T, et al. Immuno-Northern Blotting: Detection of RNA Modifications by Using Antibodies against Modified Nucleosides. PLoS One. 2015;10:e0143756.
Meyer KD. DART-seq: an antibody-free method for global m(6)A detection. Nat Methods. 2019;16:1275–80.
Saletore Y, Meyer K, Korlach J, Vilfan ID, Jaffrey S, Mason CE. The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 2012;13:175.
Vilfan ID, Tsai YC, Clark TA, Wegener J, Dai Q, Yi C, Pan T, Turner SW, Korlach J. Analysis of RNA base modification and structural rearrangement by single-molecule real-time detection of reverse transcription. J Nanobiotechnology. 2013;11:8.
Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods. 2018;15:201–6.
Smith AM, Jain M, Mulroney L, Garalde DR, Akeson M. Reading canonical and modified nucleobases in 16S ribosomal RNA using nanopore native RNA sequencing. PLoS One. 2019;14:e0216709.
Liu H, Begik O, Lucas MC, Ramirez JM, Mason CE, Wiener D, Schwartz S, Mattick JS, Smith MA, Novoa EM. Accurate detection of m(6)A RNA modifications in native RNA sequences. Nat Commun. 2019;10:4079.
Doxtader KA, Wang P, Scarborough AM, Seo D, Conrad NK, Nam Y. Structural Basis for Regulation of METTL16, an S-Adenosylmethionine Homeostasis Factor. Mol Cell. 2018;71:1001–1011 e1004.
Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer. 2017;17:332.
Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, Tavazoie SF. Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression. Cell. 2016;165:1416–27.
Lieberman J. Tapping the RNA world for therapeutics. Nat Struct Mol Biol. 2018;25:357–64.
Ge L, Zhang N, Chen Z, Song J, Wu Y, Li Z, Chen F, Wu J, Li D, Li J, et al. Level of N6-Methyladenosine in Peripheral Blood RNA: A Novel Predictive Biomarker for Gastric Cancer. Clin Chem. 2020;66:342–51.
Zhao S, Liu J, Nanga P, Liu Y, Cicek AE, Knoblauch N, He C, Stephens M, He X. Detailed modeling of positive selection improves detection of cancer driver genes. Nat Commun. 2019;10:3399.
Zhou Z, Li HQ, Liu F. DNA Methyltransferase Inhibitors and their Therapeutic Potential. Curr Top Med Chem. 2018;18:2448–57.
Mullard A. Stemming the tide of drug resistance in cancer. Nat Rev Drug Discov. 2020;19:221–3.
Jin H, Ying X, Que B, Wang X, Chao Y, Zhang H, Yuan Z, Qi D, Lin S, Min W, et al. N 6 -methyladenosine Modification of ITGA6 mRNA Promotes the Development and Progression of Bladder Cancer. EBioMedicine. 2019;47:195–207.
Xie H, Li J, Ying Y, Yan H, Jin K, Ma X, He L, Xu X, Liu B, Wang X, et al. METTL3/YTHDF2 M 6 A Axis Promotes Tumorigenesis by Degrading SETD7 and KLF4 mRNAs in Bladder Cancer. J Cell Mol Med. 2020;24:4092–104.
Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, Chen ZH, Zeng ZL, Wang F, Zheng J, et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112.
Wanna-Udom S, Terashima M, Lyu H, Ishimura A, Takino T, Sakari M, Tsukahara T, Suzuki T. The m6A methyltransferase METTL3 contributes to Transforming Growth Factor-beta-induced epithelial-mesenchymal transition of lung cancer cells through the regulation of JUNB. Biochem Biophys Res Commun. 2020;524:150–5.
Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen Z, Dinglin X, Ma S, Li D, Wu Y, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019;18:181.
Dahal U, Le K, Gupta M. RNA m6A methyltransferase METTL3 regulates invasiveness of melanoma cells by matrix metallopeptidase 2. Melanoma Res. 2019;29:382–9.
Luo G, Xu W, Zhao Y, Jin S, Wang S, Liu Q, Chen X, Wang J, Dong F, Hu DN, et al. RNA m(6) A methylation regulates uveal melanoma cell proliferation, migration, and invasion by targeting c-Met. J Cell Physiol. 2020;235:7107–19.
Miao W, Chen J, Jia L, Ma J, Song D. The m6A methyltransferase METTL3 promotes osteosarcoma progression by regulating the m6A level of LEF1. Biochem Biophys Res Commun. 2019;516:719–25.
Hua W, Zhao Y, Jin X, Yu D, He J, Xie D, Duan P. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol Oncol. 2018;151:356–65.
Liang S, Guan H, Lin X, Li N, Geng F, Li J. METTL3 serves an oncogenic role in human ovarian cancer cells partially via the AKT signaling pathway. Oncol Lett. 2020;19:3197–204.
Zhou J, Wang J, Hong B, Ma K, Xie H, Li L, Zhang K, Zhou B, Cai L, Gong K. Gene signatures and prognostic values of m6A regulators in clear cell renal cell carcinoma - a retrospective study using TCGA database. Aging (Albany NY). 2019;11:1633–47.
Wang J, Zhang C, He W, Gou X. Effect of m(6)A RNA Methylation Regulators on Malignant Progression and Prognosis in Renal Clear Cell Carcinoma. Front Oncol. 2020;10:3.
Gu C, Wang Z, Zhou N, Li G, Kou Y, Luo Y, Wang Y, Yang J, Tian F. Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N(6)-methyladenosine of Notch1. Mol Cancer. 2019;18:168.
Chen X, Xu M, Xu X, Zeng K, Liu X, Sun L, Pan B, He B, Pan Y, Sun H, et al. METTL14 Suppresses CRC Progression via Regulating N6-Methyladenosine-Dependent Primary miR-375 Processing. Mol Ther. 2020;28:599–612.
Yang X, Zhang S, He C, Xue P, Zhang L, He Z, Zang L, Feng B, Sun J, Zheng M. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46.
Gong D, Zhang J, Chen Y, Xu Y, Ma J, Hu G, Huang Y, Zheng J, Zhai W, Xue W. The m(6)A-suppressed P2RX6 activation promotes renal cancer cells migration and invasion through ATP-induced Ca(2+) influx modulating ERK1/2 phosphorylation and MMP9 signaling pathway. J Exp Clin Cancer Res. 2019;38:233.
Kong F, Liu X, Zhou Y, Hou X, He J, Li Q, Miao X, Yang L. Downregulation of METTL14 increases apoptosis and autophagy induced by cisplatin in pancreatic cancer cells. Int J Biochem Cell Biol. 2020;122:105731.
Wen L, Pan X, Yu Y, Yang B. Down-regulation of FTO promotes proliferation and migration, and protects bladder cancer cells from cisplatin-induced cytotoxicity. BMC Urol. 2020;20:39.
Zhao Y, You S, Yu YQ, Zhang S, Li PT, Ye YH, Zhao WX, Li J, Li Q, Jiao H, et al. Decreased nuclear expression of FTO in human primary hepatocellular carcinoma is associated with poor prognosis. Int J Clin Exp Pathol. 2019;12:3376–83.
Tang X, Liu S, Chen D, Zhao Z, Zhou J. The role of the fat mass and obesity-associated protein in the proliferation of pancreatic cancer cells. Oncol Lett. 2019;17:2473–8.
Kwok CT, Marshall AD, Rasko JE, Wong JJ. Genetic alterations of m(6)A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol. 2017;10:39.
Shen C, Sheng Y, Zhu AC, Robinson S, Jiang X, Dong L, Chen H, Su R, Yin Z, Li W, et al. RNA Demethylase ALKBH5 Selectively Promotes Tumorigenesis and Cancer Stem Cell Self-Renewal in Acute Myeloid Leukemia. Cell Stem Cell. 2020;27:64–80 e69.
Jin D, Guo J, Wu Y, Yang L, Wang X, Du J, Dai J, Chen W, Gong K. Miao S, et al: m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol Cancer. 2020;19:40.
Chen S, Zhou L, Wang Y. ALKBH5-mediated m(6)A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020;20:34.
Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, Takahashi H, Nishida N, Haraguchi N, Sakai D, et al. Oncogene c-Myc promotes epitranscriptome m. Oncotarget. 2018;9:7476–86.
Job B, Bernheim A, Beau-Faller M, Camilleri-Broet S, Girard P, Hofman P, Mazieres J, Toujani S, Lacroix L, Laffaire J, et al. Genomic aberrations in lung adenocarcinoma in never smokers. PLoS One. 2010;5:e15145.
Wang P, Yan Y, Yu W, Zhang H. Role of ten-eleven translocation proteins and 5-hydroxymethylcytosine in hepatocellular carcinoma. Cell Prolif. 2019;52:e12626.
Li Y, Zheng D, Wang F, Xu Y, Yu H, Zhang H. Expression of Demethylase Genes, FTO and ALKBH1, Is Associated with Prognosis of Gastric Cancer. Dig Dis Sci. 2019;64:1503–13.
Yamashita T, Higashi M, Momose S, Morozumi M, Tamaru JI. Nuclear expression of Y box binding-1 is important for resistance to chemotherapy including gemcitabine in TP53-mutated bladder cancer. Int J Oncol. 2017;51:579–86.
Campbell TM, Castro MAA, de Oliveira KG, Ponder BAJ, Meyer KB. ERalpha Binding by Transcription Factors NFIB and YBX1 Enables FGFR2 Signaling to Modulate Estrogen Responsiveness in Breast Cancer. Cancer Res. 2018;78:410–21.
Kuwano M, Shibata T, Watari K, Ono M. Oncogenic Y-box binding protein-1 as an effective therapeutic target in drug-resistant cancer. Cancer Sci. 2019;110:1536–43.
Saito Y, Kasamatsu A, Yamamoto A, Shimizu T, Yokoe H, Sakamoto Y, Ogawara K, Shiiba M, Tanzawa H, Uzawa K. ALY as a potential contributor to metastasis in human oral squamous cell carcinoma. J Cancer Res Clin Oncol. 2013;139:585–94.
Dominguez-Sanchez MS, Saez C, Japon MA, Aguilera A, Luna R. Differential expression of THOC1 and ALY mRNP biogenesis/export factors in human cancers. BMC Cancer. 2011;11:77.
Acknowledgements
We apologise for not being able to cite all the relevant publications due to space limitations. We acknowledge funding from the Spanish Ministry of Economy and Innovation (MINECO), Ministry of Science and Innovation and Agencia Estatal de Investigación (AEI) and co-financed by the European Development Regional Fund (FEDER) under grant no. SAF2016-78667-R (AEI/FEDER, UE) and PID2019-111692RB-I00. In addition we acknowledge funding from The Scientific Foundation AECC under grant no. LABAE19040BLAN and the University of Salamanca and Fundación Memoria de Samuel Solorzano Barruso (FS/21-2018). BML was supported by Junta de Castilla y León-Fondo Social Europeo (ESF) predoctoral fellowship. AUTHOR’s institution is supported by the Programa de Apoyo a Planes Estratégicos de Investigación de Estructuras de Investigación de Excelencia co-financed by the Castilla–León autonomous government and the European Regional Development Fund (CLC–2017–01).
Funding
This work was supported by Spanish Ministry of Economy and Innovation (MINECO), Ministry of Science and Innovation and Agencia Estatal de Investigación (AEI) and co-financed by the European Development Regional Fund (FEDER) under grant no. SAF2016–78667-R (AEI/FEDER, UE) and PID2019-111692RB-I00. In addition we acknowledge funding from The Scientific Foundation AECC under grant no. LABAE19040BLAN. BML was supported by Junta de Castilla y León-Fondo Social Europeo (ESF) predoctoral fellowship. AUTHOR’s institution is supported by the Programa de Apoyo a Planes Estratégicos de Investigación de Estructuras de Investigación de Excelencia co-financed by the Castilla–León autonomous government and the European Regional Development Fund (CLC–2017–01).
Author information
Authors and Affiliations
Contributions
Manuscript design: SB. Literature search and manuscript drafting: PN, BML and SB. Manuscript editing, figure design and review: SB. The author(s) read and approved the final manuscript.
Author information
PN’s, BML’s affilition is Centro de Investigación del Cáncer and Instituto de Biología Molecular y Celular del Cáncer, Consejo Superior de Investigaciones Científicas (CSIC) - University of Salamanca, 37007 Salamanca, Spain. SB’s affiliations are Centro de Investigación del Cáncer and Instituto de Biología Molecular y Celular del Cáncer, Consejo Superior de Investigaciones Científicas (CSIC) - University of Salamanca, 37007 Salamanca, Spain and Instituto de Investigación Biomédica de Salamanca (IBSAL), Hospital Universitario de Salamanca, 37007 Salamanca, Spain.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors read and give final approval of the version to be submitted and any revised version.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Nombela, P., Miguel-López, B. & Blanco, S. The role of m6A, m5C and Ψ RNA modifications in cancer: Novel therapeutic opportunities. Mol Cancer 20, 18 (2021). https://doi.org/10.1186/s12943-020-01263-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12943-020-01263-w