Abstract
Background
Clinical trials designed to test the efficacy of retinoic acid (RA) as an adjuvant for the treatment of solid cancers have been disappointing, primarily due to RA resistance. Estrogen receptor (ER)-negative breast cancer cells are more resistant to RA than ER-positive cells. The expression and subcellular distribution of two RA-binding proteins, FABP5 and CRABP2, has already been shown to play critical roles in breast cancer cell response to RA. CRABP1, a third member of the RA-binding protein family, has not previously been investigated as a possible mediator of RA action in breast cancer.
Methods
CRABP1 and CRABP2 expression in primary breast tumor tissues was analyzed using gene expression and tissue microarrays. CRABP1 levels were manipulated using siRNAs and by transient overexpression. RA-induced subcellular translocation of CRABPs was examined by immunofluorescence microscopy and immunoblotting. RA-induced transactivation of RAR was analyzed using a RA response element (RARE)-driven luciferase reporter system. Effects of CRABP1 expression and RA treatment on downstream gene expression were investigated by semi-quantitative RT-PCR analysis.
Results
Compared to normal mammary tissues, CRABP1 expression is significantly down-regulated in ER+ breast tumors, but maintained in triple-negative breast cancers. Elevated CRABP1 levels are associated with poor patient prognosis, high Ki67 immunoreactivity and high tumor grade in breast cancer. The prognostic significance of CRABP1 is attributed to its cytoplasmic localization. We demonstrate that CRABP1 expression attenuates RA-induced cell growth arrest and inhibits RA signalling in breast cancer cells by sequestering RA in the cytoplasm. We also show that CRABP1 affects the expression of genes involved in RA biosynthesis, trafficking and metabolism.
Conclusions
CRABP1 is an adverse factor for clinical outcome in triple-negative breast cancer and a potent inhibitor of RA signalling in breast cancer cells. Our data indicate that CRABP1, in conjunction with previously identified CRABP2 and FABP5, plays a key role in breast cancer cell response to RA. We propose that these three RA-binding proteins can serve as biomarkers for predicting triple-negative breast cancer response to RA, with elevated levels of either cytoplasmic CRABP1 or FABP5 associated with RA resistance, and elevated levels of nuclear CRABP2 associated with sensitivity to RA.
Similar content being viewed by others
Introduction
Current clinical management of breast cancer relies on clinicopathological features as well as expression of biological markers such as estrogen receptor (ER), progesterone receptor (PR) and epidermal growth factor receptor 2 (HER2) [1, 2]. While tamoxifen has been shown to be highly effective for the treatment of ER/PR-positive breast cancers [3], there are no specific molecular targets for tumors that don’t express ER, PR or HER2. These triple-negative tumors, which constitute 15–20 % of breast cancers [4], are more aggressive and less responsive to standard treatment than the more common ER/PR-positive breast cancers, and have a poorer prognosis [5, 6].
RA and its derivatives, collectively called retinoids, inhibit growth and induce apoptosis in a variety of epithelial cancer cells and hold great promise as chemotherapeutic agents [7–9]. Retinoids inhibit mitogen signalling [10] and induce downstream signalling pathways implicated in growth arrest, apoptosis and differentiation of precancerous and cancer cells [11–15]. However, with the exception of acute promyelocytic leukemia (APL) [14, 16, 17], clinical trials designed to test the efficacy of RA and its derivatives in the treatment of cancer have produced disappointing results, primarily because of RA-induced side effects and development of RA resistance [18, 19].
RA exerts its physiological effects by binding and activating nuclear retinoic acid receptors RARα, β and γ. RAR dimerizes with retinoid-X-receptor (RXR) and binds to retinoic acid response elements (RARE) in the promoters of target genes, regulating their transcription and function [7–9]. As intracellular transporters, the cellular RA binding proteins determine RA subcellular distribution, fate and function [20–22]. Two RA binding proteins, cellular retinoic acid binding protein 2 (CRABP2) and fatty acid-binding protein 5 (FABP5), have been shown to play opposing roles in mediating the RA cellular response by targeting RA to distinct nuclear receptors; i.e., delivery of RA to RARs by CRABP2 leads to inhibition of cell proliferation, whereas delivery of RA to peroxisome proliferator activated receptor beta (PPARβ) by FABP5 increases cell proliferation and causes RA resistance [23, 24].
We previously reported that FABP5 is preferentially expressed in ER- and triple-negative breast tumors [25], subtypes that are prone to RA resistance [26–28]. Furthermore, breast cancer cells with an elevated FABP5/CRABP2 ratio show increased resistance to RA [23, 25]. However, the FABP5/CRABP2 ratio does not always predict breast cancer cell response to RA, and RA resistance in the squamous cell carcinoma cell line COLO 16 cannot be overcome by either restoration of CRABP2 expression or an increased CRABP2/FABP5 ratio [29], indicating that other factors are involved.
Studies designed to examine the importance of CRABP1 in the clinical outcomes of various cancers have produced conflicting results [30–33]. Prior to this study, the expression and prognostic significance of CRABP1 in breast cancer had not been investigated. In light of CRABP1’s proposed role in attenuating RA activity by enhancing RA metabolism, expression of CRABP1 in breast cancer could have important implications for RA response. Here, we report the expression, clinicopathological association and function of CRABP1 in breast cancer. Our data indicate that CRABP1 is an adverse prognostic factor and a potent inhibitor of RA action in breast cancer which functions by sequestering RA in the cytoplasm rather than by enhancing RA metabolism. We propose that CRABP1 may serve as a biomarker to predict RA response and a target to optimize the efficacy of RA in breast cancer treatment.
Materials and methods
Chemicals, reagents and DNA constructs
All-trans retinoic acid was purchased from Sigma-Aldrich (Oakville, ON, Canada) and dissolved in DMSO (Sigma-Aldrich) at a concentration of 50 mM. Scrambled stealth siRNAs and gene-specific siRNAs targeting different regions of CRABP1 mRNA (nucleotides 381–405 and 484–508 of GenBank mRNA sequence NM_004378) and CRABP2 mRNA (nucleotides 418–442 and 465–489 of GenBank mRNA sequence NM_001878) were purchased from Life Technologies (Burlington, ON, Canada). The Lipofectamine RNAiMAX reagent (Life Technologies) was used for siRNA transfections. The pGL3-RARE-luciferase plasmid DNA was purchased from Addgene (Cambridge, MA, USA) and the luciferase assay system from Promega (Madison, WI, USA). Polyethylenimine (PEI) (Polysciences, Warrington, PA, USA) was used for plasmid DNA transfections. For gain-of-function studies, the entire open reading frame of CRABP1 was PCR-amplified and cloned into pcDNA3 (Life Technologies).
Cell culture and siRNA transfection
ZR-75-1, MDA-MB-468, MDA-MB-435, BT-20, T47D, BT-474, MDA-MB-231, BT-483, MCF-7, SK-Br-3, BT-549 and Hs578T breast cancer cells were cultured in Dulbecco’s modification of Eagle’s medium (DMEM) supplemented with 10 % fetal calf serum, penicillin (100 units/mL) and streptomycin (100 μg/mL). Cells were grown at 37 °C in a humidified incubator with 5 % CO2. To knockdown CRABP1 and CRABP2, MCF-7 cells were transfected with 10 nM siRNA. The medium was replaced with fresh medium 18 h after transfection and the cells were cultured for an additional 48 h. Two rounds of siRNA transfections were performed for each experiment. Hs578T, BT-549 and SK-Br-3 cells were transfected with 7 μg of empty (control) or pcDNA3 expression construct (CRABP1 or CRABP2) as previously described [34]. For cell proliferation assays, 10,000 siRNA-transfected cells were seeded in each well of 12-well plates and cultured overnight in DMEM containing 10 % FBS. The medium was then replaced with FBS-supplemented medium containing the indicated concentrations of RA (or DMSO as a vehicle control). Five days later, cells were counted using a Coulter Particle and Size Analyzer (Coulter Corporation, Mississauga, Canada).
Immunofluorescence analysis
MCF-7 cells were cultured on coverslips for 24 h and treated with 0.5 μM RA (dissolved in DMSO) or vehicle (DMSO) in serum-free DMEM medium for 6 h. Cells were then fixed in 1 % paraformaldehyde in PBS for 10 min and permeabilized in 0.5 % Triton X-100 for 5 min. Cells were immunostained with anti-CRABP1 or anti-CRABP2 antibodies, followed by Alexa 594-conjugated donkey anti-mouse (for CRABP1) or Alexa 555-conjugated donkey anti-rabbit (for CRABP2) secondary antibodies (Life Technologies). Images were acquired using a Zeiss LSM510 confocal microscope (Oberkochen, Germany) with a 40 ×/1.3 oil immersion lens.
Patient population
A total of 176 treatment-naïve primary breast cancer samples and 10 normal breast tissue samples from reduction mammoplasties were obtained from the Canadian Breast Cancer Foundation Tumor Bank and used for gene expression microarray analysis as previously described [35]. Patient material and clinical information was collected under Research Ethics Board Protocol ETH-02-86-17. Tumor tissues were frozen and histologically analysed as previously described [35].
Patients received standardized guideline-based chemo- and hormone therapies: i.e., patients with ER-positive tumors received hormone therapy, those with HER2-positive tumors received trastuzumab, high-risk node-negative disease was treated with anthracycline chemotherapy whereas anthracycline plus taxane chemotherapy was used for the treatment of node-positive disease. The 176 patients selected for this study consisted of 88 patients who experienced early relapse (<5 years after the initial treatment) and 88 patients who had not relapsed. ER, PR and HER2 status, stage and time of follow-up were balanced between the two groups. The median follow-up time for surviving patients was 4.5 years. The gene profiling data used in this publication have been deposited in NCBI [GEO Datasets: GSE22820].
RNA preparation, gene expression microarrays and RT-PCR
Total RNA was isolated from frozen human breast tumor biopsies using the TRIzol reagent (Life Technologies) and further purified with Qiagen RNeasy columns (Qiagen, Mississauga, ON, Canada). The average percent of area with tumor cells was 72.6 % and the average percent of cells that were tumor cells was 93.4 %, for all tissues analysed. Microarray hybridization was carried out as previously described [35]. Reverse transcription-polymerase chain reaction (RT-PCR) conditions were as previously described [25, 34]. The number of cycles for each primer pair was optimized for quantitative amplification within the exponential PCR product growth phase. PCR primers are listed in Additional file 1: Table S1. PCR amplification of human β-actin mRNA served as positive control as previously described [25]. For real-time quantitative RT-PCR (qRT-PCR), we used the following TaqMan FAM-labeled Gene Expression Assay primers: human CRABP1 (Hs00171635_m1), human CRABP2 (Hs00275636_m1) and human GAPDH (Hs03929097). cDNA samples were analysed in triplicate, with each cDNA undergoing 40 cycles of amplification in 96-well reaction plates (10 μL volume) using TaqMan Gene Expression Master Mix (Applied Biosystems 7900HT Real-Time PCR System).
Generation of tissue microarrays and immunohistochemical (IHC) staining
Tissue microarrays (TMA) (TMArrayer, Pathology Devices) were generated using all available formalin-fixed paraffin-embedded breast tumor tissues representing 120 patients out of the 176-patient cohort used for gene expression analysis. The TMA slides contained triplicate core tissue samples (0.6 mm in diameter) from each tumor. TMAs were immunostained with anti-CRABP1 monoclonal antibody (Sigma-Aldrich; 1:200 dilution) and anti-CRABP2 polyclonal antibody (Protein Tech Group; 1:200 dilution). The signal was detected using EnVision + anti-mouse (CRABP1) or anti-rabbit (CRABP2) secondary systems (DakoCytomation, Carpinteria, CA). Tissues were counterstained with hematoxylin. Cytoplasmic and nuclear staining were scored separately based on the average staining signal intensity throughout the tumor tissue on a scale of 0 (negative), 1 (weak), 2 (moderate) and 3 (strong). Of the 120 tumor samples tested, 105 and 106 had sufficient tissue for analysis of CRABP1 and CRABP2 immunoreactivity, respectively.
Western blotting
Cytoplasmic and nuclear extracts were prepared according to Dignam et al. [36], and whole cell lysates were prepared using Dignam buffer A plus 0.5 % SDS. Cytoplasmic protein (20 μg), nuclear protein (20 μg) and whole cell protein (40 μg) were separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes by electroblotting. Membranes were immunostained with primary antibodies in 5 % bovine serum albumin (in 1X Tris-buffered saline) at 4 °C overnight. The signal was detected with horseradish peroxidase-conjugated secondary antibodies using the ECL Western Blotting Detection Reagent (GE Healthcare Life Sciences, USA). The following primary antibodies were used for western blot analysis: anti-CRABP1 (1:1,000), anti-CRABP2 (1:1,000), β −actin (Sigma-Aldrich; 1:100,000), α-tubulin (DSHB; 1:10,000) and Lamin A/C (ThermoFisher; 1:1,000).
Luciferase reporter assay
After siRNA depletion of CRABP1 or CRABP2, MCF-7 cells were seeded in 12-well culture plates at 20,000 cells/well and transfected with the luciferase reporter construct (0.5 μg/well) under the control of a retinoic acid response element (RARE) (pGL3-RARE-luc, Addgene). Alternatively, CRABP1-negative cell lines (BT-549, SK-Br-3, Hs578T) were seeded in 12-well plates, incubated at 37 °C for 24 h and then co-transfected with the CRABP1 expression construct (0.8 μg/well) and pGL3-RARE-Luc (0.5 μg/well) diluted in 250 μL of serum-free medium containing 5 μL PEI. pcDNA3 empty vector served as the negative control for these experiments. Forty-eight h after transfection, cells were treated with RA (in DMSO) at final concentrations of 0, 0.1 and 0.5 μM for 6 h keeping the overall volume of DMSO constant in each well. Cells were then harvested and whole cell lysates prepared using the luciferase cell culture lysis reagent (CCLR, Promega). Luciferase activity was measured with the Luciferase Assay System (Promega) and quantitated using a FLUOstar OPTIMA microplate reader (BMG Labtech) following the manufacturer’s instructions. Triplicate wells were analyzed for each treatment.
Statistical analysis
Statistical analysis was performed using MedCalc Statistical Software version 12.7.2 (MedCalc Software, Ostend, Belgium) as previously described [25]. Briefly, gene profiling data for CRABP1 and CRABP2 were classified as “low” or “high” by receiver operating characteristic (ROC) curve analysis. Student t-test or chi-square test was used to examine the significance of associations between CRABP mRNA levels or immunoreactivity and clinical outcome parameters. Two-way ANOVA was used to test the significance of the effects of siRNA knockdown and RA treatment on cell proliferation. The prognostic significance of CRABP1 and CRABP2 was analyzed by logrank test on Kaplan-Meier survival curves using both gene profiling and TMA immunoassay data.
Results
CRABP1 is expressed in ER- and triple-negative breast tumors and is associated with poor clinical outcomes
Based on our gene profiling data, CRABP1 is markedly down-regulated in ER-positive breast tumors in keeping with reports indicating that CRABP1 is silenced in cancer cells [30, 31, 33, 37]. However, CRABP1 expression is maintained in ER/PR-negative breast tumors compared to normal mammary tissues (Fig. 1a and Table 1). In contrast, CRABP2 mRNA levels are significantly up-regulated in ER-positive tumors compared to normal and triple-negative tumor tissues (Fig. 1a and Table 1). Neither CRABP1 nor CRABP2 levels showed statistically significant difference between HER2-overexpressing tumors and other tissue types. These data indicate that there is an inverse relationship between CRABP1 and CRABP2 expression in triple-negative compared to ER/PR-positive tumors
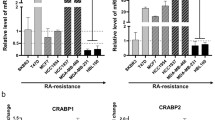
Distinct mRNA expression patterns and prognostic associations of CRABP1 and CRABP2 in breast tumors. a Comparison of CRABP1 and CRABP2 mRNA levels (based on normalized gene microarray signal intensity) in normal mammary tissues (n = 10) and human breast cancer subtypes (n = 176). b, c CRABP1 (b) and CRABP2 (c) mRNA levels in tumor tissues sampled from the 176 breast cancer patient cohort. The MT and GT numbers refer to individual patients. RNA levels were measured by real-time quantitative RT-PCR with GAPDH serving as the internal control. Expression levels from triplicate reactions are shown relative to MT633. d, e CRABP1 (d) and CRABP2 (e) mRNA levels in a panel of 11 breast cancer cell lines. RNA levels were analysed by real-time quantitative RT-PCR and shown as fold change relative to ZR-75-1. f Kaplan-Meier overall patient survival curves generated based on low and high CRABP1 mRNA levels determined by ROC analysis. g Kaplan-Meier overall patient survival curves generated based on low and high CRABP2 mRNA levels. n, denotes sample size; *, p < 0.05; **, p < 0.01
Next, we analysed CRABP expression in a sampling of ten cDNA samples from patient breast tumor tissues using real-time qRT-PCR analysis. CRABP1 mRNA was detected at elevated levels (~2 to 11-fold relative to the first tumor sample which was set at 1) in 4 out of 6 triple-negative tumors; however, none of the four non-triple negative tumors showed relative levels larger than 1 (Fig. 1b). In contrast, CRABP2 mRNA was found at higher levels in non-triple-negative tumors (~3 to 9-fold relative to MT633) compared to triple-negative tumors (with 1 out of 6 tumors showing relative levels higher than 1) (Fig. 1c).
We also screened a panel of 11 breast cancer cell lines for CRABP1 and CRABP2 expression. CRABP1 was detected at elevated levels (4.5–6.8-fold relative to ZR-75-1 which was set at 1) in two non-triple-negative breast cancer cell lines (MCF-7 and BT-483), with none of the five triple-negative cell lines having levels higher than 1 (Fig. 1d). These results suggest that CRABP1 expression is not retained in triple-negative breast cancer cells under in vitro growth conditions, perhaps as a consequence of changes in the microenvironment [38]. A similar phenomenon has been reported for other proteins and cancers, including loss of glial fibrillary acidic protein and brain fatty acid-binding protein in malignant glioma cell lines compared to tumor tissue [39–41]. CRABP2 was expressed at elevated levels (2–28-fold relative to ZR-75-1) in all six non-triple-negative breast cancer cell lines, while only two out of five triple-negative cell lines had relative CRABP2 levels higher than 1 (Fig. 1e).
We then examined the association of CRABP mRNA levels in primary breast tumors with major clinicopathological parameters (Table 1). High CRABP1 mRNA levels correlated with positive nodal status (p = 0.021), high nuclear grade (p < 0.0001), mitotic grade (p < 0.0001) and overall histological tumor grade (p < 0.0001) (Table 1). Such correlations were not observed with CRABP2 mRNA levels. Kaplan-Meier survival analysis further showed that high levels of CRABP1 mRNA were significantly (p = 0.0083) associated with lower patient survival probability (Fig. 1f), whereas high levels of CRABP2 mRNA were significantly associated with better prognosis (p = 0.0028; Fig. 1g). Again, these results suggest opposing roles for CRABP1 and CRABP2 in breast cancer clinical outcomes.
Subcellular localization of CRABP1 determines prognostic significance
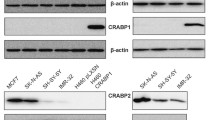
CRABPs serve as intracellular chaperones for RA and modulate its nuclear availability and biological activity [42, 43]. The subcellular localization of CRABPs is therefore a critical determinant of their function. To investigate whether there is any association between the subcellular distribution of CRABP1 and 2 and clinical outcomes, we conducted immunohistochemical analysis of a TMA containing primary tumor samples from 120 breast cancer patients. We first validated the specificity of our anti-CRABP1 and anti-CRABP2 antibodies by western blot analysis of whole cell lysates prepared from MDA-MB-435 cells (negative for both CRABP1 and 2) transfected with CRABP1 or CRABP2. No cross-immunoreactivity was observed for these two antibodies (Fig. 2a).
Immunoreactivity and subcellular distribution of CRABP1 and CRABP2 in a human primary breast tumor TMA. a Western blot of CRABP1 and CRABP2 in MDA-MB-435 cells transfected with a CRABP1 or CRABP2 expression construct, respectively. b Frequency of breast tumors with different subcellular immunoreactivity scores for CRABP1 and CRABP2: 0, negative; 1, weak; 2, intermediate; 3, strong. c Selected tissue sections from a human breast cancer TMA immunostained with anti-CRABP1 and anti-CRABP2 antibodies. Nuclear (Nuc) and cytoplasmic (Cyt) scores are indicated
We found that 34.3 % and 21.9 % of the scored tumors showed positive cytoplasmic and nuclear CRABP1 immunoreactivity, respectively (Fig. 2b and Table 1). TMA tissues with different CRABP1 and CRABP2 cytoplasmic and nuclear immunostaining intensities are shown in Fig. 2c. Elevated levels of CRABP1 in the cytoplasm, but not in the nucleus, was associated with negative ER (p = 0.0058) and PR status (p = 0.004). Both cytoplasmic and nuclear CRABP1 immunoreactivities were positively correlated with triple-negative status and high histological tumor grade (Table 1).
In contrast to CRABP1, the great majority of breast tumor tissues scored positive for CRABP2, with 90.7 % and 95.4 % of the scored samples showing cytoplasmic and nuclear CRABP2 immunoreactivity, respectively (Fig. 2b and Table 1). Close to half (43.5 %) of the tumors showed strong nuclear CRABP2 immunoreactivity (score = 3), with a strong cytoplasmic CRABP2 signal observed in 16.7 % of tumors (Fig. 2b–c). Nuclear CRABP2 protein levels were positively correlated with positive ER (p = 0.001), PR (p = 0.007) and overall non-triple negative status (p = 0.0019). A correlation was also observed between high cytoplasmic CRABP2 levels and positive PR status (p = 0.0371, Table 1). No correlations with other clinicopathological parameters were observed for both cytoplasmic and nuclear CRABP2 immunoreactivity (Table 1).
We then correlated subcellular levels of CRABPs with patient survival probabilities. We found that patients with tumors that were positive for cytoplasmic, but not nuclear, CRABP1 had a significantly lower patient survival probability (p = 0.0091) (Fig. 3a–b). In contrast, nuclear, but not cytoplasmic, CRABP2 immunoreactivity was positively associated with patient survival probability (p = 0.0127) (Fig. 3c–d). Thus, our TMA analysis shows that: (i) the clinicopathological associations of CRABP1 and 2 at the protein level are in good agreement with that of the gene profiling data, (ii) CRABP1 is more frequently found in the cytoplasm whereas CRABP2 is more abundant in the nucleus of primary breast tumors, and (iii) clinical outcome and prognostic implications can be attributed to cytoplasmic CRABP1 and nuclear CRABP2, suggesting distinct subcellular functions.
Associations of subcellular CRABP1 and CRABP2 levels with breast cancer patient survival and Ki67 immunoreactivity. CRABP1, CRABP2 and Ki67 protein levels were determined by immunohistochemical analysis of a TMA containing triplicate cores for each tumor from a 120 primary breast tumor cohort. a-b Association of cytoplasmic and nuclear CRABP1 levels with patient survival. c-d Association of cytoplasmic and nuclear CRABP2 levels with patient survival. e Positive correlation of cytoplasmic and nuclear CRABP1 protein levels with Ki67 immunoreactivity. f Positive correlation of CRABP1 and Ki67 mRNA levels based on gene microarray analysis. g No significant correlation was observed between subcellular CRABP2 levels and Ki67 immunoreactivity. h No correlation was evident between CRABP2 and Ki67 mRNA levels. HR, hazard ratio; p, statistical significance level; r, correlation coefficient. Scores: 0, negative; 1, weak; 2, intermediate; 3, strong. As the size of samples expressed CRABP1 is small in our TMAs, tumor samples were classified into “positive” and “negative” groups for survival analysis
CRABP1 correlates with tumor cell proliferation and is a potent inhibitor of RA signalling
To further understand the opposing roles of CRABP1 and CRABP2 in breast cancer progression, we immunostained our breast cancer TMA with Ki67, a cell proliferation marker (Additional file 1: Figure S1), and examined its correlation with CRABP protein levels. We found that both cytoplasmic (p = 0.024) and nuclear (p = 0.008) CRABP1 levels were positively correlated with Ki67 immunoreactivity (Fig. 3e). In contrast, neither cytoplasmic nor nuclear CRABP2 levels correlated with Ki67 protein levels (Fig. 3g). As expected, the mRNA levels of Ki67 also showed significant positive correlation with that of CRABP1, but not CRABP2 mRNA, based on gene profiling analysis (Fig. 3f, h). These results suggest that CRABP1 may affect breast cancer progression by enhancing tumor cell proliferation through modulation of RA signalling. The lack of correlation between CRABP2 and Ki67 suggests that the effect of CRABP2 on patient survival may be mediated through differentiation and apoptosis rather than proliferation, as previously reported for APL [17, 44].
CRABP1 and CRABP2 may play distinct roles in RA/RAR-mediated transcriptional activity [42, 45]. To investigate the role of CRABP1 in RA action, we used siRNAs to deplete the RA-responsive MCF-7 cells of either CRABP1 or CRABP2 (Fig. 4a–b). A small but statistically significant increase in growth inhibition was observed when CRABP1-depleted cells were treated with RA, with control cells showing 81 and 74 % relative growth at 0.1 μM and 0.5 μM RA, respectively, and CRABP1-depleted cells showing 65 and 66 % relative growth at 0.1 μM and 0.5 μM RA, respectively (Fig. 4c). A stronger growth inhibition effect was observed upon treatment of CRABP1-depleted cells with RA in the absence of serum (Additional file 1: Figure S2). In contrast to CRABP1, knockdown of CRABP2 had little effect on RA-mediated cell growth inhibition (91 % at 0.1 μM and 88 % at 0.5 μM) (Fig. 4c).
CRABP1 and CRABP2 differentially modulate RA-induced growth inhibition and RAR transcriptional activity. a, b Western blot analysis of MCF-7 cells transfected with CRABP1 or CRABP2 siRNAs using antibodies against CRABP1 or CRABP2. c Relative growth rate of MCF-7 cells treated with the indicated concentrations of RA after transfection with non-specific (control) and specific siRNAs targeting CRABP1 or CRABP2. Significance of difference was tested using two-way ANOVA. d RAR transactivation (measured by luciferase activity) of CRABP1a-depleted or CRABP2a-depleted MCF-7 cells transfected with a luciferase reporter construct under the control of a RARE. Cells were treated with DMSO (vehicle control) or RA for 6 h before harvest. Luciferase activity (a measure of RAR activation) is shown as fold change relative to cells that were cultured in the absence of RA. e The luciferase assay was repeated with a second set of siRNAs targeting CRABP1 and CRABP2 (CRABP1b, CRABP2b). f Western blots showing ectopic expression of CRABP1 in three human breast cancer cell lines. g, h, i The effects of ectopic expression of CRABP1 on RAR transactivation (measured by luciferase activity) were examined in BT-549 (g), SK-Br-3 (h) and Hs578T (i) cells co-transfected with a CRABP1 cDNA construct and a RARE-luciferase reporter construct. Cells were treated with RA for 6 h at the indicated concentrations. Luciferase activity (a measure of RAR activation) was adjusted based on protein concentrations of individual lysates and shown as fold change relative to control cells transfected with empty vector and cultured in the absence of RA. KD, denotes knockdown; *, p < 0.05; **, p < 0.01
The growth inhibitory effect of RA is believed to be mediated through activation of RARs [23, 24, 45]. We therefore examined the effect of CRABP1 and CRABP2 on RA-induced RAR activation using the luciferase reporter system. We used two different siRNAs to deplete each of the two CRABP genes in MCF-7 cells. CRABP-depleted MCF-7 cells were then transfected with a RA-responsive RARE-luciferase vector and treated with RA. An RA dose-dependent induction in luciferase activity (indicative of RAR activation) was observed for both control and siRNA transfected cells (Fig. 4d–e). However, CRABP1 depletion resulted in a ~2-fold increase in RAR activation in the presence of RA compared to control cells, whereas depletion of CRABP2 resulted in decreased RAR/RA-mediated transcriptional activity (Fig. 4d–e). These results suggest that in contrast to CRABP2 which plays a positive role in RAR activation, CRABP1 may serve as an inhibitor of RA signalling in breast cancer cells.
We then co-transfected two triple-negative breast cancer cell lines (Hs-578 T and BT-549) and one ER-negative/HER2 overexpressing cell line (SK-Br-3) with a CRABP1 expression construct and the RARE-luciferase reporter vector. CRABP1 expression in transfected cells was verified by western blotting (Fig. 4f). Endogenous CRABP1 was not detected in any of these three cell lines. Luciferase activity was induced by RA in all three cell lines in a dose-dependent manner in cells transfected with empty vector (Fig. 4g–i). A significant reduction in RA-induced RAR activation was observed in all three cell lines upon transfection with the CRABP1 expression construct. Specifically, SK-Br-3 and Hs578T cells showed reduced RAR-activation in both 0.1 μM and 0.5 μM RA-treated cells upon CRABP1 expression (Fig. 4h–i). Reduced luciferase activity was only observed at the highest concentration of RA tested in BT-549 cells (Fig. 4g).
Next, we analysed CRABP1 and CRABP2 subcellular localization in response to RA in MCF-7 cells. Both proteins were primarily found in the cytoplasm in the absence of RA (Fig. 5a). In contrast to CRABP2 which translocated to the nucleus upon RA treatment, there was no change in CRABP1 subcellular localization upon RA treatment (Fig. 5a). These results are consistent with a general role for CRABP1 in sequestering RA in the cytoplasm, rendering it unavailable for RAR activation.
a Subcellular localization of CRABP1 and CRABP2 in MCF-7 cells treated with RA. Cells were cultured in medium with serum for 24 h and then treated with 0.5 μM RA in serum-free medium for 6 h. An equivalent amount of DMSO was added to control cells. Cells were immunostained with anti-CRABP1 (red, upper panel) or anti-CRABP2 (red, lower panel) antibodies as described in Materials and Methods. DAP1 (blue) staining was used to visualize the nucleus. b Expression of RA-responsive genes in MCF-7. MCF-7 cells underwent two rounds of transfection with scrambled or CRABP1 siRNAs. Cells were treated with increasing concentrations of RA [lanes 1 to 6 (0, 5 × 10−5, 5 × 10−4, 5 × 10−3, 5 × 10−2, 5 × 10−1 μM RA)]. RNA was purified from each culture and semi-quantitative RT-PCR was carried out using gene-specific primers (Additional file 1: Table S1). c Summary of the effect of CRABP1 and RA on downstream genes and pathways. d Western blots showing the subcellular distribution of CRABP2 in SK-Br-3 cells upon CRABP1 overexpression and RA treatment. Densitometric analysis was used to quantitate CRABP2 signal intensity in the cytoplasm and nucleus relative to the cytoplasmic marker (β-tubulin) and nuclear marker (lamin A/C), respectively. Changes in band intensities are shown as fold change in relation to lane 1 (for cytoplasmic CRABP2) and lane 5 (for nuclear CRABP2)
CRABP1 attenuates RA signalling through modulation of the expression of RA responsive genes
To further explore the mechanism involved in CRABP1/RA-mediated effects in breast cancer cells, we examined the expression of critical genes implicated in RA synthesis, trafficking, metabolism and action upon CRABP1 manipulation and RA treatment. MCF-7 cells were transfected with scrambled or CRABP1 siRNAs and treated with increasing amounts of RA. Levels of CRABP2, a facilitator of RA signalling to the nucleus [46], were upregulated upon CRABP1 depletion in the absence of RA (Fig. 5b). Upon RA treatment, CRABP2 levels were induced in control cells in a dose-dependent manner but not in CRABP1-depleted cells. The transcription factor AP-2α (TFAP2A), a RA-inducible gene with tumor-suppressing activity [47, 48], was upregulated by CRABP1 depletion and RA treatment at a dose up to 5 nM. The expression of the RA metabolizing enzyme CYP26A1 was induced at high concentrations of RA (500 nM RA) in both control and CRABP1-depleted cells, with enhanced up-regulation observed in the latter. The observation that CYP26A1 levels are up-regulated to a higher degree in CRABP1-depleted compared to control cells argues against the idea that CRABP1 promotes RA metabolism [49–51]. The expression of aldehyde hydrogenase ALDH1B1 gene, which is involved in RA biosynthesis and associated with cancer cell stemness [52], was induced by RA in the control cells but not in the CRABP1-depleted cells. Interestingly, the two cellular retinol-binding protein genes (RBP1 and RBP7), encoding proteins that facilitate retinol uptake, storage and/or metabolism [53–56], exhibited opposite expression patterns upon CRABP1 knockdown and RA treatment, with RBP1 being induced by RA in both control cells and CRABP1-depleted cells, and RBP7 being down-regulated by CRABP1 depletion and suppressed by RA in a dose-dependent manner. These results (summarized in Fig. 5c) indicate that CRABP1 has an inhibitory effect on RA action through modulation of a spectrum of genes involved in various aspects of the RA network including RA biosynthesis, metabolism and intracellular trafficking.
Next, we examined the effect of CRABP1 expression on the nuclear translocation of CRABP2, which is an important indication of RA signalling to the nucleus [57]. SK-Br-3 cells were transfected with a CRABP1 expression construct and treated with RA. We observed higher levels of cytoplasmic and nuclear CRABP2 upon RA treatment in control cells (Fig. 5d). Furthermore, a marked decrease in nuclear CRABP2 levels was observed in CRABP1-overexpressing cells upon RA treatment. Taken together, these results suggest that CRABP1 acts as an inhibitor of RA action by restricting RA access to the nucleus through down-regulation of nuclear CRABP2 levels.
Our results suggest that CRABP1, in addition to the previously identified FABP5 and CRABP2, is a key factor regulating breast cancer cell response to RA. To explore the possibility that expression levels of these three RA binding proteins might be useful predictors of primary breast tumor sensitivity to RA, we analyzed their relative mRNA levels in our 176 breast cancer patient cohort based on gene microarray analysis. CRABP1 and FABP5, encoding RA binding proteins associated with RA resistance, were 5.2- and 1-fold higher, respectively, in ER-negative breast tumors compared to ER-positive tumors. On the other hand, levels of CRABP2, a positive modulator of RA signalling and activity, were 1.2-fold higher in ER-positive compared to ER-negative tumors (Fig. 6a). These results suggest that CRABP1 in particular may play an important role in the RA resistance observed in ER-negative tumors.
Schematic representation of the effects of different RA binding proteins on modulation of RA action in breast cancer. a Relative mRNA levels of FABP5, CRABP1 and CRABP2 in ER-negative (n = 64) and ER-positive (n = 112) primary breast cancer tissue samples. The mRNA levels for each gene were determined based on the normalized signal intensity of the gene microarray data and are shown relative to CRABP2 (set as 1) in the case of ER-negative tumors and FABP5 (set as 1) in the case of ER-positive tumors. These data provide insight as to the possible underlying cause of RA resistance in ER-negative tumors. b A schematic model illustrating the distinct roles of CRABP1, CRABP2 and FABP5 in modulating cellular response to RA in breast cancer cells. FABP5 channels RA to PPARδ/β, a nuclear receptor which promotes cell proliferation. CRABP1 sequesters RA in the cytoplasm. CRABP2 delivers RA to RAR, leading to cell growth inhibition. The balance between FABP5/CRABP1 and CRABP2 expression levels determines the cellular response to RA (cell growth promotion or inhibition)
Discussion
Cellular response to RA is believed to depend on two different classes of nuclear receptors, RARs and PPARs [23, 24]. RAR activation by RA results in cell growth inhibition whereas PPARδ/β activation stimulates cell proliferation. These two RA signaling pathways are in turn modulated by two intracellular RA binding proteins: CRABP2 which channels RA to the nucleus to target RAR, and FABP5 which delivers RA to the nucleus thereby activating PPARδ/β [23, 24]. We previously showed that FABP5 is preferentially expressed in ER- and triple-negative breast cancers, molecular subtypes believed to be resistant to RA treatment [25]. High levels of FABP5, as well as a low ratio of CRABP2 to FABP5, are associated with poor prognosis. In this study, we identify a third RA-binding protein, CRABP1, as an inhibitor of RA action and an adverse factor for clinical outcome in breast cancer. Like FABP5, CRABP1 is preferentially expressed in ER- and triple-negative breast cancer. We propose a model whereby CRABP1 can compensate or synergize with FABP5 to compete with CRABP2 for RA, by sequestering RA in the cytoplasm, thereby reducing RA access to RAR (Fig. 6b).
The role of CRABP1 in carcinogenesis and tumor progression is poorly understood and contradictory. For example, CRABP1 is down-regulated in some human cancers and cell lines [31–33], with DNA methylation proposed to contribute to CRABP1 silencing [33, 58–60]. DNA methylation-mediated silencing of CRABP1 has been observed in a subset of breast carcinoma tissues [60]. CRABP1 has been proposed to be a tumor suppressor in esophageal squamous cell carcinoma, with reduced CRABP1 levels associated with increased cell growth and distant lymph node metastasis [33]. Reduced levels of CRABP1 are also associated with poorer prognosis in serous (n = 40) and clear cell ovarian adenocarcinoma (n = 59) [31]. On the other hand, high levels of CRABP1 have been linked to lymph node metastasis and poor differentiation/high grade in pancreatic neuroendocrine tumors [30].
In agreement with a pro-carcinogenic role for CRABP1, we found an association between CRABP1 expression and worse clinical outcomes in breast cancer using both gene profiling and TMA analysis. Similar to a previous report indicating that CRABP1 is differentially expressed in different subtypes of pituitary adenomas [37], we found that CRABP1 is downregulated in ER+ breast tumors, but expressed in ER- and triple-negative tumors. These results suggest that downregulation of CRABP1 expression in cancer cells may be modulated by specific signaling pathways in different cancer subtypes (e.g. estrogen signalling), and that its role in tumor progression may differ between cancer types. It is noteworthy that estrogen signaling has been linked to the regulation of DNA methylation, perhaps explaining to some extent the reduced expression of CRABP1 observed in ER+ compared to ER- breast cancers [61].
CRABP1 has the highest RA binding affinity of all RA binding proteins [42]. It is generally believed that CRABP1 represses cellular response to RA by sequestering RA and/or promoting RA catabolism, reducing its availability in the nucleus for activation of RARs [51, 62–64]. Several studies have demonstrated that CRABP1 promotes RA metabolism [49–51]. However, the fact that RA metabolites can activate RAR in vitro [65], combined with the observation that a positive relationship exists between RA metabolism and cell growth inhibition in several cancer cell lines [66], suggest that CRABP1-mediated RA metabolism may not account for RA resistance. In light of our observations that: (i) cytoplasmic CRABP1 in breast tumors is an adverse factor in clinical outcomes and (ii) CRABP1 accumulates in the cytoplasm of cells treated with RA, we propose that the primary role of CRABP1 in breast cancer is to sequester RA in the cytoplasm, thereby preventing RAR activation in the nucleus (Fig. 6b). The up-regulation of the RA-metabolizing gene CYP26A1 observed upon CRABP1 depletion at high RA concentration further suggests a negative association between CRABP1 and RA metabolism.
Over five hundred genes are known to be regulated by RA, including CRABP2 which has a functional RARE in its promoter region [67, 68]. CRABP2 serves as a positive regulator of RA signalling in breast cancer cells [23, 24, 42] and its expression can be induced by RA in various cell types [67–69]. In this study, we found that CRABP1 and CRABP2 have inverse expression patterns in breast tumors and play an opposing role in the mediation of RA action in breast cancer cells. We further report that CRABP1 negatively regulates CRABP2 expression. While increased CRABP2 expression has been observed in AB1 embryonic stem cells with homozygous deletion of CRABP1 [70], this is the first report demonstrating that CRABP1 has an inhibitory effect on CRABP2 expression in cancer cells. Intriguingly, CRABP1 not only inversely regulates CRABP2 expression, but also affects nuclear translocation of CRABP2 in breast cancer cells. We postulate that CRABP1 plays a key role in attenuating RA activity in breast cancer cells, with high levels of CRABP1 reducing availability of RA in the nucleus. In turn, RA sequestration to the cytoplasm represses RA-mediated nuclear translocation of CRABP2 and induction of CRABP2 expression.
In addition to CRABP2, our data indicate that CRABP1 modulates the expression of various genes implicated in RA biosynthesis, metabolism and action. With the exception of RBP7, all these genes have previously been shown to be RA-regulated [68]. We observed that the genes encoding CYP26A1 (catalyzes RA metabolism) and ALDH1B1 (catalyzes RA biosynthesis) are up-regulated and down-regulated, respectively, in the presence of RA upon CRABP1 depletion. We speculate that this is due to a feedback process as CRABP1 depletion may increase levels of free RA (especially when cells are exposed to high doses of RA), in turn resulting in accelerated RA metabolism and reduced RA synthesis. Therefore, CRABP1 may play a role in regulating cellular levels of free RA. RBP1 and RBP7 are retinol binding proteins which facilitate retinol storage and/or retinol-RA conversion [53–56]. The different expression patterns observed for these two genes in control and CRABP1-depleted MCF-7 cells suggests opposite roles. We propose that RBP1 (upregulated by RA but not affected by CRABP1) may be involved in retinol storage whereas RBP7 (down-regulated by RA and CRABP1 depletion) may be involved in retinol metabolism producing RA.
TFAP2A is a RA-inducible gene whose expression increased upon CRABP1 knockdown in MCF-7 cells. TFAP2A encodes AP-2α, recently shown to be essential for RA action [71], has previously been reported to stimulate CRABP2 expression in mammary epithelial cells and breast cancer cells [72]. AP-2α significantly enhances RA-induced RAR activation in breast cancer cells (our unpublished data). These combined data suggest the presence of a CRABP1-AP-2α-CRABP2 axis which modulates RA action in breast cancer cells.
In summary, we show that CRABP1 expression is maintained in ER- and triple-negative breast tumors, and that elevated levels of CRABP1 is a significant indicator of high tumor grade, Ki67 immunoreactivity, and poor prognosis. Our data indicate that cytoplasmic CRABP1, like FABP5, is a potent inhibitor of RA signalling. Elevated levels of CRABP1 may lead to RA resistance in breast cancer cells through sequestration of RA in the cytoplasm thereby preventing RA-mediated induction of RAR. We further demonstrate that CRABP1 attenuates RA activity by modulating the expression of important RA-regulated genes implicated in cellular RA availability, traffic and action. Thus, both CRABP1 and FABP5 represent potential therapeutic targets to overcome RA resistance in breast cancer. The discovery that there are at least three proteins involved in RA transport in breast cancer cells (CRABP1, CRABP2 and FABP5 [23–25]), helps to address the molecular mechanism governing RA resistance in ER-negative or triple-negative breast cancer, and provides molecular tools to predict and eventually overcome RA resistance in breast cancer prevention and therapy.
Abbreviations
- ALDH1B1:
-
Aldehyde dehydrogenase 1 family, member B1
- APL:
-
Acute promyelocytic leukemia
- CRABP:
-
Cellular retinoic acid-binding protein
- CYP26A1:
-
Cytochrome P450, family 26, subfamily A, polypeptide 1
- DMEM:
-
Dulbecco’s Modified Eagle’s medium
- ER:
-
Estrogen receptor
- HER2:
-
Epidermal growth factor receptor 2
- PEI:
-
Polyethylenimine
- PPAR:
-
Peroxisome proliferator-activated receptor
- PR:
-
Progesterone receptor
- RA:
-
Retinoic acid
- RAR:
-
Retinoic acid receptor
- RARE:
-
Retinoic acid response element
- RBP:
-
Retinol binding protein
- RXR:
-
Retinoid-X-receptor
- TFAP2A:
-
Transcription factor AP-2 alpha
References
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74.
Riggs BL, Hartmann LC. Selective estrogen-receptor modulators – mechanisms of action and application to clinical practice. N Engl J Med. 2003;348:618–29.
Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, et al. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol. 2010;28:1145–53.
Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–34.
Haffty BG, Yang Q, Reiss M, Kearney T, Higgins SA, Weidhaas J, et al. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol. 2006;24:5652–7.
Bushue N, Wan YJ. Retinoid pathway and cancer therapeutics. Adv Drug Deliv Rev. 2010;62:1285–98.
Connolly RM, Nguyen NK, Sukumar S. Molecular pathways: current role and future directions of the retinoic acid pathway in cancer prevention and treatment. Clin Cancer Res. 2013;19:1651–9.
Tang XH, Gudas LJ. Retinoids, retinoic acid receptors, and cancer. Annu Rev Pathol. 2011;6:345–64.
Tighe AP, Talmage DA. Retinoids arrest breast cancer cell proliferation: retinoic acid selectively reduces the duration of receptor tyrosine kinase signaling. Exp Cell Res. 2004;301:147–57.
Faria TN, Mendelsohn C, Chambon P, Gudas LJ. The targeted disruption of both alleles of RARbeta(2) in F9 cells results in the loss of retinoic acid-associated growth arrest. J Biol Chem. 1999;274:26783–8.
Liu Y, Lee MO, Wang HG, Li Y, Hashimoto Y, Klaus M, et al. Retinoic acid receptor beta mediates the growth-inhibitory effect of retinoic acid by promoting apoptosis in human breast cancer cells. Mol Cell Biol. 1996;16:1138–49.
Seewaldt VL, Johnson BS, Parker MB, Collins SJ, Swisshelm K. Expression of retinoic acid receptor beta mediates retinoic acid-induced growth arrest and apoptosis in breast cancer cells. Cell Growth Differ. 1995;6:1077–88.
Warrell Jr RP, Frankel SR, Miller Jr WH, Scheinberg DA, Itri LM, Hittelman WN, et al. Differentiation therapy of acute promyelocytic leukemia with tretinoin (all-trans-retinoic acid). N Engl J Med. 1991;324:1385–93.
Zhuang Y, Faria TN, Chambon P, Gudas LJ. Identification and characterization of retinoic acid receptor beta2 target genes in F9 teratocarcinoma cells. Mol Cancer Res. 2003;1:619–30.
Huang ME, Ye YC, Chen SR, Chai JR, Lu JX, Zhoa L, et al. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood. 1988;72:567–72.
Petrie K, Zelent A, Waxman S. Differentiation therapy of acute myeloid leukemia: past, present and future. Curr Opin Hematol. 2009;16:84–91.
Freemantle SJ, Spinella MJ, Dmitrovsky E. Retinoids in cancer therapy and chemoprevention: promise meets resistance. Oncogene. 2003;22:7305–15.
Garattini E, Gianni M, Terao M. Retinoids as differentiating agents in oncology: a network of interactions with intracellular pathways as the basis for rational therapeutic combinations. Curr Pharm Des. 2007;13:1375–400.
Noy N. Retinoid-binding proteins: mediators of retinoid action. Biochem J. 2000;348(Pt 3):481–95.
He J, Allen JR, Collins VP, Allalunis-Turner MJ, Godbout R, Day 3rd RS, et al. CDK4 amplification is an alternative mechanism to p16 gene homozygous deletion in glioma cell lines. Cancer Res. 1994;54:5804–7.
Wolf G. Cellular retinoic acid-binding protein II: a coactivator of the transactivation by the retinoic acid receptor complex RAR.RXR. Nutr Rev. 2000;58:151–3.
Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129:723–33.
Schug TT, Berry DC, Toshkov IA, Cheng L, Nikitin AY, Noy N. Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARbeta/delta to RAR. Proc Natl Acad Sci U S A. 2008;105:7546–51.
Liu RZ, Graham K, Glubrecht DD, Germain DR, Mackey JR, Godbout R. Association of FABP5 expression with poor survival in triple-negative breast cancer: implication for retinoic acid therapy. Am J Pathol. 2011;178:997–1008.
Butler WB, Fontana JA. Responses to retinoic acid of tamoxifen-sensitive and -resistant sublines of human breast cancer cell line MCF-7. Cancer Res. 1992;52:6164–7.
Fontana JA. Interaction of retinoids and tamoxifen on the inhibition of human mammary carcinoma cell proliferation. Exp Cell Biol. 1987;55:136–44.
van der Leede BJ, Folkers GE, van den Brink CE, van der Saag PT, van der Burg B. Retinoic acid receptor alpha 1 isoform is induced by estradiol and confers retinoic acid sensitivity in human breast cancer cells. Mol Cell Endocrinol. 1995;109:77–86.
Chen NN, Li Y, Wu ML, Liu ZL, Fu YS, Kong QY, et al. CRABP-II- and FABP5-independent all-trans retinoic acid resistance in COLO 16 human cutaneous squamous cancer cells. Exp Dermatol. 2012;21:13–8.
Kainov Y, Favorskaya I, Delektorskaya V, Chemeris G, Komelkov A, Zhuravskaya A, et al. CRABP1 provides high malignancy of transformed mesenchymal cells and contributes to the pathogenesis of mesenchymal and neuroendocrine tumors. Cell Cycle. 2014;13.
Miyake T, Ueda Y, Matsuzaki S, Miyatake T, Yoshino K, Fujita M, et al. CRABP1-reduced expression is associated with poorer prognosis in serous and clear cell ovarian adenocarcinoma. J Cancer Res Clin Oncol. 2011;137:715–22.
Pfoertner S, Goelden U, Hansen W, Toepfer T, Geffers R, Ukena SN, et al. Cellular retinoic acid binding protein I: expression and functional influence in renal cell carcinoma. Tumour Biol. 2005;26:313–23.
Tanaka K, Imoto I, Inoue J, Kozaki K, Tsuda H, Shimada Y, et al. Frequent methylation-associated silencing of a candidate tumor-suppressor, CRABP1, in esophageal squamous-cell carcinoma. Oncogene. 2007;26:6456–68.
Liu RZ, Graham K, Glubrecht DD, Lai R, Mackey JR, Godbout R. A fatty acid-binding protein 7/RXRbeta pathway enhances survival and proliferation in triple-negative breast cancer. J Pathol. 2012;228:310–21.
Germain DR, Graham K, Glubrecht DD, Hugh JC, Mackey JR, Godbout R. DEAD box 1: a novel and independent prognostic marker for early recurrence in breast cancer. Breast Cancer Res Treat. 2011;127:53–63.
Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–89.
Chile T, Correa-Giannella ML, Fortes MA, Bronstein MD, Cunha-Neto MB, Giannella-Neto D, et al. Expression of CRABP1, GRP, and RERG mRNA in clinically non-functioning and functioning pituitary adenomas. J Endocrinol Invest. 2011;34:e214–8.
Rondeau G, Abedinpour P, Desai P, Baron VT, Borgstrom P, Welsh J. Effects of different tissue microenvironments on gene expression in breast cancer cells. PLoS One. 2014;9, e101160.
Ertel A, Verghese A, Byers SW, Ochs M, Tozeren A. Pathway-specific differences between tumor cell lines and normal and tumor tissue cells. Mol Cancer. 2006;5:55.
Gillet JP, Calcagno AM, Varma S, Marino M, Green LJ, Vora MI, et al. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc Natl Acad Sci U S A. 2011;108:18708–13.
Godbout R, Bisgrove DA, Shkolny D, Day 3rd RS. Correlation of B-FABP and GFAP expression in malignant glioma. Oncogene. 1998;16:1955–62.
Dong D, Ruuska SE, Levinthal DJ, Noy N. Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J Biol Chem. 1999;274:23695–8.
Sessler RJ, Noy N. A ligand-activated nuclear localization signal in cellular retinoic acid binding protein-II. Mol Cell. 2005;18:343–53.
Sakoe Y, Sakoe K, Kirito K, Ozawa K, Komatsu N. FOXO3A as a key molecule for all-trans retinoic acid-induced granulocytic differentiation and apoptosis in acute promyelocytic leukemia. Blood. 2010;115:3787–95.
Jing Y, Waxman S, Mira-y-Lopez R. The cellular retinoic acid binding protein II is a positive regulator of retinoic acid signaling in breast cancer cells. Cancer Res. 1997;57:1668–72.
Delva L, Bastie JN, Rochette-Egly C, Kraiba R, Balitrand N, Despouy G, et al. Physical and functional interactions between cellular retinoic acid binding protein II and the retinoic acid-dependent nuclear complex. Mol Cell Biol. 1999;19:7158–67.
Luscher B, Mitchell PJ, Williams T, Tjian R. Regulation of transcription factor AP-2 by the morphogen retinoic acid and by second messengers. Genes Dev. 1989;3:1507–17.
McPherson LA, Loktev AV, Weigel RJ. Tumor suppressor activity of AP2alpha mediated through a direct interaction with p53. J Biol Chem. 2002;277:45028–33.
Fiorella PD, Napoli JL. Microsomal retinoic acid metabolism. Effects of cellular retinoic acid-binding protein (type I) and C18-hydroxylation as an initial step. J Biol Chem. 1994;269:10538–44.
Napoli JL. Retinoic acid biosynthesis and metabolism. Faseb J. 1996;10:993–1001.
Won JY, Nam EC, Yoo SJ, Kwon HJ, Um SJ, Han HS, et al. The effect of cellular retinoic acid binding protein-I expression on the CYP26-mediated catabolism of all-trans retinoic acid and cell proliferation in head and neck squamous cell carcinoma. Metabolism. 2004;53:1007–12.
Ioannou M, Serafimidis I, Arnes L, Sussel L, Singh S, Vasiliou V, et al. ALDH1B1 is a potential stem/progenitor marker for multiple pancreas progenitor pools. Dev Biol. 2013;374:153–63.
Ghyselinck NB, Bavik C, Sapin V, Mark M, Bonnier D, Hindelang C, et al. Cellular retinol-binding protein I is essential for vitamin A homeostasis. EMBO J. 1999;18:4903–14.
Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, et al. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–5.
Piantedosi R, Ghyselinck N, Blaner WS, Vogel S. Cellular retinol-binding protein type III is needed for retinoid incorporation into milk. J Biol Chem. 2005;280:24286–92.
Pierzchalski K, Yu J, Norman V, Kane MA. CrbpI regulates mammary retinoic acid homeostasis and the mammary microenvironment. Faseb J. 2013;27:1904–16.
Budhu AS, Noy N. Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol Cell Biol. 2002;22:2632–41.
Lind GE, Kleivi K, Meling GI, Teixeira MR, Thiis-Evensen E, Rognum TO, et al. ADAMTS1, CRABP1, and NR3C1 identified as epigenetically deregulated genes in colorectal tumorigenesis. Cell Oncol. 2006;28:259–72.
Wu Q, Lothe RA, Ahlquist T, Silins I, Trope CG, Micci F, et al. DNA methylation profiling of ovarian carcinomas and their in vitro models identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol Cancer. 2007;6:45.
Wang F, Yang Y, Fu Z, Xu N, Chen F, Yin H, et al. Differential DNA methylation status between breast carcinomatous and normal tissues. Biomed Pharmacother. 2014;68:699–707.
Vrtacnik P, Ostanek B, Mencej-Bedrac S, Marc J. The many faces of estrogen signaling. Biochemia Medica. 2014;24:329–42.
Blaese MA, Santo-Hoeltje L, Rodemann HP. CRABP I expression and the mediation of the sensitivity of human tumour cells to retinoic acid and irradiation. Int J Radiat Biol. 2003;79:981–91.
Boylan JF, Gudas LJ. Overexpression of the cellular retinoic acid binding protein-I (CRABP-I) results in a reduction in differentiation-specific gene expression in F9 teratocarcinoma cells. J Cell Biol. 1991;112:965–79.
Boylan JF, Gudas LJ. The level of CRABP-I expression influences the amounts and types of all-trans-retinoic acid metabolites in F9 teratocarcinoma stem cells. J Biol Chem. 1992;267:21486–91.
Idres N, Marill J, Flexor MA, Chabot GG. Activation of retinoic acid receptor-dependent transcription by all-trans-retinoic acid metabolites and isomers. J Biol Chem. 2002;277:31491–8.
Takatsuka J, Takahashi N, de Luca LM. Retinoic acid metabolism and inhibition of cell proliferation: an unexpected liaison. Cancer Res. 1996;56:675–8.
Astrom A, Pettersson U, Chambon P, Voorhees JJ. Retinoic acid induction of human cellular retinoic acid-binding protein-II gene transcription is mediated by retinoic acid receptor-retinoid X receptor heterodimers bound to one far upstream retinoic acid-responsive element with 5-base pair spacing. J Biol Chem. 1994;269:22334–9.
Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res. 2002;43:1773–808.
Astrom A, Pettersson U, Voorhees JJ. Structure of the human cellular retinoic acid-binding protein II gene. Early transcriptional regulation by retinoic acid. J Biol Chem. 1992;267:25251–5.
Chen AC, Yu K, Lane MA, Gudas LJ. Homozygous deletion of the CRABPI gene in AB1 embryonic stem cells results in increased CRABPII gene expression and decreased intracellular retinoic acid concentration. Arch Biochem Biophys. 2003;411:159–73.
Bi Y, Gong M, He Y, Zhang X, Zhou X, Zhang Y, et al. AP2alpha transcriptional activity is essential for retinoid-induced neuronal differentiation of mesenchymal stem cells. Int J Biochem Cell Biol. 2014;46:148–60.
McPherson LA, Woodfield GW, Weigel RJ. AP2 transcription factors regulate expression of CRABPII in hormone responsive breast carcinoma. J Surg Res. 2007;138:71–8.
Acknowledgements
We are grateful to Lillian Cook and Cheryl Santos for their excellent technical assistance. We especially thank Dr. Kathryn Graham and Adrian Driga for management of the gene microarray database. This work was supported by grants from the Canadian Breast Cancer Foundation (R.G. and J.R.M.).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RZL conceived, carried out experiments, analysed the data and wrote the manuscript. EG, DDG and HYP carried out experiments. JRM was involved in the study design and data collection. RG was involved in the study design, data interpretation and manuscript editing. All authors reviewed and approved the manuscript.
Additional file
Additional file 1: Table S1.
Nucleotide sequences of PCR primers. Figure S1. Sample images of Ki67 immunohistochemical staining of a human breast cancer TMA. The percentages shown are the averages of the percent Ki67 positive cells in three TMA cores of the same tumor. Tumor ID numbers are indicated at the bottom. Figure S2. Effect of CRABP1 expression on RA-induced cell growth inhibition in MCF-7 cells. Cells were transfected with siRNA targeting CRABP1 (CRABP1 kd) or scrambled siRNA (control). Transfected cells were then seeded in a 96-well plate, cultured for 24 h and then treated with RA in serum-free medium at the indicated concentrations for two days. Cell proliferation was analyzed using the MTS cell proliferation assay system (Promega) following the manufacturer’s protocol. Significance of difference was analyzed by two-way ANOVA. *denotes p < 0.05 and **p < 0.01.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Liu, RZ., Garcia, E., Glubrecht, D.D. et al. CRABP1 is associated with a poor prognosis in breast cancer: adding to the complexity of breast cancer cell response to retinoic acid. Mol Cancer 14, 129 (2015). https://doi.org/10.1186/s12943-015-0380-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12943-015-0380-7