Abstract
Background
The emergence of plasmid-mediated mobile colistin resistance (mcr) gene poses a great challenge to the clinical application of polymyxins. To date, mcr-1 to mcr-10 have been found in animals, humans, and the environment. Among them, mcr-8 was first identified in Klebsiella pneumoniae (K. pneumoniae) of swine origin, and then mcr-8.1 to mcr-8.5 were successively identified. Notably, K. pneumoniae is the major host of the mcr-8 gene in both animals and humans. This study aims to explore the characteristics of K. pneumoniae strains carrying the mcr-8 gene and tmexCD1-toprJ1 gene cluster and investigate the correlation between these two antibiotic resistance genes.
Methods
The isolates from the poultry farms and the surrounding villages were identified by mass spectrometer, and the strains positive for mcr-1 to mcr-10 were screened by polymerase chain reaction (PCR). The size of the plasmid and the antimicrobial resistance genes carried were confirmed by S1-nuclease pulsed-field gel electrophoresis (S1-PFGE) and Southern hybridization, and the transferability of the plasmid was verified by conjugation experiments. Antimicrobial susceptibility testing (AST) and whole genome sequencing (WGS) were used to characterize the strains.
Results
Two K. pneumoniae isolates (KP26 and KP29) displaying polymyxin resistance were identified as mcr-8 gene carriers. Besides that, tigecycline-resistant gene cluster tmexCD1-toprJ1 was also found on the other plasmid which conferred strain resistance to tigecycline. Through epidemiological analysis, we found that the mcr-8 gene has dispersed globally, circulating in the human, animals, and the environment. Furthermore, our analysis suggests that the coexistence of mcr-8 and tmexCD1-toprJ1 on a single plasmid might evolved through plasmid recombination.
Conclusions
Although the mcr-8 and tmexCD1-toprJ1 gene clusters in the two strains of K. pneumoniae in this study were on two different plasmids, they still pose a potential threat to public health, requiring close monitoring and further study.
Similar content being viewed by others
Background
Multi-drug resistant (MDR) pathogens have become a vital global issue that caused by the overuse of antimicrobials in recent years [1]. Among MDR pathogens, Klebsiella pneumoniae is considered as the notorious recipient of multiple antimicrobial resistance (AMR) genes as hundreds of mobile AMR genes have been found in this bacterium [2]. Accumulation of AMR genes in K. pneumoniae is primarily the result of horizontal gene transfer (HGT) driven by plasmids and mobile genetic elements. The transmission of AMR genes in bacteria has posed a great threat to clinical and public health problems [3].
Polymyxins are a family of cyclic lipopeptides among which polymyxin B and polymyxin E (colistin) are the most used and studied variants. It acts as the last-resort antibiotics effective against infections caused by MDR Gram-negative bacteria in the clinical treatment. By electrostatic interacting with the negatively charged phosphate groups on lipid A of lipopolysaccharide (LPS) in the bacterial outer membrane, polymyxins inserted into the fatty acyl layer of the outer membrane and caused membrane disorganization and eventual cell death [4,5,6,7]. Since Liu et al. [8] reported for the first time the emergence of the plasmid-mediated polymyxin resistance gene mcr-1 in Enterobacteriaceae, 10 mcr gene variants (mcr-1 to mcr-10) were found in isolates sourcing from animals, environment, and humans over 20 countries [9,10,11]. The mcr gene mediates resistance by encoding a phosphoethanolamine (pEtN) transferase, which adds pEtN to the lipid A portion of LPS, thereby reducing the number of negatively charged phosphate groups on its surface. This modification results in a decreased affinity for polymyxin to bind to the target site, allowing for resistance to occur [10, 12]. A search of the GenBank database revealed that the mcr-8 gene was mainly present in K. pneumoniae of human and animal origin [13]. Following the identification of a plasmid carrying mcr-8.2 in K. pneumoniae, it has been speculated that animal-derived Klebsiella may be a reservoir for the mcr-8 gene [14]. Gene mcr-8 was identified on the transferable IncII plasmid of K. pneumoniae, and the acquisition of a single mcr-8 gene significantly increased the colistin resistance of Escherichia coli (E. coli) and K. pneumoniae [13].
Tigecycline is a tetracycline class of antibacterial drugs that can be used to treat infections caused by polymicrobial MDR including both Gram-positive and Gram-negative bacteria [15]. The emergence of tigecycline resistance has been reported worldwide though few studies are available for deciphering the molecular mechanism of resistance to tigecycline. Recently, a novel plasmid-harbored resistance-nodulation-division (RND) efflux pump gene cluster, tmexCD1-toprJ1, was reported to mediate tigecycline resistance in Gram-negative bacteria [16, 17]. The global dissemination of plasmid-borne drug resistance genes threatens to compromise the efficacy of polymyxins and tigecycline, increasing the difficulty of treatment of associated bacterial infections.
This study aimed to investigate the mechanism of plasmid-mediated colistin and tigecycline resistance by analyzing the prevalence and genetic characteristics of the mcr gene and tmexCD1-toprJ1 gene, underlining the co-transmission risk of these resistance genes.
Methods
Isolation and identification of strain and mcr genes
Samples of chicken manure, farm workers and healthy people were collected from chicken farms and their neighboring villages in Hangzhou. Fecal samples (≈1.0 g) were placed in 5 ml sterile Luria–Bertani broth and cultured overnight in an incubator at 37 ℃. The next day, the bacterial solution was coated on MacConkey agar for overnight culture. The MacConkey agar was repeatedly underlined to obtain a single colony. The species of bacteria were identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), and the mcr genes of all isolates were subsequently sequenced by PCR and Sanger sequencing using specific primers [18]. Primer pairs used in this study were listed in Additional file 1: Table S1.
Antimicrobial susceptibility testing
In accordance with Clinical and Laboratory Standards Institute (CLSI) guidelines, agar or broth microdilution is used to assess antimicrobial susceptibility. Microbroth dilution method was used to determine the MIC values of polymyxins (colistin and polymyxin B) and tigecycline, and agar dilution method was used to determine the MIC values of other antibiotics. Susceptibility results for all strains to tigecycline were interpreted according to the breakpoints of the European Committee for Antimicrobial Susceptibility Testing (EUCAST) (https://eucast.org/clinical_breakpoints/), and susceptibility results for other antibiotics were interpreted according to the CLSI guidelines. The AST was carried out using E. coli ATCC 25922 as the quality control standard.
Conjugation experiments
To verify the transfer ability of plasmids carrying the mcr-8 gene or tmexCD1-toprJ1 gene cluster, we used strains carrying mcr-8 or tmexCD1-toprJ1 as donors, and sodium azide-resistant E. coli J53 and rifampicin-resistant EC600 as the recipient strains, respectively. Mueller–Hinton agar containing 200 mg/L sodium azide and 2 mg/L polymyxin B, 200 mg/L rifampicin and 4 mg/L tigecycline were used as the screening medium for transconjugants. We then selected transconjugants on Mueller–Hinton agar with sodium azide and polymyxin B, and detected the presence of the mcr-8 gene in the transconjugants by PCR and DNA sequencing.
S1-PFGE and southern hybridization
To identify the number and size of plasmids in K. pneumoniae carrying the mcr-8 gene, we performed the S1-PFGE and Southern hybridization experiments. Salmonella H9812 was used as control strain and size marker [19]. After electrophoresis, it was hybridized with a digoxigenin-labeled mcr-8 probe to determine the location of the mcr-8 gene, and finally detected using the nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP) color detection kit (Roche, catalog no. 11745832910).
WGS and bioinformatic analysis
The genomic DNA of mcr-8 positive isolates was extracted by Gentra Puregene yeast /Bact kit (Qiagen, CA, USA). WGS was then performed on the Nanopore PromethION platform (Nanopore, Oxford, UK) in accordance with the 10-kbp library protocol. Paired-end libraries detection was performed using the Illumina Novaseq 6000 system (Illumina, San Diego, CA, USA). Hybrid assembly of Illumina short reads and PromethION long reads using Unicycler v0.4.8. WGS data was annotated using rast (https://rast.nmpdr.org/). The assembled contigs were analyzed using the Center for Genomic Epidemiology website(https://www.genomicepidemiology.org/) to identify acquired antimicrobial resistance (AMR) genes, plasmid incompatibility types, and perform Multi Locus Sequence Typing (MLST). The analysis of virulence genes was performed on Virulence Factor Database (http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi).
The strains of K. pneumoniae carrying mcr-8 were screened from NCBI nucleotide database by BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed 29 March 2023), the K. pneumoniae harboring mcr-8 in GenBank are shown in Additional file 2: Table S2. The phylogenetic tree was generated using Prokka and Roary. The transposon and insertion sequence were detected using the ISFinder database (https://isfinder.biotoul.fr/about.php). Finally, Easyfig.2.0 was used for linear comparison of multiple plasmids carrying mcr-8 and tmexCD1-toprJ1.
Results
Characteristics of antimicrobial susceptibility of the mcr-positive K. pneumoniae strain
We collected fecal samples from chicken manure, chicken farm workers, and healthy workers in chicken farms and residents nearby villages around Hangzhou neighborhood. A total of 112 strains of Enterobacteriaceae were identified, among which 60 strains were isolated from chicken, 5 strains were from chicken farm workers, and 47 strains were from healthy residents. Colony PCR was conducted to screen mcr genes using primers as described in materials and methods. Two K. pneumoniae KP26 and KP29 from chicken manure were found to harbor mcr-8 gene. Antimicrobial susceptibility analysis of KP26 and KP29 were performed and interpreted according to CLSI guidelines (Table 1). The result showed that KP26 and KP29 exhibited low-level resistance to polymyxin B (KP26, MIC = 4 mg/L; KP29, MIC = 4 mg/L) and colistin (KP26, MIC = 8 mg/L; KP29, MIC = 4 mg/L). Besides that, KP26 and KP29 also displayed resistance to tigecycline (KP26, MIC = 8 mg/L; KP29, MIC = 8 mg/L). MIC data revealed that KP26 and KP29 exhibited resistance to cefazolin (> 128 mg/L), levofloxacin (> 32 mg/L), aztreonam (64 mg/L), ceftriaxone (> 64 mg/L), cefepime (> 64 mg/L), amikacin (> 128 mg/L), ceftazidime (64 mg/L), cefepime-sulbactam (64/32 mg/L), and ciprofloxacin (> 32 mg/L), but is susceptible to carbapenem antibiotics, including ertapenem (0.25 mg/L), imipenem (0.125 mg/L), and meropenem (0.03 mg/L) (Table 1).
Conjugation experiments showed that the colistin resistance gene mcr-8 of KP26 and KP29 could be successfully transferred to the recipient E. coli J53, making the conjugants acquire colistin resistance phenotype. Results of S1-PFGE and Southern hybridization showed that KP26 carried 4 plasmids and KP29 carried 5 plasmids, with mcr-8 gene located at plasmid 101.2 kb (Fig. 1).
S1-PFGE profile (left) and Southern hybridization (right) analysis using mcr-8-specific probes to detect mcr-8-harboring isolates. Salmonella strain H9812 was used as a control strain and size marker. The names of the isolates are shown in the first line. The arrows indicated the locations of mcr-8 harboring plasmids according to the Southern hybridization experiment
Genomic analysis and molecular type of K. pneumonia KP26 and KP29
The complete genomes of isolates KP26 and KP29 were sequenced by WGS. KP26 consists of a 5,252,651 bp chromosome and four plasmids (pKP26-tmexCD1, pKP26-mcr8, pKP26-3, pKP26-4) (Table 2). KP29 consists of a 5,253,463 bp chromosome and five plasmids (pKP29-tmexCD1, pKP29-2, pKP29-mcr8, pKP29-4, pKP29-5) (Table 2). MLST analysis showed that the KP26 and KP29 strains belong to ST11.
Antibiotic resistance determinants blaSHV-182, fosA, oqxA and oqxB were both detected on chromosomes of KP26 and KP29. Most of the resistance genes are located on plasmids, especially on the large plasmids pKP26-tmexCD1 and pKP29-tmexCD1, which are 271,379 bp. They include aph(6)-Id, aph(3')-Ia, aph(3'')-Ib, aadA1, aadA2b, armA, blaDHA-1, msr(E), mph(E), cmlA1, qnrB4, sul1, sul3, tmexD1, toprJ1, and tmexC1. Only one resistance gene, mcr-8, has been identified in pKP26-mcr8 and pKP29-mcr8. Other resistance genes were identified on two other plasmids of KP26 and three other plasmids of KP29.
Characterization of mcr-8-harboring plasmids
Plasmid pKP26-mcr8 was of type IncFIA/IncFII with a total length of 101,185 bp and contained only one antimicrobial resistance gene, mcr-8.1. It was 100% identical to plasmid pKP29-mcr8. Therefore, we only describe the characterization of pKP26-mcr8. The mcr-8.1 gene on pKP26-mcr8 was associated with the genetic context of IS903B-orf-dgkA-baeS-copR-mcr-8.1-orf-IS903B. The pKP26-mcr8 was shown to be conjugative under laboratory conditions. We performed comparative genomic analysis based on DNA sequences of pKP26-mcr8 and five plasmids with different sources in NCBI (Fig. 2). The results showed that pKP26-mcr8, pKP46-mcr8 and pKP91 [13] isolated from animals had similar genetic background around the mcr-8 gene IS903B-orf-dgkA-baeS-copR-mcr-8.1-orf-IS903B. In clinical plasmid pMCR8_095845, p18-29mcr-8.2 and environmental plasmid pZZW20-88 K, ISKpn26 took the position of IS903B, which was located in the downstream region of mcr-8 gene.
Genetic features of the mcr-8-carrying plasmid in K. pneumoniae strain KP26 and KP29. The linear genetic environment surrounding mcr-8 is depicted. Arrows indicate the direction of transcription for each gene, and different colors represent different genes. The mcr-8 gene is shown in red, genes associated with mobile elements are shown in green, the three genes in the figure are shown in black, and hp and other functional genes are shown in yellow. Regions with high homology are represented by gray shading
Evolutionary analysis of isolates carrying mcr-8
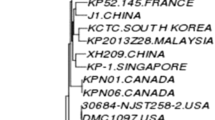
We retrieved genome sequences of 138 mcr-8 positive K. pneumoniae from the NCBI database. Among these isolates, 122 strains had definite source. There are 63 strains isolated from animals, 56 strains from human, and 3 from environment (Fig. 3). Twenty strains did not have complete genome, so we constructed a phylogenetic tree consisting of 118 K. pneumoniae isolates, K. pneumoniae KP26 and KP 29 based on single nucleotide polymorphisms (SNPs) of core genomes. The 120 K. pneumoniae isolates were clustered in 3 clonal groups (Fig. 4). These isolates were identified in North America, Asia and Africa. Most of them were detected in China, followed by Thailand and Vietnam, indicating that Asia is the main prevalent continent for mcr-8 positive K. pneumoniae. The most common MLST type among the 114 strains was ST43 (17/114), followed by ST11 (16/114), which is also the dominant epidemic strain in China [20]. K. pneumoniae KP26 and KP29 were belong to cluster II and closed to KP_SAMN23139063. It is worth noting that isolates in cluster III were from different countries around world isolated from humans, animals and environment. This implies that mcr-8 positive K. pneumoniae are undergoing evolution during the process of dissemination.
Among all 138 isolates containing mcr-8 in the NCBI database, 131 of them had geographical distribution with well-defined locations. Different hosts of the isolates are marked with different colors, with red, yellow, blue and black indicating human, animal, environmental and unknown origin respectively. The number of isolates harboring mcr-8 in each country is plotted under the country name on a world map. And the world map was created using the corresponding map data of the R package ggplot2 v3.3.5
Phylogenetic tree generated from the core genome sequences of the mcr-8-harboring isolates identified in this study and other mcr-8-harboring K. pneumoniae isolates in the NCBI database. Each isolate has a sample identifier (ID), location, MLST, and host. The figure shows important antimicrobial resistance genes (ARGs) and major virulence-associated genes (VGs)
In order to investigate the virulence genes in mcr-8-positive K. pneumoniae, the whole genome sequence of mcr-8-positive K. pneumoniae were compared with virulence factor database VFDB. The results showed that the virulence genes included iron-absorption virulence gene (entA, entB, fepC), invasive virulence genes (ompA), adhesive virulence gene (yagV/ecpE, yagW/ecpD, yagX/ecpC, yagY/ecpB, yagZ/ecpA, ykgK/ecpR), and enterotoxin gene (astA). Among the 120 strains of K. pneumoniae, only 1 strain was found to carry both iroB and iucA.
Characterization of the tmexCD1-toprJ1-harboring plasmid
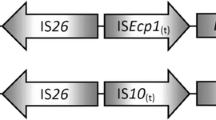
The plasmid type of pKP26-tmexCD1 was IncFIB(Mar)/IncHI1B, with a length of 271,379 bp. Resistance genes include aph(6)-Id, aph(3’)-Ia, aph(3’)-Ib, aadA1, aadA2b, armA, blaDHA-1, msr(E), mph(E), cmlA1, qnrB4, sul1, and sul3. The sequence of pKP29-tmexCD1 was 100% identical to pKP26-tmexCD1. To investigate the core genetic environment of tmexCD1-toprJ1, we analyzed the tmexCD1-toprJ1-containing regions from four tmexCD1-toprJ1-bearing plasmids in Klebsiella spp. isolated from animals, humans and environment. The core genetic structures of tmexCD1-toprJ1 in Klebsiella spp. were divided into two types (Fig. 5). The type I genetic context is IS903B-strB-strA-tnpR-∆tnpA-toprJ1-tmexD1-tmexC1-tnfxB1-∆hp-IS26. Four tmexCD1-toprJ1-bearing plasmids (pHKU57_1, pKP26-tmexCD1, pKP46-3, and pKPT698-tmexCD) were belong to type I, and slightly different is that the pHKU57_1 lacks an IS903B upstream. They possessed two insertion sequence (IS) elements IS26 located on the flank of tmexCD1-toprJ1, which conferred the ability to acquire and mobilize tmexCD1-toprJ1, resulting the transmission in Klebsiella. The core genetic environment of type II was strB-strA-tnpR-ΔtnpA-toprJ1-tmexD1-tmexC1-tnfxB1-hp in plasmid pHNAH8I-1[16]. It lacked the flanked IS26 element compared with type I, which indicates that the mobilization of tmexCD1-toprJ1 might be driven by IS26 from the plasmid pHNAH8I-1[21]. The IS elements play an important role in the spreading of tmexCD1-toprJ1.
Genetic features of the tmexCD1-toprJ1-carrying plasmid in K. pneumoniae strain KP26 and KP29. The linear genetic environment surrounding tmexCD1-toprJ1 is depicted. Arrows indicate the direction of transcription of each gene, and different genes are shown in different colors. The tmexCD1-toprJ1 gene is shown in red, genes associated with mobile elements are shown in green, the three genes in the figure are shown in black, hp and other functional genes are shown in yellow, and int genes are shown in purple
Discussion
In this study, we found that two strains of K. pneumoniae isolated from chickens were identified as belonging to multi-drug resistant ST11 type and carried both the mcr-8 gene and the tmexCD1-toprJ1 gene cluster on two different plasmids. The complete genome sequences of two strains were obtained by WGS and compared with the genome of other K. pneumoniae isolates carrying the mcr-8 gene and tmexCD1-toprJ1 retrieved from NCBI database. Our results showed that mcr-8 gene is globally dispersed and has been circulating in the human, animals, and the environment, urging us to pay close attention to the convergence and co-transmission of mcr-8 and the tmexCD1-toprJ1 gene in K. pneumoniae.
In recent years, the discovery of mcr genes carried by plasmids has attracted a great deal of public attention. To date, a total of ten mcr genes (mcr-1 to mcr-10) have been identified [10]. The mcr-8 gene was initially detected on an IncFII-type plasmid carrying carbapenemase-encoding gene blaNDM in swine-origin K. pneumoniae. Although mcr-8 gene and its variants have been identified in humans, animals, and environmental sources of K. pneumoniae, Raoultella ornithinolytica, and Klebsiella quasipneumoniae, K. pneumoniae is still considered as the primary host for the mcr-8 gene in both animals and human [13, 14, 22, 23]. In this study, we collected 112 strains of Enterobacteriaceae isolated from chicken manure and identified two mcr-8-positive K. pneumoniae strains. WGS revealed that these two strains also carried the tigecycline resistance gene cluster tmexCD1-toprJ1. The genetic context of mcr-8 in pKP26-mcr8 and pKP29-mcr8 is flanked with IS903B. This arrangement is consistent with genetic context of the mcr-8 gene in pKP91 [13]. But in the environmental isolate pZZW20-88 K, a single IS903B is located upstream of the mcr-8 gene, while ISKpn26 is located downstream. Similarly, in the human isolate pMCR8_095845, ISKpn26 lies in the downstream region of mcr-8 gene. Therefore, we speculated that the insertion sequence flanked with mcr-8 gene is variable and both IS903B and ISKpn26 can mediate the transference of mcr-8 gene.
We also analyzed the nucleotide and protein sequences of genes that are reported to be involved in polymyxin resistance, including mgrB, phoP/phoQ, pmrA/pmrB, crrA/crrB, qseB/qseC, yciM and lpxM in KP26 and KP29. In addition to crrA/crrB genes which were not exist, two type of point mutations were found in the lpxM of KP26 and KP29 (I7N and S229T). Whether lpxM mutations affects the polymyxins resistance in KP26 and KP29 needs further investigation.
The tnfxB1-tmexCD1-toprJ1 gene cluster was found on the transposon Tn5393. There are two IS26 located upstream and downstream of this gene cluster, indicating that the tmexCD1-toprJ1 gene cluster was transferred via IS26 transposition [21]. In addition, we found that pKP26-tmexCD1 contain a umuC gene serving as the integration site for the mobilization of tmexCD1-toprJ1 gene cluster mediated by int gene. However, attempts to transfer the plasmid carrying this gene cluster into EC600 through conjugation experiments or electroporation, were unsuccessful. We inferred that the tmexCD1-toprJ1-carrying plasmid might have a preference for the host. This might explain the widespread presence of this gene cluster in K. pneumoniae, which is rare in other Enterobacteriaceae species [24]. Apart from chicken in this study, the tmexCD1-toprJ1 was also detected in ST6265 K. pneumoniae strains from patient sources [25]. This result suggests that strains carrying tmexCD1-toprJ1 are capable of transmitting between human and animals. We also investigated the existence of iron-absorption virulence gene, invasive virulence gene and adhesive virulence gene in KP26 and KP29 through WGS analysis. However, hypervirulence genes were not found in these two strains. This indicates that KP26 and KP29 are not hypervirulent strains.
Although our study found that the mcr-8 and tmexCD1-toprJ1 gene were located on the two different plasmids, it has been reported that the mcr-8 gene and the tmexCD1-toprJ1 gene cluster can coexist on the same plasmid and undergo co-transfer during transmission. Plasmid recombination have occurred during the conjugation between the mcr-8-carrying plasmid and tmexCD1-toprJ1-carrying plasmid mediated by IS26 [26]. Using colistin could accelerate the process of plasmid fusion and formation of tigecycline-resistant strains. Through phylogenetic analysis of KP26, KP29, and mcr-8-carrying K. pneumoniae strains from the NCBI database, it was observed that the co-occurrence of the tmexCD1-toprJ1 gene cluster was found in 35% (42/120) of the strains. Therefore, it is highly possible that IS26 can mediate the plasmid fusion between pKP26-tmexCD1 and pKP26-mcr8 to carry more antibiotic resistance genes. The emergence of isolates resistant to both colistin and tigecycline is inevitable due to the use of these drugs as last-resort antibiotic treatments. This, in turn, facilitates the spreading of mcr-8 and tmexCD1-toprJ1 within the same strain.
Conclusions
In conclusion, we reported the presence of the mcr-8 and tmexCD1-toprJ1 gene cluster in two animal-derived K. pneumoniae strains in the chicken farm in China. These two resistance genes were located on two different plasmids. By screening mcr-8-carrying K. pneumoniae isolates from the NCBI database, we found mcr-8 positive K. pneumoniae strains have distributed around the world, spreading in different hosts. The acquisition of colistin and tigecycline resistance genes by K. pneumoniae might pose a serious threat to the public health. Therefore, we emphasize the importance of a “One Health” approach, particularly enhancing surveillance for the coexistence of mcr-8 and tmexCD1-toprJ1 in the same K. pneumoniae strain.
Availability of data and materials
The complete sequence of KP29 and KP26 has been submitted to GeneBank under accession no. CP140760 to CP140770.
Abbreviations
- K. pneumoniae :
-
Klebsiella pneumoniae
- E. coli :
-
Escherichia coli
- PCR:
-
Polymerase chain reaction
- S1-PFGE:
-
S1-nuclease pulsed-field gel electrophoresis
- AST:
-
Antimicrobial susceptibility testing
- WGS:
-
Whole genome sequencing
- MALDI-TOF–MS:
-
Matrix-assisted laser desorption ionization time-of- flight mass spectrometry
- CLSI:
-
Clinical and Laboratory Standards Institute
- EUCAST:
-
European Committee on Antimicrobial Susceptibility Testing
- MIC:
-
Minimum inhibitory concentration
- MDR:
-
Multi-drug resistant
- AMR:
-
Antimicrobial resistance
- RND:
-
Resistance-Nodulation-Division
- LPS:
-
Lipopolysaccharide
- MLST:
-
Multi Locus Sequence Typing
- NBT-BCIP:
-
Nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate
References
Cerceo E, et al. Multidrug-resistant gram-negative bacterial infections in the hospital setting: overview, implications for clinical practice, and emerging treatment options. Microb Drug Resist. 2016;22(5):412–31.
Wyres KL, Holt KE. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr Opin Microbiol. 2018;45:131–9.
Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18(6):344–59.
Dixon RA, Chopra I. Leakage of periplasmic proteins from Escherichia coli mediated by polymyxin B nonapeptide. Antimicrob Agents Chemother. 1986;29(5):781–8.
Falagas ME, Karageorgopoulos DE, Nordmann P. Therapeutic options for infections with Enterobacteriaceae producing carbapenem-hydrolyzing enzymes. Future Microbiol. 2011;6(6):653–66.
LaPlante K, Cusumano J, Tillotson G. Colistin for the treatment of multidrug-resistant infections. Lancet Infect Dis. 2018;18(11):1174–5.
Li J, et al. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. 2006;6(9):589–601.
Liu YY, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–8.
Baron S, et al. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents. 2016;48(6):583–91.
Hussein NH, et al. Mobilized colistin resistance (mcr) genes from 1 to 10: a comprehensive review. Mol Biol Rep. 2021;48(3):2897–907.
Rhouma M, et al. Colistin in pig production: chemistry, mechanism of antibacterial action, microbial resistance emergence, and one health perspectives. Front Microbiol. 2016;7:1789.
Son SJ, et al. MCR-1: a promising target for structure-based design of inhibitors to tackle polymyxin resistance. Drug Discov Today. 2019;24(1):206–16.
Wang X, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7(1):122.
Yang X, et al. Emergence of mcr-8.2-bearing Klebsiella quasipneumoniae of animal origin. J Antimicrob Chemother. 2019;74(9):2814–7.
Yaghoubi S, et al. Tigecycline antibacterial activity, clinical effectiveness, and mechanisms and epidemiology of resistance: narrative review. Eur J Clin Microbiol Infect Dis. 2022;41(7):1003–22.
Lv L, et al. Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in klebsiella pneumoniae. Mbio. 2020;11:2.
He R, et al. Characterization of a plasmid-encoded resistance-nodulation-division efflux pump in klebsiella pneumoniae and klebsiella quasipneumoniae from patients in China. Antimicrob Agents Chemother. 2021;65:2.
Woodman ME, et al. Direct PCR of intact bacteria (Colony PCR). 2016. Curr Protoc Microbiol. https://doi.org/10.1002/cpmc.14.
Guo X, et al. Emergence of IMP-8-producing comamonas thiooxydans causing urinary tract infection in China. Front Microbiol. 2021;12: 585716.
Wang Q, et al. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis. 2018;67(2):S196-s205.
Harmer CJ, Moran RA, Hall RM. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. Mbio. 2014;5(5):e01801-14.
Farzana R, et al. Emergence of mobile colistin resistance (mcr-8) in a highly successful Klebsiella pneumoniae sequence type 15 clone from clinical infections in Bangladesh. Msphere. 2020;5:2.
Wang X, et al. Emergence of colistin resistance gene mcr-8 and its variant in Raoultella ornithinolytica. Front Microbiol. 2019;10:228.
Peng K, et al. Plasmids shape the current prevalence of tmexCD1-toprJ1 among Klebsiella pneumoniae in food production chains. Msystems. 2021;6:5.
Sun L, et al. Distribution and spread of the mobilized RND efflux pump gene cluster tmexCD-toprJ in Klebsiella pneumoniae from different sources. Microbiol Spectr. 2023;11(4): e0536422.
Wang X, et al. Co-transfer of mcr-8 with bla(NDM-1) or tmexCD1-toprJ1 by plasmid hybridisation. Int J Antimicrob Agents. 2022;60(2): 106619.
Acknowledgements
Not applicable.
Funding
This work was supported by The National Key Research and Development Programme of China (2022YFC2303900), the Natural Science Foundation of Zhejiang province, China (No. LY24H190002). And this work was also supported by the Fundamental Research Funds of Hangzhou Medical College (KYYB202004) to FW.
Author information
Authors and Affiliations
Contributions
FW and QL designed the scheme of this experiment, XM was the main contributor to conducting experiments and writing this paper, LX did some analysis of the data, HF, JY, KD, HZ, JF were responsible for collecting samples. FW and QL review and finalize. All authors contributed to the article and agreed to the submitted version.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Mo, X., Zhang, H., Fan, J. et al. Co-existence of two plasmids harboring transferable resistance-nodulation-division pump gene cluster, tmexCD1-toprJ1, and colistin resistance gene mcr-8 in Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob 23, 67 (2024). https://doi.org/10.1186/s12941-024-00727-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-024-00727-x