Abstract
Background
Plasmodium knowlesi is recognized as the fifth Plasmodium species causing malaria in humans. It is morphologically similar to the human malaria parasite Plasmodium malariae, so molecular detection should be used to clearly discriminate between these Plasmodium species. This study aimed to quantify the rate at which P. knowlesi is misidentified as P. malariae by microscopy in endemic and non-endemic areas.
Methods
The protocol of this systematic review was registered in the PROSPERO International Prospective Register of Systematic Reviews (ID = CRD42020204770). Studies reporting the misidentification of P. knowlesi as P. malariae by microscopy and confirmation of this by molecular methods in MEDLINE, Web of Science and Scopus were reviewed. The risk of bias in the included studies was assessed using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS). The pooled prevalence and 95% confidence interval (CI) of the misidentification of P. knowlesi as P. malariae by microscopy were estimated using a random effects model. Subgroup analysis of the study sites was performed to demonstrate any differences in the misidentification rates in different areas. Heterogeneity across the included studies was assessed and quantified using Cochran’s Q and I2 statistics, respectively. Publication bias in the included studies was assessed using the funnel plot, Egger’s test and contour-enhanced funnel plot.
Results
Among 375 reviewed studies, 11 studies with a total of 1569 confirmed P. knowlesi cases in humans were included. Overall, the pooled prevalence of the misidentification of P. knowlesi as P. malariae by microscopy was estimated at 57% (95% CI 37–77%, I2: 99.3%). Subgroup analysis demonstrated the highest rate of misidentification in Sawarak, Malaysia (87%, 95% CI 83–90%, I2: 95%), followed by Sabah, Malaysia (85%, 95% CI 79–92%, I2: 85.1%), Indonesia (16%, 95% CI 6–38%), and then Thailand (4%, 95% CI 2–9%, I2: 95%).
Conclusion
Although the World Health Organization (WHO) recommends that all P. malariae-positive diagnoses made by microscopy in P. knowlesi endemic areas be reported as P. malariae/P. knowlesi malaria, the possibility of microscopists misidentifying P. knowlesi as P. malariae is a diagnostic challenge. The use of molecular techniques in cases with malariae-like Plasmodium with high parasite density as determined by microscopy could help identify human P. knowlesi cases in non-endemic countries.
Similar content being viewed by others
Background
Plasmodium knowlesi was first recognized as a cause of simian malaria in long-tailed or pig-tailed macaques [1]. It was also recognized as a cause of human malaria in 1965 [2]. The large focus on P. knowlesi as a cause of human malaria was reported in Sarawak, Malaysia, in 2004 [3]. Since then, P. knowlesi malaria in humans has been reported throughout Malaysia [4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22] and other Southeast Asian countries including Thailand [23,24,25,26], Indonesia [27,28,29,30], Singapore [31, 32], Brunei [33], Cambodia [34, 35], Laos [36, 37], Myanmar [38], the Philippines [39], and Vietnam [40]. Moreover, P. knowlesi malaria has also been reported in travellers returning from endemic countries [41,42,43,44,45,46,47,48,49,50,51,52,53,54].
Although most P. knowlesi infections are asymptomatic, approximately 19% of infected patients develop severe infections, including acute kidney injury (AKI) (45.6%), jaundice (42%), and hyperparasitaemia (32.5%), as the common clinical manifestations [55]. Deaths from P. knowlesi infection have been linked to delayed parenteral treatment [56]. In the endemic country of Malaysia, early intravenous artesunate treatment is now recommended for all severe malaria cases to prevent mortality, resulting in a lower death rate during 2010–2014 [8]. The risk factors associated with P. knowlesi infection include older age, male sex, plantation work, sleeping outside, and travelling in areas where monkeys live [21, 55, 57]. A recent study also suggested that the transmission of P. knowlesi malaria between humans might occur with mosquitoes as vectors, given the presence of family clustering [14].
The identification or detection of malaria parasites relies on the results of analysis based on microscopy, the standard for malaria diagnosis. However, the use of microscopy to diagnose P. knowlesi malaria is inaccurate since the morphological features of the early trophozoites of P. knowlesi resemble those of Plasmodium falciparum, and the growing trophozoites are similar to the band-form trophozoites of Plasmodium malariae [1, 58]. In this study, the rate at which P. knowlesi is misidentified as P. malariae by microscopy was estimated and quantified to clarify the inherent disadvantage of solely utilizing microscopy to identify P. knowlesi infection in endemic and non-endemic areas.
Methods
This study followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) (Additional file 1: Checklist S1) [59]. The protocol was registered in the PROSPERO International Prospective Register of Systematic Reviews (ID = CRD42020204770).
Search strategy
Searches of potentially relevant articles published from January 1, 1993, to August 17, 2020 were performed in MEDLINE, Scopus and Web of Science. The search terms used were (Plasmodium OR malaria) AND knowlesi AND (microscopy OR microscopic OR blood film OR “blood film” OR “thick film” OR “thin film”) AND (PCR OR “polymerase chain reaction”). The searches aimed to find original articles in any language and ended on August 17, 2020.
Eligibility criteria
Original research articles were eligible to be included in the present study if they were on retrospective or prospective cross-sectional studies and reported: (1) the misidentification of P. knowlesi as P. malariae as identified by microscopy and (2) the confirmation of P. knowlesi cases by molecular methods. Studies/papers were excluded for the following reasons: absence of P. malariae or P. knowlesi as determined by microscopy, absence of P. knowlesi as determined by PCR, microscopic findings of P. malariae/P. knowlesi, P. knowlesi in macaques, submicroscopic P. knowlesi, unextractable data, case–control studies, case reports or case series, clinical trials, conference abstracts, experimental research, guidelines, letters to the editor, test performances, review articles, systematic reviews and use of the same participants or data set as in another study.
Study selection and data extraction
The selection of the included studies according to the eligibility criteria was performed by two of the authors (MK and AM). Any discrepancies between these two authors were resolved by discussion in order to reach a consensus. For each study that was included in the analysis, the following information was extracted: name of the first author, year of publication, study area (years of the survey), study design, age range (years) of the participants, sex (male, %) of the participants, PCR detection for Plasmodium spp., target genes, number of P. malariae and P. knowlesi identified by microscopy (including mixed infections), number of P. malariae and P. knowlesi identified by PCR (including mixed infections), and number of species discordances. Raw data from each study were stored in a standardized datasheet before data synthesis. Data selection and extraction were managed using Endnote Software X7 (Clarivate Analytics, Philadelphia, USA).
Quality of the included studies
The risk of bias for each study was assessed using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS) [60]. This tool comprises four domains: patient selection, index test, reference standard, and flow and timing [60].
Statistical analysis
Data from the included studies were analysed using the STATA Statistical Software Version 15.0 (StataCorp LLC, Texas, USA). The number of cases of Plasmodium species discordance (P. knowlesi as P. malariae) as identified by microscopy and the number of P. knowlesi cases identified by PCR were used to analyse the pooled prevalence of the misidentification of P. knowlesi as P. malariae. The pooled prevalence of discordance of the misidentification of P. knowlesi as P. malariae was estimated by a random effects model using the numerator in the prevalence calculation as the number of discordances, and the denominator as the number of PCR-positive malaria cases. The pooled prevalence and 95% confidence interval (CI) of the misidentification were estimated using a random effects model. Subgroup analysis of the study sites was performed to demonstrate any differences in the pooled prevalence in both endemic and non-endemic countries. The existence and level of heterogeneity across the included studies were assessed using Cochrane Q and I2 statistics, respectively. Publication bias was assessed using funnel plot asymmetry and Egger’s test for asymmetry.
Results
Search results
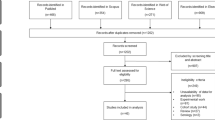
A total of 375 potentially relevant studies were identified from the searched databases. Among these, 109 were duplicates and removed. The papers on the remaining 266 studies were subjected to title and abstract screening. After this step, 146 papers were examined for their full text. Among these, 135 were excluded for the following reasons: no discordance between microscopy and PCR (n = 2), no P. malariae (n = 5), no P. knowlesi (n = 10), unextractable data (n = 8), case–control studies (n = 4), case reports or case series (n = 30), clinical trial (n = 1), conference abstract (n = 1), experimental studies (n = 21), guidelines (n = 2), letter to the editor (n = 1), microscopy findings reported as P. malariae/P. knowlesi (n = 6), P. knowlesi in macaques (n = 2), test performances (n = 13), review articles (n = 24), submicroscopic P. knowlesi (n = 2), systematic review (n = 1), and use of the same participants or data set (n = 1) (Fig. 1). Finally, a total of 11 studies [3, 13,14,15,16,17,18,19, 23, 24, 27] met the inclusion criteria and were included in the qualitative and quantitative synthesis.
Characteristics of the included studies
The characteristics of the included studies are presented in Table 1. The largest proportion of the included studies (5/11, 45.5%) were conducted in Sabah [14, 15, 17,18,19], while two (18.2%) were in Sawarak [3, 19], two (18.2%) in Thailand [23, 24], one (9.1%) in Malaysia (nine states) [16], and one (9.1%) in Aceh, Indonesia [27]. Most of the included studies (6/11, 54.5%) were retrospective in nature, while five (5/11, 45.5%) were prospective. Seven studies (45.5%) [3, 14,15,16,17, 19, 23] reported information on the age and sex of patients infected with P. knowlesi. All of these seven studies reported mean/median age in the range of 11–91 years, and the proportion of males was between 70 and 77.9%. Seven studies [3, 13,14,15,16, 19, 27] enrolled malaria positive samples for the analysis, while four studies [17, 18, 23, 24] enrolled patients suspected of having malaria. In most of the studies [3, 13,14,15,16,17, 19, 23, 24, 27], a test of nested PCR amplifying 18S rRNA was performed to identify P. knowlesi, with the exception of the study by Goh et al. [18], in which a Hexaplex PCR test was performed. Based on the 11 included studies, 1894 malaria cases were identified by microscopy, while 7953 malaria cases were identified by PCR. Microscopy identified 1425 P. malariae cases, while PCR identified 45. Also, microscopy identified 182 P. knowlesi cases, while PCR identified 1569.
Quality of the included studies
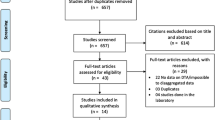
The risk of bias in each study was assessed using QUADAS. The results of the quality assessment are presented in Fig. 2 and Additional file 2. Seven studies (7/11, 63.6%) [3, 13,14,15,16, 19, 27] introduced bias in the selection of malaria positive samples for analysis.
Prevalence of the misidentification of P. knowlesi as P. malariae by microscopy
The total number of instances in which P. knowlesi was misidentified as P. malariae by microscopy was 1170. Based on the 11 included studies, the pooled prevalence of the misidentification of P. knowlesi as P. malariae by microscopy was 57% (37–77%, I2: 99.3%) (Fig. 3). The highest rate of misidentification of P. knowlesi as P. malariae was demonstrated in the study by Anderios et al. (25/25, 100%) [13], while the lowest rate was by Jongwutiwes et al. (1/33, 3.03%) [24].
Pooled prevalence of the misidentification of P. knowlesi as P. malariae. The pooled prevalence of discordance of the misidentification of P. knowlesi as P. malariae was estimated by a random effects model using the numerator in the prevalence calculation as the number of discordances, and the denominator as the number of PCR-positive malaria cases. ES estimated proportion, CI confidence interval, random random effects model
Subgroup analysis of the misidentification of P. knowlesi as P. malariae
Subgroup analysis of the study sites was performed to demonstrate the differences in the geographical distribution of the misidentification (Fig. 3). The results of the subgroup analysis demonstrated that the pooled prevalence of the misidentification was highest in Sawarak, Malaysia (87%, 95% CI 83–90%, I2: 95%, 2 studies), and Sabah, Malaysia (85%, 95% CI 75–92%, I2: 85.1%, 5 studies). The study by Yusof et al. [16] demonstrated 27% (95% CI 22–33%) misidentification in nine states of Malaysia. A low prevalence of the misidentification of P. knowlesi as P. malariae was demonstrated in Thailand (4%, 95% CI − 2 to 9%, I2: 95%, 2 studies) and Aceh, Indonesia (16%, 95% CI 6–38%).
Publication bias
The funnel plot could not be generated because few studies were included in the present study. Egger’s test demonstrated that no small-study effect was found (p: 0.126, coefficient: − 11.6, standard error: 6.79), indicating no publication bias among the included studies.
Discussion
Although P. knowlesi is well documented in Malaysia, the pooled quantification of the misidentification of this species as P. malariae has not been previously described. In this study, such misidentification was quantified using 11 studies [3, 13,14,15,16,17,18,19, 23, 24, 27], and it was found that the rate of this misidentification from 2000 to 2015 was 57%, with high heterogeneity among the included studies. Subgroup analysis of the study sites demonstrated a large difference in the misidentification rates. The highest prevalence of misidentification was demonstrated in two states of Malaysian Borneo, Sabah and Sawarak, where P. knowlesi was endemic in the last decade. In these areas, the number of P. knowlesi cases was not available until PCR testing was used to identify it in 2004 [3]. Molecular detection assay, nested PCR assay and real-time PCR test have been described for P. knowlesi targeting 18S rRNA gene targets [3, 61], with sensitivity of between 1 and 6 parasites/µl of blood [62]. From the time that nested PCR was implemented for diagnosis, the number of cases of P. knowlesi significantly increased, while a small number of P. malariae cases were still observed by PCR in Sabah during 2008–2011 [14, 15, 18] and in Sawarak during 2001–2006 [19]. This indicated that the highest number of P. malariae cases identified by microscopy in the last decade was caused by the emergence of P. knowlesi malaria, as these two species are morphologically similar and difficult to distinguish from each other using microscopy. While the highest prevalence of the misidentification of P. knowlesi as P. malariae occurred in Sabah and Sawarak, microscopically misdiagnosed cases of P. malariae were not found in other parts of Malaysia, such as Pahang and Kelantan [16]. This explained why the prevalence of the misidentification of P. knowlesi as P. malariae in the study by Yusof et al. [16] in nine states of Malaysia was lower than in studies conducted in Sabah and Sawarak [3, 13,14,15, 17,18,19].
In areas in which P. knowlesi was endemic, it was also frequently misidentified as P. falciparum or P. vivax malaria by microscopy [10]. The similarity of P. knowlesi and P. falciparum occurs at the stage of young rings of both species, which contain double chromatin dots, multiple-infected erythrocytes, and applique forms [63]; while the similarity of P. knowlesi and P. malariae occurs in the trophozoite, schizont, and gametocyte stages [63]. The recent decrease in diagnostic discrepancies by microscopy was due to the increased awareness and recognition among microscopists of P. knowlesi infections in endemic areas. Moreover, P. malariae is less endemic in Southeast Asia, where the presence malarial parasites with morphology similar to that of P. malariae coupled with high parasitaemia has been reported as P. knowlesi infection by default [16]. Moreover, the World Health Organization (WHO) recommends that all P. malariae-positive diagnoses by microscopy in P. knowlesi endemic areas be reported as P. malariae/P. knowlesi [64].
In areas where P. knowlesi is not endemic, such as Thailand and Indonesia, a low prevalence of the misidentification of P. knowlesi as P. malariae by microscopy was observed. Only one case of P. knowlesi misidentified as P. malariae from 33 confirmed cases of P. knowlesi was recorded by Jongwutiwes et al. during 2008–2009 [24], and only one such case among ten confirmed cases of P. knowlesi was recorded by Putaporntip et al. during 2006–2007 [23]. There was also a low prevalence of such misidentification by microscopy during 2014–2015 in Aceh, Indonesia [27], as only three cases of P. knowlesi were misdiagnosed from 19 confirmed cases of P. knowlesi as recorded by Coutrier et al. [27]. In addition, P. knowlesi was also misidentified as P. falciparum and P. vivax, as reported by studies in both Thailand and Indonesia [24, 27]. This indicated that microscopists were unable to recognize P. knowlesi because its ring forms were similar to those of P. falciparum, or sometimes its growing trophozoites were similar to those of P. vivax. Misidentification, such as the misdiagnosis of P. falciparum as P. knowlesi, might cause the administration of chloroquine, and the resistance of P. falciparum to chloroquine can increase the likelihood of patient mortality. Further, the misidentification of severe P. knowlesi as P. vivax malaria may lead to treatment failure, such as missed parenteral treatments as per national guidelines, which have been reported to be associated with fatal outcomes [56]. In addition to the misidentification of P. knowlesi mono-infection, mixed-infections of P. knowlesi combined with other Plasmodium species were also recorded in Thailand and Indonesia, such as mixed-infections with P. falciparum or mixed-infections with P. vivax malaria, which microscopists reported as P. falciparum or P. vivax mono-infections [23, 24, 27]. Severe complications due to P. knowlesi malaria in those co-infected patients in non-endemic countries such as Thailand and Indonesia were less likely since low parasite density of P. knowlesi was observed [23, 24], and P. knowlesi was responsive to chloroquine treatment in cases of mixed infections with P. vivax malaria. In addition, severe adverse events from unnecessary primaquine treatments were not experienced among co-infected patients [27].
The present study had some limitations. First, high heterogeneity among the included studies was observed, although subgroup analysis was performed; therefore, the results of the pooled prevalence needed to be interpreted carefully. Second, a low number of included studies were used for pooled analysis; therefore, the pooled prevalence might not have been precisely estimated. Third, studies reporting on P. knowlesi/P. malariae as determined by microscopy were not included in the present study since the number of misidentifications could not be estimated.
Conclusion
Although the WHO recommends that all P. malariae-positive diagnoses made by microscopy in P. knowlesi endemic areas be reported as P. malariae/P. knowlesi, the possibility of the misidentification of P. knowlesi as P. malariae by microscopists is a diagnostic challenge in both endemic and non-endemic countries. Assuming the low incidence of P. malariae in Malaysia and Southeast Asia, cases of symptomatic malaria with hyperparasitaemia and parasite morphology resembling that of P. malariae should be diagnosed as P. knowlesi/P. malariae by microscopy, so that severe complications among patients infected by P. knowlesi can be reduced.
Availability of supporting data
All data related to the present study are available.
References
Singh B, Daneshvar C. Human infections and detection of Plasmodium knowlesi. Clin Microbiol Rev. 2013;26:165–84.
Chin W, Contacos PG, Coatney GR, Kimball HR. A naturally acquited quotidian-type malaria in man transferable to monkeys. Science. 1965;149:865.
Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–24.
Barber BE, William T, Jikal M, Jilip J, Dhararaj P, Menon J, et al. Plasmodium knowlesi malaria in children. Emerg Infect Dis. 2011;17:814–20.
Jiram AI, Ooi CH, Rubio JM, Hisam S, Karnan G, Sukor NM, et al. Evidence of asymptomatic submicroscopic malaria in low transmission areas in Belaga district, Kapit division, Sarawak, Malaysia. Malar J. 2019;18:156.
Jiram AI, Hisam S, Reuben H, Husin SZ, Roslan A, Ismail WRW. Submicroscopic evidence of the simian malaria parasite, Plasmodium knowlesi, in an orang asli community. Southeast Asian J Trop Med Public Health. 2016;47:591–9.
William T, Jelip J, Menon J, Anderios F, Mohammad R, Awang Mohammad TA, et al. Changing epidemiology of malaria in Sabah, Malaysia: increasing incidence of Plasmodium knowlesi. Malar J. 2014;13:390.
Rajahram GS, Barber BE, William T, Grigg MJ, Menon J, Yeo TW, et al. Falling Plasmodium knowlesi malaria death rate among adults despite rising incidence, Sabah, Malaysia, 2010–2014. Emerg Infect Dis. 2016;22:41–8.
William T, Rahman HA, Jelip J, Ibrahim MY, Menon J, Grigg MJ, et al. Increasing incidence of Plasmodium knowlesi malaria following control of P. falciparum and P. vivax malaria in Sabah, Malaysia. PLoS Negl Trop Dis. 2013;7:e2026.
Barber BE, William T, Grigg MJ, Yeo TW, Anstey NM. Limitations of microscopy to differentiate Plasmodium species in a region co-endemic for Plasmodium falciparum, Plasmodium vivax and Plasmodium knowlesi. Malar J. 2013;12:8.
Vythilingam I, Lim YAL, Venugopalan B, Ngui R, Leong CS, Wong ML, et al. Plasmodium knowlesi malaria an emerging public health problem in Hulu Selangor, Selangor, Malaysia (2009–2013): epidemiologic and entomologic analysis. Parasites Vectors. 2014;7:14.
Barber BE, Bird E, Wilkes CS, William T, Grigg MJ, Paramaswaran U, et al. Plasmodium knowlesi malaria during pregnancy. J Infect Dis. 2015;211:1104–10.
Anderios F, Mohamed Z, Ratnam S, Ibrahim MY, Awang TAM. Detection of malaria parasites in Sabah by nested polymerase chain reaction: a focus of naturally acquired Plasmodium knowlesi infections. Sains Malays. 2008;37:137–41.
Barber BE, William T, Dhararaj P, Anderios F, Grigg MJ, Yeo TW, et al. Epidemiology of Plasmodium knowlesi malaria in north-east Sabah, Malaysia: family clusters and wide age distribution. Malar J. 2012;11:401.
Naing DKS, Anderios F, Lin Z. Geographic and ethnic distribution of P. knowlesi infection in Sabah, Malaysia. Int J Collab Res Intern Med Public Health. 2011;3:391–400.
Yusof R, Lau YL, Mahmud R, Fong MY, Jelip J, Ngian HU, et al. High proportion of knowlesi malaria in recent malaria cases in Malaysia. Malar J. 2014;13:168.
Joveen-Neoh WF, Chong KL, Wong CM, Lau TY. Incidence of malaria in the interior division of Sabah, Malaysian Borneo, based on nested PCR. J Parasitol Res. 2011;2011:104284.
Goh XT, Al Lim Y, Vythilingam I, Chew CH, Lee PC, Ngui R, et al. Increased detection of Plasmodium knowlesi in Sandakan division, Sabah as revealed by PlasmoNexTM. Malar J. 2013;12:264.
Cox-Singh J, Davis TM, Lee KS, Shamsul SS, Matusop A, Ratnam S, et al. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis. 2008;46:165–71.
Grigg MJ, William T, Drakeley CJ, Jelip J, Von Seidlein L, Barber BE, et al. Factors that are associated with the risk of acquiring Plasmodium knowlesi malaria in Sabah, Malaysia: a case–control study protocol. BMJ Open. 2014;4:e006004.
Grigg MJ, Cox J, William T, Jelip J, Fornace KM, Brock PM, et al. Individual-level factors associated with the risk of acquiring human Plasmodium knowlesi malaria in Malaysia: a case–control study. Lancet Planet Health. 2017;1:e97–104.
William T, Menon J, Rajahram G, Chan L, Ma G, Donaldson S, et al. Severe Plasmodium knowlesi malaria in a tertiary care hospital, Sabah, Malaysia. Emerg Infect Dis. 2011;17:1248–55.
Putaporntip C, Hongsrimuang T, Seethamchai S, Kobasa T, Limkittikul K, Cui L, et al. Differential prevalence of Plasmodium infections and cryptic Plasmodium knowlesi malaria in humans in Thailand. J Infect Dis. 2009;199:1143–50.
Jongwutiwes S, Buppan P, Kosuvin R, Seethamchai S, Pattanawong U, Sirichaisinthop J, et al. Plasmodium knowlesi malaria in humans and macaques, Thailand. Emerg Infect Dis. 2011;17:1799–806.
Sermwittayawong N, Singh B, Nishibuchi M, Sawangjaroen N, Vuddhakul V. Human Plasmodium knowlesi infection in Ranong province, southwestern border of Thailand. Malar J. 2012;11:36.
Ngernna S, Rachaphaew N, Thammapalo S, Prikchoo P, Kaewnah O, Manopwisedjaroen K, et al. Case series of human Plasmodium knowlesi infection on the southern border of Thailand. Am J Trop Med Hyg. 2019;101:1397–401.
Coutrier FN, Tirta YK, Cotter C, Zarlinda I, Gonzalez IJ, Schwartz A, et al. Laboratory challenges of Plasmodium species identification in Aceh Province, Indonesia, a malaria elimination setting with newly discovered P. knowlesi. PLoS Negl Trop Dis. 2018;12:e0006924.
Ompusunggu S, Dewi RM, Yuliawaty R, Sihite BA, Ekowatiningsih R, Siswantoro H, et al. First finding of human Plasmodium knowlesi malaria cases in central Kalimantan. Bull Penelit Kesehat. 2015;43:63–76.
Herdiana H, Irnawati I, Coutrier FN, Munthe A, Mardiati M, Yuniarti T, et al. Two clusters of Plasmodium knowlesi cases in a malaria elimination area, Sabang Municipality, Aceh, Indonesia. Malar J. 2018;17:186.
Lubis IND, Wijaya H, Lubis M, Lubis CP, Divis PCS, Beshir KB, et al. Contribution of Plasmodium knowlesi to multispecies human malaria infections in North Sumatera, Indonesia. J Infect Dis. 2017;215:1148–55.
Ong CW, Lee SY, Koh WH, Ooi EE, Tambyah PA. Monkey malaria in humans: a diagnostic dilemma with conflicting laboratory data. Am J Trop Med Hyg. 2009;80:927–8.
Jeslyn WP, Huat TC, Vernon L, Irene LM, Sung LK, Jarrod LP, et al. Molecular epidemiological investigation of Plasmodium knowlesi in humans and macaques in Singapore. Vector Borne Zoonotic Dis. 2011;11:131–5.
Koh GJ, Ismail PK, Koh D. Occupationally acquired Plasmodium knowlesi malaria in Brunei Darussalam. Saf Health Work. 2019;10:122–4.
Imwong M, Madmanee W, Suwannasin K, Kunasol C, Peto TJ, Tripura R, et al. Asymptomatic natural human infections with the simian malaria parasites Plasmodium cynomolgi and Plasmodium knowlesi. J Infect Dis. 2019;219:695–702.
Khim N, Siv S, Kim S, Mueller T, Fleischmann E, Singh B, et al. Plasmodium knowlesi infection in humans, Cambodia, 2007–2010. Emerg Infect Dis. 2011;17:1900–2.
Pongvongsa T, Culleton R, Ha H, Thanh L, Phongmany P, Marchand RP, et al. Human infection with Plasmodium knowlesi on the Laos-Vietnam border. Trop Med Health. 2018;46:33.
Iwagami M, Nakatsu M, Khattignavong P, Soundala P, Lorphachan L, Keomalaphet S, et al. First case of human infection with Plasmodium knowlesi in Laos. PLoS Negl Trop Dis. 2018;12:e0006244.
Jiang N, Chang Q, Sun X, Lu H, Yin J, Zhang Z, et al. Co-infections with Plasmodium knowlesi and other malaria parasites, Myanmar. Emerg Infect Dis. 2010;16:1476–8.
Luchavez J, Espino F, Curameng P, Espina R, Bell D, Chiodini P, et al. Human infections with Plasmodium knowlesi, the Philippines. Emerg Infect Dis. 2008;14:811–3.
Van den Eede P, Van HN, Van Overmeir C, Vythilingam I, Duc TN, le Hung X, et al. Human Plasmodium knowlesi infections in young children in central Vietnam. Malar J. 2009;8:249.
Hoosen A, Shaw MT. Plasmodium knowlesi in a traveller returning to New Zealand. Travel Med Infect Dis. 2011;9:144–8.
Kroidl I, Seilmaier M, Berens-Riha N, Bretzel G, Wendtner C, Löscher T. Monkey malaria (Plasmodium knowlesi infection) after travelling to Thailand. Dtsch Med Wochenschr. 2015;140:815–7 (in German).
Link L, Bart A, Verhaar N, van Gool T, Pronk M, Scharnhorst V. Molecular detection of Plasmodium knowlesi in a Dutch traveler by real-time PCR. J Clin Microbiol. 2012;50:2523–4.
Mackroth MS, Tappe D, Tannich E, Addo M, Rothe C. Rapid-antigen test negative malaria in a traveler returning from Thailand, molecularly diagnosed as Plasmodium knowlesi. Open Forum Infect. 2016;3:ofw039.
Mali S, Steele S, Slutsker L, Arguin PM. Malaria surveillance—United States, 2008. MMWR. 2010;59:1–15.
Nowak SP, Zmora P, Pielok Ł, Kuszel Ł, Kierzek R, Stefaniak J, et al. Case of Plasmodium knowlesi malaria in Poland linked to travel in southeast Asia. Emerg Infect Dis. 2019;25:1772–3.
Orth H, Jensen BO, Holtfreter MC, Kocheril SJ, Mallach S, MacKenzie C, et al. Plasmodium knowlesi infection imported to Germany, January 2013. Euro Surveill. 2013;18:4–6.
Ozbilgin A, Cavus I, Yildirim A, Gunduz C. The first monkey malaria in Turkey: a case of Plasmodium knowlesi. Mikrobiyol Bul. 2016;50:484–90 (in Turkish).
Pan B, Pei FQ, Ruan CW, Lin RX, Cen YZ, Liu MR, et al. Diagnosis and treatment of the first imported case of Plasmodium knowlesi infection in China. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2016;34:513–6 (in Chinese).
Ranaweera AD, Danansuriya MN, Pahalagedera K, Gunasekera W, Dharmawardena P, Mak KW, et al. Diagnostic challenges and case management of the first imported case of Plasmodium knowlesi in Sri Lanka. Malar J. 2017;16:126.
Tang THT, Salas A, Ali-Tammam M, Del Carmen MM, Lanza M, Arroyo E, et al. First case of detection of Plasmodium knowlesi in Spain by real time PCR in a traveller from southeast Asia. Malar J. 2010;9:219.
Tanizaki R, Ujiie M, Kato Y, Iwagami M, Hashimoto A, Kutsuna S, et al. First case of Plasmodium knowlesi infection in a Japanese traveller returning from Malaysia. Malar J. 2013;12:128.
Zheng H, Zhu HM, Ning BF, Li XY. Molecular identification of naturally acquired Plasmodium knowlesi infection in a human case. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2006;24:273–6 (in Chinese).
Zhu HM, Li J, Zheng H. Human natural infection of Plasmodium knowlesi. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2006;24:70–1 (in Chinese).
Kotepui M, Kotepui KU, Milanez GD, Masangkay FR. Prevalence of severe Plasmodium knowlesi infection and risk factors related to severe complications compared with non-severe P. knowlesi and severe P. falciparum malaria: a systematic review and meta-analysis. Infect Dis Poverty. 2020;9:106.
Rajahram GS, Barber BE, William T, Menon J, Anstey NM, Yeo TW. Deaths due to Plasmodium knowlesi malaria in Sabah, Malaysia: association with reporting as Plasmodium malariae and delayed parenteral artesunate. Malar J. 2012;11:284.
Grigg MJ, William T, Barber BE, Rajahram GS, Menon J, Schimann E, et al. Age-related clinical spectrum of Plasmodium knowlesi malaria and predictors of severity. Clin Infect Dis. 2018;67:350–9.
Knowles R, Gupta BMD. A study of monkey-malaria, and its experimental transmission to man. Ind Med Gaz. 1932;67:301–20.
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097.
Yerlikaya S, Campillo A, Gonzalez IJ. A systematic review: performance of rapid diagnostic tests for the detection of Plasmodium knowlesi, Plasmodium malariae, and Plasmodium ovale monoinfections in human blood. J Infect Dis. 2018;218:265–76.
Divis PC, Shokoples SE, Singh B, Yanow SK. A TaqMan real-time PCR assay for the detection and quantitation of Plasmodium knowlesi. Malar J. 2010;9:344.
Cox-Singh J, Mahayet S, Abdullah MS, Singh B. Increased sensitivity of malaria detection by nested polymerase chain reaction using simple sampling and DNA extraction. Int J Parasitol. 1997;27:1575–7.
Lee KS, Cox-Singh J, Singh B. Morphological features and differential counts of Plasmodium knowlesi parasites in naturally acquired human infections. Malar J. 2009;8:73.
WHO. Informal consultation on the public health importance of Plasmodium knowlesi meeting report [press release]. Manila, Philippines: World Health Organization Regional Office for the Western Pacific Press; 2011.
Acknowledgements
The authors would like to thank the New Strategic Research (P2P) project, Walailak University, Thailand for providing partial fund for this study.
Funding
This research was partially supported by the New Strategic Research (P2P) project, Walailak University, Thailand. The funders had a role in the collection, analysis, and interpretation of the data.
Author information
Authors and Affiliations
Contributions
MK and AM carried out the study design, study selection, data extraction, and statistical analysis; and drafted the manuscript. FRM, KUK and GDM participated in the study selection and data extraction, and drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there is no conflict of interest regarding the publication of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Checklist S1.
PRISMA Checklist S1.
Additional file 2.
Methodological quality summary.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Mahittikorn, A., Masangkay, F.R., Kotepui, K.U. et al. Quantification of the misidentification of Plasmodium knowlesi as Plasmodium malariae by microscopy: an analysis of 1569 P. knowlesi cases. Malar J 20, 179 (2021). https://doi.org/10.1186/s12936-021-03714-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-021-03714-1