Abstract
Background
Non-human primates (NHPs) as a source for Plasmodium infections in humans are a challenge for malaria elimination. In Brazil, two species of Plasmodium have been described infecting NHPs, Plasmodium brasilianum and Plasmodium simium. Both species are infective to man. Plasmodium brasilianum resembles morphologically, genetically and immunologically the human quartan Plasmodium malariae. Plasmodium brasilianum naturally infects species of non-human primates from all New World monkey families from a large geographic area. In the family Callitrichidae only the genus Saguinus has been described infected so far. The present study describes the natural infection of P. brasilianum in tamarins and marmosets of the genera Callithrix, Mico and Leontopithecus in the Atlantic forest.
Methods
One hundred and twenty-two NHPs of the family Callitrichidae housed in the Primate Centre of Rio de Janeiro (CPRJ) were sampled in June 2015, and January and July 2016. The CPRJ is located in the Atlantic forest in the Guapimirim municipality, in the Rio de Janeiro state, where human autochthonous cases of malaria have been reported. The samples were screened for the presence of Plasmodium using optical microscopy and nested PCR for detection of 18S small subunit rRNA gene. The amplicon was sequenced to confirm the molecular diagnosis.
Results
The frequency of Plasmodium infections detected by nested PCR in New World monkeys of the family Callitrichidae was 6.6%. For the first time, Callitrichidae primates of genera Callithrix, Mico and Leontopithecus were found naturally infected with P. brasilianum. Infection was confirmed by sequencing a small fragment of 18S rRNA gene, although no parasites were detected in blood smears.
Conclusions
The reported P. brasilianum infection in NHP species maintained in captivity suggests that infection can be favoured by the presence of vectors and the proximity between known (and unknown) hosts of malaria. Thus, the list of potential malaria reservoirs needs to be further explored.
Similar content being viewed by others
Background
Many efforts are currently being undertaken towards control of malaria around the world. However, despite a 37% reduction in global malaria incidence and 60% in deaths in the last 15 years (2000–2015), the disease still remains an important public health problem in many tropical and sub-tropical countries [1]. It is a mosquito-borne disease of humans and other vertebrates caused by parasites of the genus Plasmodium. In the context of the malaria Eradication Research Agenda (malERA), non-human primates (NHPs) have increased their importance as source for Plasmodium infections in humans [2–4]. Therefore, there is now a consensus that some species of Plasmodium that typically infect NHPs, such as Plasmodium cynomolgi and Plasmodium knowlesi in Asia and Plasmodium brasilianum and Plasmodium simium in the Americas, may under special conditions infect humans [4–7].
Plasmodium brasilianum and P. simium are considered to be morphologically, genetically and immunologically highly similar to the human malaria parasites Plasmodium malariae and Plasmodium vivax, respectively [4, 8–13]. Plasmodium brasilianum was first described in Cacajao calvus (bald-uakari) from the Amazonas state in Brazil. This parasite is widely spread throughout Central and South America; it has been found in the Amazon forest of Panama, Venezuela, Colombia, French Guiana, Peru and Brazil, as well as in the Atlantic forest in the Brazilian Southern and Southeastern regions [4, 11, 14–19]. It has a remarkable plasticity, being able to infect a large number of species belonging to all families of New World monkeys, i.e., families Aotidae, Atelidae, Callitrichidae, Cebidae and Pitheciidae [11, 19–25]. Although the family Callitrichidae has six genera: Callithrix, Cebuella, Leontopithecus, Mico, Saguinus and Callibella [26, 27], P. brasilianum has been reported naturally infecting only the genus Saguinus (S. geoffroyi, S. midas and S. niger) [11, 21, 28].
Until the 1950s, malaria was a nationwide problem in Brazil. After successive and often successful campaigns, the disease was controlled in the majority of the national territory, remaining endemic only in the Amazon basin (Northern Brazil) [29, 30]. However, autochthonous cases of the disease have been described in the extra-Amazon region, most of them in the Southern and Southeastern Atlantic forest. Between 2006 and 2014 sixty-one malaria autochthonous cases were reported in Rio de Janeiro (RJ) state in the Southeastern with an average of 6.8 cases per year [31]. Between January 2015 and July 2016, 49 patients diagnosed with malaria reported the infection location as the Atlantic forest in RJ state [32]. Many studies suggest that the maintenance of autochthonous malaria in the Atlantic forest may involves the presence of infected NHPs acting as reservoirs of the disease. In these environments, malaria transmission would be carried out by the bromeliad mosquito Anopheles (Kerteszia) cruzii, the natural vector of both human and NHP malaria in the Atlantic forest [11]. Therefore, malaria in these mountain valleys may be a zoonosis [23, 33–35].
The report of autochthonous malaria cases in the RJ state has motivated the search for infected NHPs in its Atlantic forest region [35, 36]. The present study describes for the first time the natural infection of P. brasilianum in the genera Callithrix, Mico and Leontopithecus (family Callitrichidae) using molecular approaches.
Methods
Study area, ethics and monkey blood samples
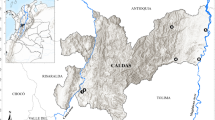
NHPs from the Primate Centre of Rio de Janeiro (CPRJ, according to its initials in Portuguese) were studied in this survey. The CPRJ, part of the Brazilian Institute of Environment and renewable natural resources (Register Number 458460), is a unit for wild monkey conservation, which occupies an area of 259.54 hectares. It is located in the municipality of Guapimirim (between latitudes 22°27′ and 22°32′ S and longitudes 42°50′ and 42°56′ W), almost in the centre of RJ state, at the Serra dos Órgãos slopes, in the Atlantic forest (Fig. 1). The study had the authorization of the Fiocruz Research Ethical Committee. The Fiocruz Ethics Committee for the Use of Laboratory Animals agreed to the protocol for sample collection. The handling of monkeys was exclusively done by CPRJ technicians.
A total of 122 NHPs of the family Callitrichidae were sampled in June 2015 (n = 4), January 2016 (n = 115) and July 2016 (n = 4). The origin of all animals, sex, species and date of sample collection are shown in Additional file 1: Table S1. Of these animals, 10 belonged to the genus Callithrix (five Geoffroy marmosets—Callithrix geoffroyi, two common marmosets—Callithrix jacchus, and three hybrids); 23 of the genus Saguinus (nine golden-handed tamarins—Saguinus midas, eight pied bare-face tamarins—S. bicolor, 3 black-handed tamarins—Saguinus niger, one ochraceous bare-face tamarin—Saguinus martinsi ochraceus, one Martin’s bare-face tamarin—Saguinus martinsi martinsi and one hybrid); 85 of the genus Leontopithecus (eight golden lion tamarins—Leontopithecus rosalia, four black lion tamarins—Leontopithecus chrysopygus and 73 golden-headed lion tamarins—Leontopithecus chrysomelas); and five belonged to the genus Mico (four black-and-white tassel-ear marmosets—Mico humeralifer and one Maués marmoset—Mico mauesi) (Additional file 1: Table S1). The species were determined by the veterinaries from CPRJ based on Rylands et al. and Van Roosmalen et al. [26, 27]. Seventy-five NHPs (61%) were captive from conservation centers (CPRJ and Renabra nursery) or Zoos (Belo Horizonte Zoo, Mario Nardelli Zoo, Niteroi Zoo and São Carlos Zoo) and 47 (39%) were wild caught in different areas, including eight in the Brazilian Amazon (Additional file 1: Table S1). It is important to highlight that the wild animals from Serra da Tiririca Park in Niteroi (RJ state) were housed in the CPRJ Centre for a short period of time before transfer to be released in conservation areas of Bahia state or other states. No animal was splenectomized at the time of sample collection. Approximately 5 mL of blood were taken in vacuum tubes containing EDTA by femoral vein puncture. Packed red blood cells were separated by centrifugation, frozen and sent to the malaria laboratory at CPqRR, Fiocruz Minas.
Parasitological and molecular tests
Thick and thin blood smears stained with Giemsa’s solution were examined under 100× optical microscopy for morphological analysis by two expert microscopists using standard methods [37].
DNA extraction from blood samples was performed using the QIAamp DNA mini Kit (Qiagen, Venlo, The Netherlands) according to manufacturer’s recommendations. The DNA was eluted in 45 μL of Buffer AE and stored at −20 °C until it was used. The samples were subjected to nested PCR using primers for identification of human Plasmodium species (P. malariae/P. brasilianum, P. vivax/P. simium and Plasmodium falciparum), targeting the small subunit of 18S ribosomal RNA (18S SSU rRNA) gene [38]. Briefly, all PCR reactions were performed in 20 μL final volume containing 250 μM each oligonucleotide primer, 10 μL of master mix (Promega) (0.3 units of Taq Polymerase, 200 μM each deoxyribonucleotide triphosphates and 1.5 mM MgCl2) and 1 μL DNA (5–10 ng). For all positive samples the DNA extraction was repeated and PCR performed using 2 μL whenever the use of 1 μL was negative. The PCR assays were performed using an automatic thermocycler (PTC-100TM v.7.0) (MJ Research Inc, USA) and the following cycling parameters were used: an initial denaturation at 95 °C for 5 min followed by 24 cycles of annealing at 58 °C for 2 min, extension at 72 °C for 2 min and denaturation at 94 °C for 1 min followed by a final annealing incubation at 58 °C for 2 min and extension at 72 °C for 5 min. The temperature was then reduced to 4 °C until the samples were removed from the cycler. The second round of PCR was performed with the same parameters of the first reaction, using 1 μL of the amplified DNA from the first reaction and 29 cycles of amplification. It is important to highlight that DNA extraction and master mix preparation were performed in “parasite DNA-free rooms” distinct from each other with different sets of pipettes and using plugged pipette tips to prevent cross-contamination. Every PCR reaction included positive controls for each pair of primers and a negative control with no DNA. The sources of genomic DNA samples that served as positive controls in the nested PCR assays were: i) DNA of P. brasilianum of MR4 (Malaria Research and Reference Reagent Resource Center—ATCC, USA); (ii) DNA of patient with high parasitaemia for P. vivax; (iii) P. falciparum DNA, strain 3D7 maintained in the malaria laboratory (CPqRR-Fiocruz Minas).
The amplified fragments were visualized using electrophoresis on 2% agarose gels in 1× TAE buffer (40 mM Tris–acetate, 1 mM EDTA) with 5 μg/mL ethidium bromide (Invitrogen) in a horizontal system (Bio-Rad) at 100 V for about 30 min. The gels were examined under a UV transilluminator and the image captured under digital system (UVP—Bio-Doc System it). Electrophoresis was performed in a room specifically assigned for analysis of amplified DNA, with appropriate sets of micropipettes and plugged pipette tips.
Sequencing of PCR-amplified DNA
Two microlitre of PCR products were amplified using 2.0 μM of forward species-specific primer and 1 μL of Big Dye terminator kit for DNA sequencing. The following cycling parameters were used: 96 °C for 1 min, 35 cycles of 96 °C for 15 s, followed by the 58 °C for 15 s and 60 °C for 15 s. The fragments were precipitated using ammonium acetate, suspended in formamide HI-DI (Applied Biosystems) and electrophoretically separated in ABI 3730 DNA automatic sequencer.
Data analysis
The sequence data were analyzed using the Blast program and the sequences aligned using the Clustal W programs in Bioedit package v7.2.5 [39].
Results
A total of 122 non-human primates of the family Callitrichidae housed in CPRJ (Fig. 1) were sampled (Additional file 1: Table S1). The NHPs belonged to different species of the genera Callithrix (n = 10), Saguinus (n = 23), Leontopithecus (n = 84) and Mico (n = 5) (Table 1). Eight samples (6.6%) were positive by nested PCR for P. brasilianum/P. malariae: three Saguinus (one Saguinus martinsi ochraceus; one Saguinus martinsi martinsi and one hybrid), three Leontopithecus (one Leontopithecus rosalia and two Leontopithecus chrysomelas), one Callithrix (Callithrix geoffroyi) and one Mico (Mico humeralifer) (Table 1). The highest PCR positivity rates were shown for the genera Mico (20%) and Saguinus (13%), while a very low frequency of infection was recorded for Leontopithecus (3.6%). It is important to highlight that some PCRs were positive only using the double amount of DNA template and in about 25% of samples the PCR showed inconsistent results (sometimes positive, sometimes negative). Negative controls never showed any amplified fragments and positive controls showed fragments with the expected sizes. Importantly, no Giemsa’s solution-stained thick or thin blood smear was positive by optical microscopy.
A species-specific fragment of 18S SSU rRNA from P. brasilianum was sequenced to confirm the infection. The sequences obtained were compared to sequences of P. brasilianum and P. malariae available at Genbank (Fig. 2). A high similarity was shown by alignment between P. brasilianum and P. malariae. In addition, two distinct variants among P. brasilianum were identified.
Multiple alignment of Plasmodium 18S SSU rRNA gene partial sequences showing two variants (V1 and V2) among Brazilian P. brasilianum. The 18S SSU rRNA gene partial sequences (second amplicon from Snounou’s PCR) obtained herein from parasites isolated from Atlantic forest NHPs: Callitrix geoffroyi (2294), Mico humeralifer (2390), Leontopithecus chrysomelas (2453 and 5427), Leontopithecus rosalia (2457) and Saguinus hybrid (2898); and from Amazonian NHPs: Saguinus martinsi martinsi (2200) and Saguinus martinsi ochraceus (2546). These sequences were compared to P. brasilianum and P. malariae sequences from GenBank (accession number)
Discussion
Plasmodium brasilianum infects a large number of species belonging to all families of New World monkeys, in contrast with the majority of other primate-infective Plasmodium species, which tend to infect hosts within the same taxonomic family [11, 19]. Within the family Callitrichidae 1193 specimens from 28 species were previously surveyed for Plasmodium infection. Only three species of Saguinus were naturally infected (Saguinus niger, Saguinus midas and Saguinus geoffroyi) (Additional file 2: Table S2). Infection of only one Saguinus geoffroyi by P. brasilianum was described in 1971 [28]. In 1985, the first description of malaria infection in Saguinus niger was reported; among 109 Callitrichidae only 4 (3.7%) were found infected by optical microscopy [21]. In 1992, a review of data from five different studies, including 509 Callitrichidae specimens from the genera Callithrix, Cebuella, Leontopithecus and Saguinus showed an infection rate of 0.8% among this family (4 Saguinus niger) also using only optical microscopy [11]. In 2000, one out of 90 Saguinus midas infected with P. brasilianum by microscopy and 3 out of 54 by PCR were reported in a survey in French Guiana [19]. Here, P. brasilianum/P. malariae DNA was identified in two new species of Saguinus: Saguinus martinsi martinsi and Saguinus martinsi ochraceus. Moreover, samples of other 3 genera: Callithrix, Leontopithecus and Mico, were found positive by PCR for P. brasilianum/P. malariae DNA. Therefore, Plasmodium DNA was detected by nested PCR in 6.6% of individuals (8/122). Considering the absence of parasite detection in blood smears, the detection limit of the nested PCR protocol used here as <3 parasites/μL blood for human samples, and PCR inconsistencies, it is suggested that the parasitaemia is even lower than expected for infected NHPs [11, 40, 41]. Four out of nine Plasmodium infection surveys in Callitrichidae were based only on microscopy [11, 21, 22, 28]. Interestingly, Deane mentioned that Prof. Mauro Barreto had already found Plasmodium in one Callithrix geoffroyi examined for Trypanosoma cruzi, however this information has never been published [11]. Corroborating the findings presented here, splenectomized common marmosets (Callithrix jacchus) were experimentally infected with P. brasilianum [42–44]. Moreover, the use of a low copy number target in the nested PCR might result in an underestimation of the infection rate, so it needs to be further investigated using more abundant genomic targets.
A previous survey of wild Leontopithecus chrysomelas from Niteroi, RJ state (n = 268) based on microscopy and PCR failed to detect Plasmodium infection [45]. This survey was conducted during animal transfer from an urban park named Serra da Tiririca (Atlantic forest fragment) in Niteroi, to Bahia state (place of natural occurrence for this species of NHP). After capture, the animals were kept in quarantine at CPRJ, where they were examined for Plasmodium infection immediately after arrival. Here were examined 31 wild animals from Niteroi that were kept longer at CPRJ and, interestingly, one of them was found infected. This infected animal had arrived at CPRJ in September 2015, and it was examined in January 2016 being positive for P. brasilianum infection. None of these animals were surveyed before. Taking into account that 100% of the wild Leontopithecus chrysomelas from Niteroi were negative in the previous very large sampling [45], it was suggested that the infected animal had acquired the infection at CPRJ during the 4 months of housing there.
Of the eight animals found infected in this study, two were wild captured in Manaus surroundings (Amazonia state) and transferred to CPRJ more than 6 years ago (Saguinus martinsi ochraceus and Saguinus martinsi martinsi) (Additional file 1: Table S1). As these animals were never tested before, it is uncertain whether they came infected from Amazonia (where human malaria is endemic in Brazil and where NHP infection is also prevalent) or they became infected at the CPRJ. Our hypothesis is that transmission occurs in the area where the animals are maintained in captivity based on: (i) the majority of infected NHPs were born at CPRJ (Additional file 1: Table S1); (ii) vectors for Plasmodium exist in the Atlantic forest area where the CPRJ is located [46–48]; (iii) many potential susceptible NHPs were housed in close proximity to animals found infected [36]; (iv) all positive animals were housed near each other and close to the forest (less than 1 km); (v) stress caused by captivity could result in increased susceptibility to infection and vulnerability to higher parasitaemias, thus favouring transmission [49]; and (vi) autochthonous cases of human malaria have been reported in this municipality [35, 37]. Therefore, in these conditions, infection could spread among NHPs, including species never found infected before as well as human beings. In agreement with this hypothesis, analysis of 18S SSU rRNA gene partial sequences, although of very short length for evolutionary analysis, identified two variants of P. brasilianum circulating among NHPs: one shared between Atlantic forest and Amazonian NHPs and a second one identified only in Atlantic forest NHPs.
Our previous identification at CPRJ of new NHP species infected by P. simium [36], a much more host restrictive simian parasite than P. brasilianum supports the results reported here. Moreover, host specificity of ape malaria parasites could also be affected by captivity, as evidenced by reported transfers of Plasmodium across chimpanzees and gorillas in Africa [50].
Conclusions
Identification of new NHP species infected with Plasmodium in captivity suggests that a much large number of potential hosts, such as tamarins and marmosets, could act as malaria reservoirs in the Atlantic forest. However, whether the DNA detected by PCR reflects red blood cells infection, as well as the role of these new potential hosts in malaria transmission in the Atlantic forest need to be further determined, including evaluation of infected vectors. Once autochthonous human cases of malaria are reported in the regions where the monkeys were found infected, the presence of potential wild reservoirs may have important implications for public health by compromising malaria control and eradication efforts. Health governmental agencies need therefore to take into account zoonotic malaria for development of appropriate control strategies in these regions.
Abbreviations
- CPRJ:
-
Primate Centre of Rio de Janeiro
- NHP:
-
non-human primate
- PCR:
-
polymerase chain reaction
- 18S SSU rRNA:
-
18S small subunit of ribosomal RNA
References
WHO. World malaria report 2015. Geneva: World Health Organization; 2015.
Alonso PL, Brown G, Arevalo-Herrera M, Binka F, Chitnis C, Collins F, et al. A research agenda to underpin malaria eradication. PLoS Med. 2011;8:e1000406.
Ramasamy R. Zoonotic malaria—global overview and research and policy needs. Front Public Health. 2014;2:123.
Lalremruata A, Magris M, Vivas-Martínez S, Koehler M, Esen M, Kempaiah P, et al. Natural infection of Plasmodium brasilianum in humans: man and monkey share quartan malaria parasites in the Venezuelan Amazon. EBioMedicine. 2015;2:1186–92.
Deane LM, Deane MP, Ferreira Neto J. Studies on transmission of simian malaria and on the natural infection of man with Plasmodium simium in Brazil. Bull World Health Organ. 1966;35:805–8.
de Arruda M, Nardin EH, Nussenzweig RS, Cochrane AH. Sero-epidemiological studies of malaria in Indian tribes and monkeys of the Amazon Basin of Brazil. Am J Trop Med Hyg. 1989;41:379–85.
Ta TH, Hisam S, Lanza M, Jiram AI, Ismail N, Rubio JM. First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar J. 2014;13:68.
Coatney GR, Collins WE, Warren M, Contacos P. The primate malarias. Washington: Gov. Print Office; 1972. p. 366.
Cochrane AH, Barnwell JW, Collins WE, Nussenzweig RS. Monoclonal antibodies produced against sporozoites of the human parasite Plasmodium malariae abolish infectivity of sporozoites of the simian parasite Plasmodium brasilianum. Infect Immun. 1985;50:58–61.
Lal AA. Circumsporozoite protein gene from Plasmodium brasilianum. Animal reservoirs for human malaria parasites? J Biol Chem. 1988;263:5495–8.
Deane LM. Simian malaria in Brazil. Mem Inst Oswaldo Cruz. 1992;87(Suppl 3):1–20.
Leclerc M, Hugot J, Durand P, Renaud F. Evolutionary relationships between 15 Plasmodium species from new and old world primates (including humans): an 18S rDNA cladistic analysis. Parasitology. 2004;129:677–84.
Duarte AM, Porto MA, Curado I, Malafronte RS, Hoffmann EH, de Oliveira SG, et al. Widespread occurrence of antibodies against circumsporozoite protein and against blood forms of Plasmodium vivax, P. falciparum and P. malariae in Brazilian wild monkeys. J Med Primatol. 2006;35:87–96.
Dunn FL, Lambrecht FL. The hosts of Plasmodium brasilianum Gonder and von Berenger-Gossler. J Parasitol. 1908;49:316–9.
Porter JA Jr, Johnson CM, De Sousa L. Prevalence of malaria in Panamanian primates. J Parasitol. 1966;52:669–70.
Serrano JA. Natural infection of a monkey, Alaouatta seniculus straminea, by Plasmodium brasilianum in Venezuela. Acta Cient Venez. 1967;18:13–5.
Marinkelle CJ, Grose ES. Plasmodium brasilianum in Colombian monkeys. Trop Geogr Med. 1968;20:276–80.
Collins WE, Skinner JC, Huong AY, Broderson JR, Sutton BB, Mehaffey P. Studies on a newly isolated strain of Plasmodium brasilianum in Aotus and Saimiri monkeys and different anophelines. J Parasitol. 1985;71:767–70.
Fandeur T, Volney B, Peneau C, De Thoisy B. Monkeys of the rainforest in French Guiana are natural reservoirs for P. brasilianum/P. malariae malaria. Parasitology. 2000;120:11–21.
Garnham PCC. Malaria parasites and other haemosporidia. Oxford: Blackwell; 1966.
De Arruda ME. Presença do Plasmodium brasilianum em macacos na área de enchimento do reservatório da usina hidrelétrica de Tucuruí. Pará. Mem Inst Oswaldo Cruz. 1985;68:367–9.
Lourenço-de-Oliveira R, Deane LM. Simian malaria at two sites in the Brazilian Amazon. I—the infection rates of Plasmodium brasilianum in non-human primates. Mem Inst Oswaldo Cruz. 1995;90:331–9.
Duarte AM, Malafronte RS, Cerutti C, Curado I, de Paiva BR, Maeda AY, et al. Natural Plasmodium infections in Brazilian wild monkeys: reservoirs for human infections? Acta Trop. 2008;107:179–85.
Araújo MS, Messias MR, Figueiró MR, Gil LH, Probst CM, Vidal NM, et al. Natural Plasmodium infection in monkeys in the state of Rondônia (Brazilian Western Amazon). Malar J. 2013;12:180.
Bueno MG, Rohe F, Kirchgatter K, Di Santi SM, Guimarães LO, Witte CL, et al. Survey of Plasmodium spp. in free-ranging neotropical primates from the Brazilian Amazon region impacted by anthropogenic actions. EcoHealth. 2013;10:48–53.
Rylands AB, Schneider H, Langguth A, Mitermeier RA, Groves CP, Rodriguez-Luna E. An assessment of the diversity of New World Primates. Neotrop Primates. 2000;8:61–93.
Van Roosmalen MGM, Van Roosmalen T. The description of a new marmoset genus, Callibella (Callitrichidae-Primates) including its molecular phylogenetic status. Neotrop Primates. 2003;11:1–10.
Baerg DC. A naturally acquired infection of Plasmodium brasilianum in the marmoset, Saguinus geoffroyi. J Parasitol. 1971;57:8.
Loiola CC, da Silva CJ, Tauil PL. Malaria control in Brazil: 1965 to 2001. Rev Panam Salud Publica. 2002;11:235–44.
Oliveira-Ferreira J, Lacerda MV, Brasil P, Ladislau JL, Tauil PL, Daniel-Ribeiro CT. Malaria in Brazil: an overview. Malar J. 2010;9:115.
Pina-Costa A. An outbreak of autochthonous malaria in Atlantic Forest areas of the Rio de Janeiro state, Brazil. 2016 (in press).
Ministerio da Saude, Serviço de vigilância em saúde, SIVEP—malária. http://200.214.130.44/sivep_malaria/.
Coatney GR. The simian malarias: zoonoses, anthroponoses, or both? Am J Trop Med Hyg. 1971;20:795–803.
Yamasaki T, Summa ME, Neves DV, de Oliveira SG, Gomes AC. Natural Plasmodium infections in Brazilian wild monkeys: reservoirs for human infections? Acta Trop. 2008;107:179–85.
Pina-Costa A, Brasil P, Santi SM, Araujo MP, Suárez-Mutis MC, Santelli AC, et al. Malaria in Brazil: what happens outside the Amazonian endemic region. Mem Inst Oswaldo Cruz. 2014;109:618–33.
Alvarenga DA, de Pina-Costa A, de Sousa TN, Pissinatti A, Zalis MG, Suarez-Mutis MC, et al. Simian malaria in the Brazilian Atlantic forest: first description of natural infection of capuchin monkeys (Cebinae subfamily) by Plasmodium simium. Malar J. 2015;14:81.
Ministério da Saúde, Serviço de Vigilância em Saúde, Brazil. Manual de diagnostico laboratorial da malaria. Brasilia/DF; 2009.
Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol. 1993;58:283–92.
BioEdit package. Carlsbad; 2013. http://www.mbio.ncsu.edu/bioedit/page2.html
Tangpukdee N, Duangdee C, Wilairatana P, Krudsood S. Malaria diagnosis: a brief review. Korean J Parasitol. 2009;47:93–102.
Costa DC, Madureira AP, Amaral LC, Sanchez BA, Gomes LT, Fontes CJ, et al. Submicroscopic malaria parasite carriage: how reproducible are polymerase chain reaction-based methods? Mem Inst Oswaldo Cruz. 2014;109:21–8.
Taliaferro WH, Taliaferro LG. Superinfection and protective experiments with Plasmodium brasilianum in monkeys. Am J Epidemiol. 1934;20:60–72.
Deane LM, Silveira LPS. Infecção experimental do sagui Callitrix jacchus pelo Plasmodium brasilianum. Rev Paul Med. 1966;68:119.
Wedderburn N, Mitchell GH, Davies DR. Plasmodium brasilianum in the common marmoset Callithrix jacchus. Parasitology. 1985;90:573–8.
Aitken EH, Bueno MG, Dos Santos Ortolan L, Alvaréz JM, Pissinatti A, Kierulff MC, et al. Survey of Plasmodium in the golden-headed lion tamarin (Leontopithecus chrysomelas) living in urban Atlantic forest in Rio de Janeiro, Brazil. Malar J. 2016;15:93.
Flores-Mendoza C, Lourenço-de-Oliveira R. Bionomics of Anopheles aquasalis Curry 1932, in Guaraí, State of Rio de Janeiro, southeastern Brazil-I. Seasonal distribution and parity rates. Mem Inst Oswaldo Cruz. 1996;91:265–70.
Azevedo AL 1997. Aspectos da epidemiologia da malária e da biologia de Anopheles (Kerteszia) cruzii Dyar & Knab em vales montanhosos do sistema de Mata Atlântica, Rio de Janeiro (RJ), MsD Thesis, Instituto Oswaldo Cruz-Fiocruz, Rio de Janeiro, p. 150.
CEPA—Centro de Estudos e Pesquisa em Antropozoonoses do Estado do Rio de Janeiro Máximo da Fonseca Filho 2009. I Encontro Macrorregional de Malária do Rio de Janeiro. http://sms.rio.rj.gov.br/coe/Apresentacoes/Malaria/I_Encontro_Macrorregional_de_Malaria_RJ_DEZ_2009.pdf.
Costa DC, da Cunha VP, de Assis GM, de Souza Junior JC, Hirano ZM, de Arruda ME, et al. Plasmodium simium/Plasmodium vivax infections in southern brown howler monkeys from the Atlantic Forest. Mem Inst Oswaldo Cruz. 2014;109:641–53.
Ngoubangoye B, Boundenga L, Arnathau C, Mombo IM, Durand P, Tsoumbou TA, et al. The host specificity of ape malaria parasites can be broken in confined environments. Int J Parasitol. 2016;46:737–44.
Authors’ contributions
CTDR, CFAB, PB, APC and DAMA conceived the idea and participated in the study design. DAMA, AP, SBM, CBJ and APC were responsible for sample collection. DAMA and CFAB carried out the experiments and analysed the data. CFAB, CTDR and DAMA drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors thank the PDTIS sequencing facilities of Fiocruz and the team of the Primate Centre of Rio de Janeiro (CPRJ/INEA). They also thank the Greater Los Angeles Zoo Association, the American Society of Primatologist, the Conservation International (CI), the Parc Zoologique et Botanique Ville de Mulhouse, France; to Jean Marc Lernould from the Zoological Society for the Conservation of Species and Populations—München Germany; and to Dr Roland Wirth and the Conservation International (CI), for cooperation in the program for reproduction of Sapajus from the Brazilian Atlantic Forest. The authors are indebted to Sidnei Silva for parasitological diagnosis assistance and to Hermano Albuquerque for drawing the map. CFAB and CTDR are supported by the CNPq (through a Research Productivity Fellowship) and CTDR also receives a fellowship from the FAPERJ, as a Cientista do Nosso Estado. APC receives a fellowship from the Coordenação de Pessoal de Ensino Superior (Capes) and DAMA is supported by a fellowship from the CPqRR.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All raw data presented in this manuscript are archived and available in the Centro de Pesquisas René Rachou.
Consent for publications
All authors and the Oswaldo Cruz Foundation provided consent to publishing this manuscript.
Funding
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG), Fundação de Amparo a Pesquisa do estado do Rio de Janeiro (FAPERJ—Proc. E-26/171.271/2006) and the Secretary for Health Surveillance (SVS) of the Brazilian Ministry of Health, through the Global Fund (Agreement IOC-005-Fio-13).
Author information
Authors and Affiliations
Corresponding author
Additional files
12936_2017_1724_MOESM2_ESM.docx
Additional file 2: Table S2. Review of description of natural infection by Plasmodium brasilianum in family Callitrichidae.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Alvarenga, D.A.M., Pina-Costa, A., Bianco, C. et al. New potential Plasmodium brasilianum hosts: tamarin and marmoset monkeys (family Callitrichidae). Malar J 16, 71 (2017). https://doi.org/10.1186/s12936-017-1724-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-017-1724-0