Abstract
Avian haemosporidians of the genera Plasmodium and Haemoproteus are a group of widely distributed blood parasites that can negatively affect the fitness of their hosts. Colombia contains the greatest diversity of birds on the planet, but knowledge about the associations between haemosporidian and its avifauna is scarce and fragmented. We collected blood samples from 255 birds (203 residents and 52 neotropical migrants) belonging to 27 families and 108 species. The study was conducted in six localities in the inter-Andean valleys of the Cauca and Magdalena rivers. Parasites of the genera Plasmodium and Haemoproteus were identified in the samples by morphological and molecular analysis of a fragment of the mitochondrial gene cyt b. Among the samples, 9.3% (n = 24) were positive for Plasmodium or Haemoproteus. Co-infection with Plasmodium and Haemoproteus was found in Red-eyed Vireo. Seventeen haemosporidian lineages were identified, five of which were reported for the first time in resident birds (Common Ground Dove, Checker-throated Stipplethroat, Tropical Kingbird, Pale-breasted Thrush, and Ruddy-breasted Seedeater) and one in the Summer Tanager (neotropical migrant). The research results confirm the wide diversity of haemosporidian present in tropical lowlands and the possible role of neotropical migratory birds in dissemination on haemosporidian along their migratory routes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Haemosporidian (Apicomplexa: Haemosporida) are a group of hemoparasites that infect a wide range of vertebrate hosts globally, including reptiles, amphibians, mammals, and birds (Ricklefs and Fallon 2002; Valkiūnas 2005; Lacorte et al. 2013; Ellis et al. 2020). Birds are susceptible to infection by mosquito-borne Plasmodium genera (Culicidae) and Haemoproteus (subgenera Parahaemoproteus and Haemoproteus), transmitted by Ceratopogonidae and Hippoboscidae flies, respectively (Valkiūnas 2005; Santiago-Alarcon et al. 2012; Toscani Field et al. 2018). Haemosporidians of the genera Plasmodium and Haemoproteus have complex life cycles with sexual reproductive stages in arthropod vectors and asexual reproduction in their avian hosts (Valkiūnas 2005). However, asexual reproduction may be disrupted in a non-competent avian host, and the mature forms of the parasite (gametocytes), which are infectious to the vector, are not generated (i.e., abortive infection) (Valkiūnas 2005; Valkiūnas and Iezhova 2017). Infections caused by haemosporidian can have adverse effects on birds’ reproductive success and survival, due to the damage caused in tissues and organs such as the spleen, liver, and brain, and consequently, affect their populations (Palinauskas et al. 2008, 2015; Risely et al. 2018).
Over the past few decades, the implementation of morphological and molecular analyses has allowed the identification of a wide diversity of avian haemosporidian morphospecies and lineages in the Neotropical region (Bensch et al. 2009; González et al. 2015; Fecchio et al. 2018; 2019; Ellis et al. 2020). In the neotropics, there have been reports of resident and migratory birds infected by the genera Plasmodium and Haemoproteus, which could play a significant role in the dispersal of haemosporidian throughout the Americas (Valkiūnas 2005; DeBrock et al. 2021; Lotta-Arévalo et al. 2023). In Colombia, 140 neotropical migratory bird species are recorded during the spring and fall migrations (Echeverry-Galvis et al. 2022). Infection by Plasmodium (Haemamoeba) cathemerium, Haemoproteus (Parahaemoproteus) vireonis, or Haemoproteus (Parahaemoproteus) coatneyi has been detected in birds of the Vireonidae, Turdidae, or Cardinalidae families (González et al. 2015; Pulgarín-R et al. 2019). In Colombia, there is a limited and fragmented knowledge about haemosporidians and their interaction with wild birds, with studies mainly conducted in localities of the Caribbean, Orinoco, and High Andean regions (González et al. 2015; Alvarez-Londoño et al. 2022; Lotta-Arévalo et al. 2023). This lack of information hinders the understanding of birds’ role in the epidemiology of Plasmodium and Haemoproteus.
In this context, and considering that the regions encompassing the inter-Andean valleys of the Cauca and Magdalena rivers are include in the migration routes of at least 83 species of neotropical migratory birds (Fierro-Calderón and Eusse 2010; Naranjo et al. 2012), this region could serve as a “transmission zone” where resident-migratory bird species and haemosporidians of South and North American origin converge (Moens and Pérez-Tris 2016; Fecchio et al. 2018). This research aimed to determine the diversity and prevalence of Plasmodium and Haemoproteus in neotropical resident and migratory wild birds at six locations in the inter-Andean valleys of the Cauca and Magdalena rivers.
Materials and methods
Study area
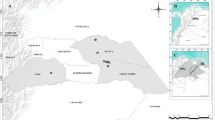
The study was conducted in six localities within the department of Caldas, Colombia (Latitude: 4.07194, Longitude: − 75.95722). The localities are situated in the inter-Andean valleys of the Cauca (Municipality of Palestina) and Magdalena rivers (Municipalities of La Dorada, Samaná and Victoria) (Fig. 1). The six localities are found within an elevation range of 126 and 1050 m a.s.l. They experience a bimodal rainfall pattern (average annual precipitation ranging from 2000 to 4000 mm). The highest rainfall occurs between March and June and between September and December. The average temperature ranges between 20 and 27 °C (Martínez-Sánchez et al. 2018). The sampled localities in the inter-Andean valley of the Magdalena River are farms dedicated to the production of avocado (Persea americana Mill.) or cocoa (Theobroma cacao L.), or buffalo rearing (Bubalus bubalis L.) and two secondary forests. The two locations that were sampled in the inter-Andean valley of the Cauca River correspond to two farms that produce cocoa (T. cacao) or citrus (Citrus spp.) production (Table 1).
Study area. A Map of Colombia depicting department of Caldas (black). B Sampled localities in Cauca River Valley, (1) Granja Luker and (2) Granja Montelindo; sampled localities in Magdalena River Valley, (3) Corregimiento Berlín, (4) Finca El Edén, (5) Hacienda Alcaparrosa, and (6) Charca de Guarinocito
Collection and processing of bird blood samples
Wild birds were captured from September 2021 to February 2022 using six mist nets (12 × 2.5 m × 36 mm) at each location for five consecutive days, with a total sampling intensity of 1980 h/net. The nets were operated between 06:00 and 17:00 h. Captured birds were marked with a small cut on the first rectrix to avoid recounts and were subsequently released at the location where they were captured (Martínez-Sánchez et al. 2018). The bird taxonomy followed the nomenclature of Remsen et al. (2023). Residency status (neotropical migratory or resident) was determined according to Echeverry-Galvis et al. (2022). A blood sample (approximately 20 to 50 μl) was obtained from each captured bird through brachial vein puncture using 25G and 27G gauge needles (Busi et al. 2020). To obtain the hummingbird blood sample, a small cut was made near the fingernail (Owen 2011). The samples were used to create blood smears (at least two per bird), fixed in absolute methanol for 5 min, and stained with 5% GIEMSA solution, pH 7.2 for 45 min (Alvarez-Londoño et al. 2022). The remaining blood was also deposited on FTA cards (Flinders Technology Associates) for subsequent molecular analysis.
Morphological analysis
For morphological identification of haemosporidians, blood smears were analyzed using a light microscope, specifically an Olympus BX43 optical microscope, with 40 × and 100 × objectives; approximately 100–150 fields were visualized per objective (Valkiūnas 2005). The parasites were identified using specialized taxonomic guides (Valkiūnas 2005; Valkiūnas and Iezhova 2018; 2022). Images of the parasites were captured using an Olympus DP28 digital camera and edited with Olympus CellSens Standard v3.22.11 software.
Molecular analysis
DNA extraction from blood samples stored on FTA cards was performed using the Wizard Genomic DNA Purification kit (Promega Corporation, Madison, USA) following the manufacturer’s instructions. To detect Plasmodium and Haemoproteus DNA, nested Polymerase Chain Reaction (PCR) testing was conducted on a fragment of a cytochrome b (cyt b) gene. The initial PCR utilized primers AE064/AE066 to amplify a 1109 bp fragment for all three genera of haemosporidians (Plasmodium, Haemoproteus, and Leucocytozoon) (Pacheco et al. 2018). One microliter of the initial PCR was utilized for the second PCR, utilizing the HaemF/HaemR2 primers that amplify a 480 bp fragment for Plasmodium and Haemoproteus (Hellgren et al. 2004). To determine Plasmodium DNA, a nested PCR was conducted using the initial PCR amplicon and primers AE983/AE985 which amplify a 580 bp fragment (Pacheco et al. 2018). Similarly, a nested PCR was performed using the initial PCR amplicon and primers AE980/AE982 to determine Haemoproteus DNA, resulting in amplification of a 346 bp fragment (Pacheco et al. 2018). Samples positive by PCR with primers AE983/AE985 and primers AE980/AE982 were considered coinfections. All PCR reactions were conducted with both a negative control (H2Odd) and positive control (Haemoproteus columbae) (Alvarez-Londoño et al. 2022). To visualize PCR products, horizontal electrophoresis was carried out using 1% agarose gels with 1X TBE buffer, stained with SYBR SAFE (Thermo Fisher Scientific, Waltham, USA), and visualized in UV photodocumenter. Positive samples for HaemF/HaemR2 were sent to Macrogen (Seoul, South Korea) for purification and Sanger sequencing. The obtained sequences were edited in the Geneious Prime program (2023.0.4. https://www.geneious.com/) and aligned in the MEGA11 software (Tamura et al. 2021). To identify Plasmodium and Haemoproteus genetic lineages, we compared our sequences with sequences deposited in the public databases MalAvi (http://130.235.244.92/Malavi/, Bensch et al. 2009) and GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequences that had at least one nucleotide difference with the MalAvi sequences were considered new lineages and were named according to the nomenclature proposed by Bensch et al. (2009).
Phylogenetic reconstruction was conducted using Bayesian inference on the identified lineages of Plasmodium and Haemoproteus. The alignments included our sequences and sequences of the cyt b gene obtained from the MalAvi database which were linked to morphospecies, and a sequence of Leucocytozoon buteonis BUTREG01 [DQ177264] was used as an outgroup. The total length of the alignments was 480 bp. The General Time Reversible model with invariant sites and gamma distribution (GTR + I + G) was selected according to the corrected Akaike information criterion using jModeltest v.2.1.6 (Darriba et al. 2012). Bayesian analysis was carried out using MrBayes v3.2.7a (Ronquist and Huelsenbeck 2003) via the CIPRESS Science Gateway v3.3 (Miller et al. 2010). Two independent Markov and Monte Carlo chains (MCMC) were used simultaneously for a total of 15 million generations, with four chains sampled every 1000 generations. Convergence was evaluated by calculating the mean and standard deviation of the frequencies divided between the two runs below 0.01 and graphically using Tracer (Rambaut and Drummond 2007). A total of 25% of the trees were discarded as run periods, and the remaining trees were used to create a consensus tree applying a 50% majority rule. The phylogeny was visualized using FigTree v1.3.1 (Rambaut 2013). All obtained sequences were deposited in MalAvi and GenBank. The percent prevalence of Plasmodium or Haemoproteus in birds was determined using the following equation [(number of infected individuals / number of individuals examined) × 100] (Bush et al. 1997). Molecularly positive samples that did not present forms of the parasite in the blood were not included in the prevalence estimation (Alvarez-Londoño et al. 2022).
Results
A total of 255 birds belonging to 108 species and 27 families (79% resident, 20% neotropical migrants, and 1% introduced) were examined (Tables 2 and 3). Twelve percent of the bird species are neotropical migrants of Tyrannidae, Vireonidae, Turdidae, Parulidae, and Cardinalidae (Tables 2 and 3). The 68.5% of the examined birds were captured in agricultural areas, while 31.5% were found in secondary forests. Parasites from the genera Plasmodium and Haemoproteus were identified in 20 passerine and non-passerine species (Table 2). The prevalence of Plasmodium was 4.3% (11/255), while the prevalence of Haemoproteus was 3.5% (9/255). Seventy-one percent of the haemosporidian-infected birds were residents, while 29% were neotropical migrants. Neotropical migratory birds Red-eyed Vireo (Vireo olivaceus), Gray-cheeked Thrush (Catharus minimus), Swainson’s Thrush (Catharus ustulatus), and Northern Waterthrush (Parkesia noveboracensis) were found infected by haemosporidians. These birds were captured during the fall migration (between October and November 2021) (Table 2). At the same time, the Summer Tanager (Piranga rubra) and Scarlet Tanager (Piranga olivacea) were captured in winter (February 2022) (Table 2). Morphologically, Plasmodium (Novyella) unalis infection (trophozoites, meronts, and gametocytes) was identified in the Pale-breasted Thrush (Turdus leucomelas) (Fig. 2) and Haemoproteus (Haemoproteus) paramultipigmentatus infection in the Common Ground Dove (Columbina passerina), Haemoproteus (Parahaemoproteus) witti in the Rufous-tailed Hummingbird (Amazilia tzacatl), Haemoproteus (Parahaemoproteus) nucleocentralis in the Ruddy-breasted Seedeater (Sporophila minuta), and Haemoproteus (Parahaemoproteus) erythrogravidus in the Blue-necked Tanager (Stilpnia cyanicollis) in the Magdalena Valley (Fig. 3). Haemoproteus (Parahaemoproteus) tyranni was detected in a Tropical Kingbird (Tyrannus melancholicus) in the Cauca River Valley (Table 2; Fig. 3).
Parasites of the genera Plasmodium found in the study. a Erythrocytic meront of Plasmodium sp. (VIOLI03) in the Red-eyed Vireo. b Macrogametocyte of Plasmodium sp. (SETAUD23) in the House Wren. c Trophozoite of Plasmodium (H.) matutinum (LINN1) in the Gray-cheeked Thrush. d Trophozoite of Plasmodium unalis (TULEU09) in the Pale-breasted Thrush. e Erythrocytic meront of Plasmodium unalis (TULEU09) in the Pale-breasted Thrush. f Microgametocyte of Plasmodium unalis (TULEU09) in the Pale-breasted Thrush. g–h Macrogametocyte of Plasmodium unalis (TULEU09) in the Pale-breasted Thrush. i Trophozoite of Plasmodium sp. (PIRUB04) in the Summer Tanager. j Macrogametocyte of Plasmodium sp. (EMBHER01) in the Sooty Ant-Tanager. k Young erythrocytic meront of Plasmodium sp. (PADOM09) in the Blue-gray Tanager. Short black arrow pigment granules. Black long arrow merozoite. Black arrowhead parasite nucleus. White long arrow vacuole. Scale bar = 10 μm
Parasites of the genera Haemoproteus found in the study. a–d Macrogametocytes of H. (H.) paramultipigmentatus in the Common Ground Dove. e, f, h Microgametocytes of H. (P.) witti in the Rufous-tailed Hummingbird. g Macrogametocyte of H. (P.) witti in Rufous-tailed Hummingbird. i–l Macrogametocytes of H. (P.) tyranni in the Tropical Kingbird. m, p Macrogametocytes of H. (P.) nucleocentralis in the Ruddy-breasted Seedeater. n, o Microgametocytes of H. (P.) nucleocentralis in the Ruddy-breasted Seedeater. q, s, t Macrogametocytes of H. (P.) erythrogravidus in the Blue-necked Tanager. r Microgametocyte of H. (P.) erythrogravidus in the Blue-necked Tanager. Black arrowhead parasite nucleus. Short black arrow pigment granules. White arrowhead protrusions of infected erythrocyte envelope. Scale bar = 10 μm
Molecularly, 17 haemosporidian lineages were determined (10 Plasmodium and seven Haemoproteus). Lineages that were previously reported in the MalAvi database of Plasmodium were recorded in the Red-eyed Vireo (VIOLI03), the House Wren (Troglodytes aedon) (SETAUD23), the Gray-cheeked Thrush (LINN1), the Swainson’s Thrush (BT7), the Scarlet Tanager (TACTHA01), the Sooty Ant-Tanager (Habia gutturalis) (EMBHER01), the Bananaquit (Coereba flaveola) (PADOM09), and the Blue-gray Tanager (Thraupis episcopus) (PADOM09). However, Haemoproteus lineages were detected in the Rufous-tailed Hummingbird (EUPMAC01), the Tropical Kingbird (MYMAC03), the Orange-billed Sparrow (Arremon aurantiirostris) (ATLBRU01), and the Blue-necked Tanager (TANCYA01) (Table 2; Fig. 4). Some of these lineages were documented for the first time in Colombia within resident or neotropical migratory birds (SETAUD23, TACTHA01, VIOLI03, EMBHER01, EUPMAC01, ATLBRU01, TANCYA01, and LINN1). Three new Plasmodium lineages were detected, PIRUB04 infecting the Summer Tanager, TULEU09 infecting the Pale-breasted Thrush, and EPIFUL01 infecting the Checker-throated Stipplethroat (Epinecrophylla fulviventris). Additionally, three new Haemoproteus lineages were found, COLPAS11 infecting the Common Ground Dove, TYRMEL03 infecting the Tropical Kingbird, and SPOMIN01 infecting the Ruddy-breasted Seedeater (Table 2; Fig. 4). Notably, the new H. (P.) nucleocentralis lineage SPOMIN01 was closely related (posterior probability = 1) to the TANDES01 lineage, with an evolutionary distance of 2.4% (Table S1, Fig. 4). The GenBank accession codes obtained for the cyt b gene in this study are [OR654036–OR654050, OR767283–OR767285, OR805347]. Likewise, the sequences of the new lineages were deposited in the MalAvi database.
Bayesian phylogeny of Haemoproteus spp. and Plasmodium spp. based on 117 lineages of cytochrome b gene. Morphological species names followed by MalAvi lineage, GenBank accession number within square brackets, and the country where the sequences are reported. The colored rectangles correspond to the geographic regions where the lineages from the MalAvi database have been found. Lineages and sequences reported in this study are shown in bold. The sequence of Leucocytozoon buteonis BUTREG01 was used as an outgroup. The scale bar represents the number of nucleotide substitutions per site. Nodal support values indicate posterior probabilities
However, no parasitic forms of Plasmodium were found in blood smears of the Northern Waterthrush, the Scarlet Tanager, and the Gray-headed Tanager (Eucometis penicillata) (Table 2). Only one individual of the Red-eyed Vireo was co-infected with Plasmodium sp. (VIOLI03) and Haemoproteus (P.) sp. (Table 2). The blood smears obtained in this study are located in the Genetics Laboratory of the Universidad de Caldas.
Discussion
The prevalence of Plasmodium spp. or Haemoproteus spp. was similar to that established in other localities in Colombia (Rodríguez and Matta 2001; Valkiūnas et al. 2003; Basto et al. 2006; Alvarez-Londoño et al. 2022). It has been suggested that the prevalence of these haemosporidians in birds seems to be determined by habitat characteristics (e.g., lentic water bodies, ponds, or forest type) that support the development, abundance, and diversity of their vectors or by ecophysiological traits of their hosts (González et al. 2015; Alvarez-Londoño et al. 2022). Particularly, one of the locations with the highest number of Plasmodium spp. infected birds was the Granja Luker. On this farm, pools are formed as a result of the crop drainage network, and there are bromeliads attached to trees that serve as habitat for mosquitoes of the genus Wyeomyia, recognized vectors of avian Plasmodium (Bensch et al. 2009; Morcillo et al. 2023).
Seventy percent of the Plasmodium lineages reported in our study have previously been detected in America, Europe, Asia, or Australia (Bensch et al. 2009; Fig. 4). However, some of these Plasmodium lineages were documented for the first time in Colombia within resident or neotropical migratory birds. In relation to this, we consider it as an extension of the geographic distribution range across the Americas for the SETAUD23 and TACTHA01 lineages of Plasmodium sp. detected in House Wren and Scarlet Tanager, respectively. Both lineages have been previously reported in North America in neotropical migratory birds wintering in Colombia (Bensch et al. 2009; Fig. 4). We recorded the geographic expansion in the South American distribution of the VIOLI03 and EMBHER01 lineages of Plasmodium sp., as well as the EUPMAC01 lineages of H. (P.) witti, ATLBRU01 of Haemoproteus sp., TANCYA01 of H. (P.) erythrogravidus. These lineages had previously been recorded only in other countries of the Americas, such as the USA, Guyana, Peru, or Brazil, in both passerine and non-passerine birds (Beadell et al. 2006; Durrant et al. 2006; Bensch et al. 2009; Fecchio et al. 2019). Likewise, we report the presence of P. (H.) matutinum LINN1 in Colombia infected to a migratory Gray-cheeked Thrush. This Plasmodium morphospecies has previously been recorded in North America, Europe, Asia, and Australia, infecting birds of various orders including Gruiformes, Charadriiformes, Strigiformes, or Passeriformes (Bensch et al. 2009; Fig. 4). In particular, P. (H.) matutinum has been observed in birds on the east coast of the USA (states of Michigan and New York) (Bensch et al. 2009). These localities are part of the migration routes of the Gray-cheeked Thrush from their breeding grounds in Asia (northeastern Siberia) and North America (from Alaska to the East Coast) (Udvardy 1994). This suggests that neotropical migratory birds may play a key role in the dispersal of parasites of the genus Plasmodium between temperate and tropical regions of the Americas (Hellgren et al. 2007; Fecchio et al. 2018). We reported six new lineages of haemosporidians infecting resident and migratory birds, of which three lineages of Haemoproteus (COLPAS11, TYRMEL03, and SPOMIN01) and one lineage of Plasmodium (TULEU09) were related to a morphospecies through morphological analysis. The TULEU09 lineage of P. (N.) unalis recorded in the Pale-breasted Thrush appears to be related to the TFUS06 lineage of P. (N.) unalis that has been previously documented in the Great Thrush (Turdus fuscater) in Colombia, as well as four other species of the genus Turdus in the Atlantic forest of Brazil (Turdus rufiventris, T. leucomelas, Turdus albicollis, and Turdus flavipes) (Mantilla et al. 2013; Tostes et al. 2018; Fig. 4). In this regard, it has been suggested that there is a high degree of intraspecific polymorphism, and a wide diversity of host species are present within this parasite species (Mantilla et al. 2013; Vanstreels et al. 2015; Tostes et al. 2018). The newly discovered EPIFUL01 lineage of Plasmodium sp. detected in the Checker-throated Stipplethroat is part of a polytomy of diverse Plasmodium morphospecies (Fig. 4). Therefore, this new lineage could be another Plasmodium species, but further studies on this host are required to confirm. The new Plasmodium sp. lineage PIRUB04 found in the Summer Tanager was related to the lineages EMBHER01 in the Sooty-ant Tanager, SETAUD23 in the House Wren, TACTHA01 in the Scarlet Tanager, and PADOM09 in the Bananaquit (evolutionary distances of < 1.7%; Table S1). The latter lineages were related to the Plasmodium (H.) cathemerium morphospecies (ZONCAP15), which was previously documented to infect the Rufous-collared Sparrow (Zonotrichia capensis) in Ecuador (Fig. 4). Moreover, the new COLPAS11 lineage of H. (H.) paramultipigmentatus was detected in Common-ground Dove in the Magdalena River Valley. This lineage grouped with the COLPAS03 and COLBUC01 lineages belonging to the morphospecies H. (H.) paramultipigmentatus and Haemoproteus (Haemoproteus) multipigmentatus, respectively (Fig. 4). These lineages are closely associated with birds from the Columbidae family, with species distributed in Mexico, Ecuador, Brazil, Venezuela, and recently reported in Colombia (Valkiūnas et al. 2010; Lotta-Arévalo et al. 2023). Another new lineage, TYRMEL03 of H. (P.) tyranni, was detected in a Tropical Kingbird in the Cauca River Valley. This lineage was associated with the MYMAC03 lineage reported in this study in another individual from the Tropical Kingbird (Fig. 4). The MYMAC03 lineage had previously been detected only through molecular methods in another member of the Tyrannidae family (Myiodynastes maculatus) in Brazil (Ferreira et al. 2017). The morphospecies H. (P.) tyranni does not have an associated lineage in the MalAvi database; however, it has a significantly higher number of records in North America (the USA and Canada), while in the rest of the Americas, it has only been previously recorded in Panama, Brazil, and Venezuela, infecting mainly birds of the family Tyrannidae (e.g., Pitangus sulphuratus and T. melancholicus) (Valkiūnas 2005; Valera et al. 2019; da Silva et al. 2022). Therefore, we consider that the record of H. (P.) tyranni increases its distribution range in South America and provides a lineage associated with this morphospecies. The new lineage SPOMIN01 of H. (P.) nucleocentralis was closely related to the lineage TANDES01 of the same morphospecies (detected only in Brazil). These two lineages have only been reported in two species of the Thraupidae family (Sporophila minuta and Tangara desmaresti) (Anjos et al. 2021). This study contributes to the knowledge of the diversity of Plasmodium and Haemoproteus lineages and species present in neotropical resident and migratory birds in tropical lowlands.
Data availability
No datasets were generated or analysed during the current study.
References
Alvarez-Londoño J, Cardona-Romero M, Martínez-Sánchez ET, Ossa-López PA, Pérez-Cárdenas JE, Gonzalez AD, Rivera-Páez FA, Castaño-Villa GJ (2022) Avian haemosporidian (Haemosporida: Plasmodium and Haemoproteus) in the department of Arauca, Colombian Orinoquia region. Parasitol Res 121:1775–1787. https://doi.org/10.1007/s00436-022-07511-w
Anjos CC, Chagas CRF, Fecchio A, Schunck F, Costa-Nascimento MJ, Monteiro EF, Mathias BS, Bell JA, Guimarães LO, Comiche KJM, Valkiūnas G, Kirchgatter K (2021) Avian malaria and related parasites from resident and migratory birds in the brazilian atlantic forest, with description of a new Haemoproteus species. Pathog 10:1–21. https://doi.org/10.3390/pathogens10020103
Basto N, Rodríguez OA, Marinkelle CJ, Gutiérrez R, Matta NE (2006) Haematozoa in birds from La Macarena National Natural Park (Colombia). Caldasia 28:371–377
Beadell JS, Ishtiaq F, Covas R, Melo M, Warren BH, Atkinson CT, Bensch S et al (2006) Global phylogeographic limits of Hawaii’s avian malaria. Proc Royal Soc: Biol Sci 273:2935–2944. https://doi.org/10.1098/rspb.2006.3671
Bensch S, Hellgren O, PÉrez -Tris J (2009) MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour 9:1353–1358. https://doi.org/10.1111/j.1755-0998.2009.02692.x
Bush AO, Lafferty KD, Lotz JM, Shostak AW (1997) Parasitology meets ecology on its own terms: Margolis et al. revisited. J Parasitol 83:575-583. https://doi.org/10.2307/3284227
Busi A, Cardona-Salazar LJ, Castillo DG, Ossa-López PA, Rivera-Páez FA, Vásquez RA, Castaño-Villa GJ (2020) Morphological differences in a population of rufous-collared sparrow (Zonotrichia capensis, statius müller, 1776) (passerine, emberizidae) at different elevations in the tropical andes. Biota Neotropica 20. https://doi.org/10.1590/1676-0611-bn-2019-0867
da Silva BR, Vanstreels RET, Serafini PP, Fontana CS, da Silva TW, Chiarani E, Carvalho AM et al (2022) Blood parasites of passerines in the Brazilian Pampas and their implications for a potential population supplementation program for the endangered Yellow Cardinal (Gubernatrix cristata). Parasitol Res 121:3203–3215. https://doi.org/10.1007/s00436-022-07638-w
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. https://doi.org/10.1038/nmeth.2109
DeBrock S, Cohen E, Balasubramanian S, Marra PP, Hamer SA (2021) Characterization of the Plasmodium and Haemoproteus parasite community in temperate-tropical birds during spring migration. Int J Parasitol: Parasites Wildl 15:12–21. https://doi.org/10.1016/j.ijppaw.2021.03.013
Durrant KL, Beadell JS, Ishtiaq F, Graves GR, Olson SL, Gering E, Peirce MA, Milensky CM, Schmidt BK, Gebhard C, Fleischer RC (2006) Avian hematozoa in south america: a comparison of temperate and tropical zones. Ornithol Monogr 60:98–111. https://doi.org/10.2307/40166831
Echeverry-Galvis MÁ, Acevedo-Charry O, Avendaño JE, Gómez C, Stiles FG, Estela FA, Cuervo AM (2022) Checklist of the birds of Colombia 2022: additions, taxonomic changes, and status update. Ornitol Colomb 2022:25–51
Ellis VA, Fecchio A, Ricklefs RE (2020) Haemosporidian parasites of Neotropical birds: causes and consequences of infection. Auk 137:1–16. https://doi.org/10.1093/auk/ukaa055
Fecchio A, Pinheiro R, Felix G, Faria IP, Pinho JB, Lacorte GA, Braga EM et al (2018) Host community similarity and geography shape the diversity and distribution of haemosporidian parasites in Amazonian birds. Ecography 41:505–515. https://doi.org/10.1111/ecog.03058
Fecchio A, Bell JA, Pinheiro RBP, Cueto VR, Gorosito CA, Lutz HL, Gaiotti MG et al (2019) Avian host composition, local speciation and dispersal drive the regional assembly of avian malaria parasites in South American birds. Mol Ecol 28:2681–2693. https://doi.org/10.1111/mec.15094
Ferreira FCJ, Rodrigues RA, Ellis VA, Leite LO, Borges MA, Braga EM (2017) Habitat modification andseasonality influence avian haemosporidian parasite distributions in southeastern Brazil. PLoSOne 12:e0178791. https://doi.org/10.1371/journal.pone.0178791
Fierro-Calderón E, Eusse D (2010) Estado de Conocimiento de las Aves en el Departamento de Caldas: Prioridades de Conservación y Vacíos de Información. Asoc Calidris: Cali, Colombia
González AD, Lotta IA, García LF, Moncada LI, Matta NE (2015) Avian haemosporidians from Neotropical highlands: evidence from morphological and molecular data. Parasitol Int 64:48–59. https://doi.org/10.1016/j.parint.2015.01.007
Hellgren O, Waldenström J, Bensch S (2004) A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J Parasitol 90:797–802. https://doi.org/10.1645/GE-184R1
Hellgren O, Waldenström J, Peréz-Tris J, Szöll Ösi E, Hasselquist D, Krizanauskiene A, Ottosson U, Bensch S (2007) Detecting shifts of transmission areas in avian blood parasites—a phylogenetic approach. Mol Ecol 16:1281–1290. https://doi.org/10.1111/j.1365-294X.2007.03227.x
Lotta-Arévalo IA, González AD, Gamboa-Suárez BA, Pacheco MA, Escalante AA, Moreno C, Rodríguez-Fandíño O, Cuervo A, Matta NE (2023) Haemosporidians in non-passerine birds of Colombia: an overview of the last 20 years of research. Diversity 15:57. https://doi.org/10.3390/d15010057
Mantilla JS, González AD, Valkiūnas G, Moncada LI, Matta NE (2013) Description and molecular characterization of Plasmodium (Novyella) unalis sp. nov. from the Great Thrush (Turdus fuscater) in highland of Colombia. Parasitol Res 112:4193–4204. https://doi.org/10.1007/s00436-013-3611-0
Martínez-Sánchez ET, Cardona-Romero M, Rivera-Páez FA, Pérez-Cárdenas JE, Castaño-Villa GJ (2018) Contribution of agroecosystems to the conservation of bird diversity in the department of Caldas. Rev Fac Nac Agron Medellín 71:8445–8457
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the gateway computing environments workshop (GCE), New Orleans, LA pp 1–8. https://doi.org/10.1109/GCE.2010.5676129
Moens MAJ, Pérez-Tris J (2016) Discovering potential sources of emerging pathogens: South America is a reservoir of generalist avian blood parasites. Int J Parasitol 46:41–49. https://doi.org/10.1016/j.ijpara.2015.08.001
Morcillo AC, Martínez-Sánchez ET, Ospina-Bautista F, González R, Rivera-Páez FA, Estévez-Varón JV (2023) Mosquitos (Diptera:Culicidae) Asociados con Tillandsia elongata (Bromeliaceae) en un Agroecosistema de los Andes Colombianos. Bol Cient Cent Mus Hist Nat 27:167–177. https://doi.org/10.17151/bccm.2023.27.1.12
Naranjo LG, Amaya JD, Eusse-González D, Cifuentes-Sarmiento Y (2012) Guía de las Especies Migratorias de la Biodiversidad en Colombia. Aves (1st edn), Ministerio de Ambiente y Desarrollo Sostenible /WWF, Bogotá
Owen JC (2011) Collecting, processing, and storing avian blood: a review. J Field Ornithol 82:339–354. http://www.jstor.org/stable/41409786. Accessed 20 Feb 2024
Pacheco MA, Cepeda AS, Bernotienė R, Lotta IA, Matta NE, Valkiūnas G, Escalante AA (2018) Primers targeting mitochondrial genes of avian haemosporidians: PCR detection and differential DNA amplification of parasites belonging to different genera. Int J Parasitol 48:657–670. https://doi.org/10.1016/j.ijpara.2018.02.003
Palinauskas V, Valkiunas G, Bolshakov CV, Bensch S (2008) Plasmodium relictum (lineage P-SGS1): effects on experimentally infected passerine birds. Exp Parasitol 120:372–380. https://doi.org/10.1016/j.exppara.2008.09.001
Palinauskas V, Žiegyte R, Ilgunas M, Iezhova TA, Bernotiene R, Bolshakov C, Valkiūnas G (2015) Description of the first cryptic avian malaria parasite, Plasmodium homocircumflexum n. sp., with experimental data on its virulence and development in avian hosts and mosquitoes. Int J Parasitol 45:51–62. https://doi.org/10.1016/j.ijpara.2014.08.012
Pulgarín-R PC, Gómez C, Bayly NJ, Bensch S, FitzGerald AM, Starkloff N, Kirchman JJ, González-Prieto AM, Hobson KA, Ungvari-Martin J, Skeen H, Castaño MI, Cadena CD (2019) Migratory birds as vehicles for parasite dispersal? Infection by avian haemosporidians over the year and throughout the range of a long-distance migrant. J Biogeogr 46:83–96. https://doi.org/10.1111/jbi.13453
Rambaut A, Drummond AJ (2007) Tracer v1.5 MCMC trace analyses tool. http://tree.bio.ed.ac.uk/software/tracer/. Accessed 10 Feb 2024
Rambaut A (2013) FigTree: tree fgure drawing too, version1.3.1. In: Institute of Evolutionary Biology, U.o.E. (ed). http://tree.bio.ed.ac.uk/software/fgtree/. Accessed 10 Feb 2024
Remsen JV, Areta JI, Bonaccorso E, Claramunt S, Jaramillo A, Pacheco JF, Robbins MB, Stiles FG, Stotz DF, Zimmer JK (2023) A classifcation of the bird species of South America. Am Ornithol Soc. http://www.museum.lsu.edu/~Remsen/SACCB aseline.htm (30 Jul. 2023)
Ricklefs RE, Fallon SM (2002) Diversification and host switching in avian malaria parasites. Proc Royal Soc: Biol Sci 269:885–892. https://doi.org/10.1098/rspb.2001.1940
Risely A, Klaassen M, Hoye BJ (2018) Migratory animals feel the cost of getting sick: a meta-analysis across species. J Anim Ecol 87:301–314. https://doi.org/10.1111/1365-2656.12766
Rodríguez OA, Matta NE (2001) Blood parasites in some birds from eastern plains of Colombia. Mem Inst Oswaldo Cruz 96:1173–1176. https://doi.org/10.1590/S0074-02762001000800026
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinform 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Santiago-Alarcon D, Palinauskas V, Schaefer HM (2012) Diptera vectors of avian Haemosporidian parasites: untangling parasite life cycles and their taxonomy. Biol Rev 87:928–964. https://doi.org/10.1111/j.1469-185X.2012.00234.x
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Toscani Field J, Weinberg J, Bensch S, Matta NE, Valkiūnas G, Sehgal RNM (2018) Delineation of the genera Haemoproteus and Plasmodium using RNA-Seq and multi-gene phylogenetics. J Mol Evol 86:646–654. https://doi.org/10.1007/s00239-018-9875-3
Tostes R, Dias RJP, De Oliveira L, Senra MVX, Massard CL, D’Agosto M (2018) Molecular and morphological characterization of a Brazilian lineage of Plasmodium (Novyella) Unalis in Turdus spp. (Passeriformes) of the Atlantic forest, with remarks on new hosts and high genetic variation. J Parasitol 104:70–78. https://doi.org/10.1645/16-189
Udvardy MD (1994) National audubon society field guide to North American Birds: Western Edition Alfred A. Knopf, New York
Valera KR, Silva-Sánchez CJ, Agelvis K, Campos H, Fajardo A, Romero-Palmera J (2019) Morphometric description of Haemoproteus (Parahaemoproteus) tyranni in Tyrannus melancholicus from Venezuela. Bol Malar Salud Ambient 59:122–129
Valkiūnas G, Iezhova TA (2017) Exo-erythrocytic development of avian malaria and related haemosporidian parasites. Malar J 16:101. https://doi.org/10.1186/s12936-017-1746-7
Valkiūnas G, Iezhova TA (2018) Keys to the avian malaria parasites. Malar J 17:212. https://doi.org/10.1186/s12936-018-2359-5
Valkiūnas G, Iezhova TA (2022) Keys to the avian Haemoproteus parasites (Haemosporida, Haemoproteidae). Malar J 21:269. https://doi.org/10.1186/s12936-022-04235-1
Valkiūnas G, Salaman P, Iezhova TA (2003) Paucity of hematozoa in Colombian birds. J Wildl Dis 39:445–448. https://doi.org/10.7589/0090-3558-39.2.445
Valkiūnas G, Santiago-Alarcon D, Levin II, Iezhova TA, Parker PG (2010) A new Haemoproteus species (Haemosporida: Haemoproteidae) from the endemic galapagos dove Zenaida galapagoensis, with remarks on the parasite distribution, vectors, and molecular diagnostics. J Parasitol 96:783–792. https://doi.org/10.1645/GE-2442.1
Valkiūnas G (2005) Avian malaria parasites and other haemosporidia, (1st edn). CRC Press. https://doi.org/10.1201/9780203643792
Vanstreels RET, Da Silva-Filho RP, Kolesnikovas CKM, Bhering RCC, Ruoppolo V, Epiphanio S, Amaku M et al (2015) Epidemiology and pathology of avian malaria in penguins undergoing rehabilitation in Brazil. Vet Res 46:30. https://doi.org/10.1186/s13567-015-0160-9
Acknowledgements
To the research groups Genética, Biodiversidad y Manejo de Ecosistemas—GEBIOME (Universidad de Caldas) and Ecosistemas Tropicales (Universidad de Caldas); and the owners of localities (Granja Montelindo, Granja Luker, Hacienda Alcaparrosa, and Finca El Edén) and their farm managers (J. Tobar, M. Salazar Velásquez, D. Gómez Londoño, Vicente, and J. A. Aguirre) in which the study was conducted.
Funding
Open Access funding provided by Colombia Consortium. This research was funded by the Vicerrectoría de Investigación y Posgrados, Universidad de Caldas-VIPUCa—Project: “Hemoparásitos asociados a aves silvestres residentes y migratorias en los valles de los ríos Magdalena y Cauca (departamento de Caldas)” [Code: 0311321] and Project: “Hemosporidios (Leucocytozoon, Plasmodium y Haemoproteus) presentes en aves silvestres residentes y migratorias en la Universidad de Caldas” [Code: 0776822].
Author information
Authors and Affiliations
Contributions
Maria Camila Hernández-Ospina: formal analysis, investigation, writing—review and editing. Diego Chitan-Guerrero: formal analysis, investigation, writing—review and editing. Johnathan Alvarez-Londoño: conceptualization, formal analysis, investigation, writing—review and editing. Mauricio Bohada-Murillo: formal analysis, investigation. Estefani T. Martínez-Sánchez: formal analysis, investigation, resources. Fredy A. Rivera-Páez: conceptualization, funding acquisition, investigation, formal analysis, writing—review. Gabriel J. Castaño-Villa: conceptualization, funding acquisition, supervision, project administration, investigation, writing—review and editing.
Corresponding author
Ethics declarations
Ethics approval
This research was conducted under the framework permit granted to the Universidad de Caldas by the Autoridad Nacional de Licencias Ambientales de Colombia (ANLA, according to resolution 00519 of March 03, 2022) and approved by the bioethics committee of the Facultad de Ciencias Agropecuarias of the Universidad de Caldas of July 28, 2021.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Section Editor: Nawal Hijjawi
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hernández-Ospina, M.C., Chitan-Guerrero, D., Alvarez-Londoño, J. et al. Avian haemosporidians of the genera Plasmodium and Haemoproteus from resident and Neotropical migrant birds in Colombia. Parasitol Res 123, 252 (2024). https://doi.org/10.1007/s00436-024-08260-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00436-024-08260-8