Abstract
It is commonly assumed that gastrointestinal cancer is the most common form of cancer across the globe and is the leading contributor to cancer-related death. The intricate mechanisms underlying the growth of GI cancers have been identified. It is worth mentioning that both non-coding RNAs (ncRNAs) and certain types of RNA, such as circular RNAs (circRNAs), long non-coding RNAs (lncRNAs), and microRNAs (miRNAs), can have considerable impact on the development of gastrointestinal (GI) cancers. As a tumour suppressor, in the group of short non-coding regulatory RNAs is miR-34a. miR-34a silences multiple proto-oncogenes at the post-transcriptional stage by targeting them, which inhibits all physiologically relevant cell proliferation pathways. However, it has been discovered that deregulation of miR-34a plays important roles in the growth of tumors and the development of cancer, including invasion, metastasis, and the tumor-associated epithelial-mesenchymal transition (EMT). Further understanding of miR-34a’s molecular pathways in cancer is also necessary for the development of precise diagnoses and effective treatments. We outlined the most recent research on miR-34a functions in GI cancers in this review. Additionally, we emphasize the significance of exosomal miR-34 in gastrointestinal cancers.
Similar content being viewed by others
Introduction
Cancer’s incidence and fatality rate have skyrocketed in recent years, making it one of the primary reasons for people’s passing away all around the globe [1]. Gastrointestinal (GI) cancers affects the GI systems, including the liver, rectum, colon, stomach, pancreas, and esophagus [2]. Both in industrialized and developing countries, gastrointestinal cancers are the most frequent form of the disease [3], In 2020, 3.5 million new instances of gastrointestinal cancer will be diagnosed, resulting in 2.2 million deaths worldwide [1]. Multiple GI malignancies are included in GLOBOCAN 2020’s 10 most common and deadly malignancies [1]. Approximately 10% of all cancer fatalities are a result of colorectal cancer. Colorectal cancer (CRC) is frequently seen in countries of the Western World. Meanwhile, both esophageal squamous cell carcinoma (ESCC) and gastric cancer (GC) are more common in East Asia; the combination of these two types of cancer is responsible for nearly 15% of all cancer fatalities worldwide. In younger individuals from industrialized nations, esophageal cancer and adenocarcinoma are on the rise, whereas rates of colorectal, pancreatic, and biliary cancer are stable or falling [1]. Some advanced gastrointestinal malignancies need systemic therapy, Molecular markers may improve patient outcomes by guiding therapy decisions [4,5,6]. MicroRNAs, often known as miRNAs, are short endogenous noncoding RNAs that range in length from 20 to 24 nucleotides that cause mRNA cleavage or impede coding for proteins by Watson-Crick base combining with target 3′-UTRs [7, 8]. MiRNAs govern 60% of the human protein-coding transcriptome, according to some research [9]. Aberrant expression of a single miRNA affects cellular pathways to cause cancer, differentiation, apoptosis, and survival signaling [7, 8]. Multiple studies have implicated miRNAs with carcinogenesis [10]. Studies have revealed that the members of the miR-34 family, miR-34c, miR-34b, and miR-34a, can prevent the forming of tumors and spur cell death in cancer cells. Figure 1 shows the sequence alignment of the mature miR-34a, miR-34b, and miR-34c. The seed-sequences are high-lighted in bold. Although the seed region in miR-34 family is exactly the same, but because the majority of the reviewed sources are related to miR-34a, the focus of this study is on this miRNA. The important thing is that by checking the tissueatlas2 database, we will find that the expression of miR-34a is much higher in our target tissues compared to the other two members of this family, and in other words, these two members can be ignored and the observed effects related to has-miR-34a-5p (miR-34a) (https://ccb-web.cs.uni-saarland.de/tissueatlas2/patterns/hsa/mirna/hsa-miR-34b-5p/; https://ccb-web.cs.uni-saarland.de/tissueatlas2/patterns/hsa/mirna/hsa-miR-34c-5p/; https://ccb-web.cs.uni-saarland.de/tissueatlas2/patterns/hsa/mirna/hsa-miR-34a-5p/ ).
Certain gene sequences are chosen by the microRNAs, specifically those associated with the cell cycle (CDK4, CDK6, and CCND1) and those meant to stop cells from dying (BCL2, SIRT1). This selection is intentional and is to serve the functions of the microRNAs. That’s why they’re so important as tumor-suppressing miRNAs [11]. Tumor suppressor miRNAs play a crucial role in preventing the initiation of cancer by regulating the expression of genes that code for oncoproteins [12, 13]. Numerous research studies have indicated a correlation between the decrease in certain miRNAs and the advancement of cancer. Yet, only specific tumor suppressor miRNAs that are frequently diminished in cancerous cells are responsible for causing a strong effect. This phenomenon has been observed in multiple miRNA families that have gained significant recognition, such as the well-known tumor-suppressing let-7, miR-15/16, and miR-200. These miRNAs act as suppressors of tumors by silencing their targeted oncogenic mRNA network, ultimately hindering the process of tumorigenesis. MiR-34 relatives (miR-34a, miR-34b/c) are spread out over two different chromosomal locations [11], This suggests they may be controlled by somewhat distinct transcriptional and epigenetic processes. P53 directly regulates miR-34a, and CpG methylation deactivate it in many malignancies [14, 15]. . One kind of miRNA is called microRNA-34a (miR-34a) that is shown get aberrant expression in many types of cancer, including gastric cancer (GC) [16], esophageal cancer (EC) [17], hepatocellular carcinoma (HCC) [18], colorectal cancer (CRC) [19], pancreatic cancer (PC) [20], gallbladder cancer (GBC) [21], and others [22]. Fresh research suggests a link between miR-34a and the development of gastrointestinal cancer [23], invasion [24] and metastasis [25], showing that miR-34a plays crucial physiologic functions in cellular signaling networks such MAPK/Ras [26], Wnt/-Catenin [27], PI3K/Akt [28], SIRT1/p53 [29], and FoxM1/c-Myc pathway [30].
Some classes of miRNAs function as Cancer-fighting genes also may block carcinogenesis by Reduce oncogenes, suggesting that miRNAs is promising for potential use in cancer therapy as a diverse disease [31]. miRNAs have a synergistic therapeutic impact in cancer because they may target numerous oncogenes or biological pathways all at once [32]. Additionally, in comparison to plasmid DNA-based gene therapy and protein-based pharmaceuticals, miRNAs as exogenous antisense nucleotides shown much less immunotoxicity [33]. Accordingly, Cancer therapy may benefit greatly from the study of microRNAs. It’s well-known that miRNAs are known to affect prognosis, diagnosis, therapy, and medication resistance of GI cancer [34,35,36,37,38,39]. We outlined the most recent research on miR-34a functions in GI cancers in this review. Additionally, we emphasize the significance of exosomal miR-34 in gastrointestinal cancers.
MircroRNA-34 and pancreatic cancer
Pancreatic ductal adenocarcinoma (PDAC) has been revealed to contain approximately 70 distinct miRNAs that are aberrantly expressed, influencing metastasis, proliferation, EMT, metabolism, and apoptosis. Tp53 gene dysfunction and an active mutation of the oncogene Kras are the most major genetic changes driving PDAC. Mir34a may be of utmost importance in the control of pancreatic ductal adenocarcinoma (PDAC) as it is specifically targeted by the Tp53 protein (Fig. 2) [40]. 90% of pancreatic ductal adenocarcinoma (PDAC) cases are caused by a mutation in the KRAS oncogene that turns the gene permanently on [41]; G12D substitution accounts for 41%, G12V for 34%, and G12R for 16% of these occurrences, respectively [42]. When KrasG12D is expressed in the epithelium, paracrine signaling is initiated. All lesion types that are generated by the normal exocrine pancreas, including pancreatic ductal adenocarcinoma, ductal metaplasia, and pancreatic intraepithelial neoplasia, are due to a fibro-inflammatory setting induced by desmoplasia associated with pancreatic ductal adenocarcinoma [43]. Progression of the illness is also fueled by the inability of tumor suppressor genes including p53, p16, and SMAD4 to operate [44].
The cycle of interactions involving the TP53 gene that regulates miR-34a levels. This figure adapted from [79]
An in vivo analysis of Mir34a’s tumor suppressive effect in PDAC utilizing genetically altered mice models is presented by Hidalgo-Sastre et al. [45]. Mice with a mutation in the oncogene Kras (KrasG12D; Mir34a/) in the pancreas were bred to test the idea that Mir34a performs inhibitor of PDAC development in vivo. Compared to KrasG12D controls, KrasG12D; Mir34a/ mice developed PDAC more quickly and had more advanced pre-neoplastic lesions, as shown by histological investigation. An investigation revealed that the increased expression of the pro-inflammatory cytokines TNFA and IL6 in regular acinar cells stimulates the accelerated phenotype, possibly caused by the migration of immune cells. Their research indicates that Mir34a suppresses PDAC growth through controlling the immunological environment of the tumor, suggesting that reactivating Mir34a could be a helpful therapeutic strategy for slowing the disease’s progression [45].
The location of the SERPINE1 gene, which codes for a 50 kDa single-chain glycoprotein, has been identified at 7q21.2-q22. One of the significant parts of the plasminogen activator system is the SERPINE1 gene (PAs). This gene is responsible for delaying the activation of plasminogen into the protease enzyme plasmin by suppressing the action of tissue-type plasminogen activator (tPA) and urokinase-type plasminogen activator (uPA) [46]. The SERPINE1 gene encodes Plasminogen activator inhibitor-1 (PAI-1), which contributes to the control of the migration and the penetration of tumor cells [47, 48]. There are two possible states for the PAI-1 protein: an active and a dormant one. The conformation of PAI-1 is significant because it activates Multiple cell signaling pathways contribute to malignant progression [49]. SERPINE1 has been identified as a miR-34a target in colorectal [50] and non-small cell lung cancer by prior studies [51].
To identify WT-primary TP53’s targets in PDAC, Akula et al. investigated the expression in MIA-PaCa-2 cells of 440 PDAC-derived proteins [49]. Due to the TP53 and KRAS mutations in MIA-PaCa-2 cells, PDAC can be effectively researched in vitro utilizing these cells. In cells lacking wild-type TP53, RPPA detected the synthesis of tumor-promoting proteins. By utilizing a vector to express Wild-Type TP53 (WT-TP53) or through administration of berberine or a modified form of berberine (BBR), these impaired states were significantly ameliorated. When compared to cells expressing mutated TP53 (mTP53), the cells expressing WT-TP53 had higher expression of the miR-34a-associated signaling. Their in vivo research, examining human PDAC specimens, miR-34a and related signaling was reduced considerably in the pancreatic cancer tissues compared to the non-cancerous specimens, which is consistent with the findings from their in vitro analysis. Their research also indicated that SERPINE1 is a miR-34a target that plays a crucial role in PDAC biology. By uncovering the key target (TP53/miR-34a axis), they created a useful biomarker (SERPINE1) to aid in the early detection of pancreatic cancer [49].
Previous investigations have revealed a relationship between Notch signaling and the epithelial-mesenchymal transition (EMT) and its relationship to the origination and evolution of pancreatic cancer [52,53,54]. EMT is a program that can be reversed and is characterized by a process wherein epithelial cells become mesenchymal cells [55]. In addition to its involvement in tissue regeneration, cancer metastasis and embryonic development all rely on EMT. E-cadherin protein is lost during the EMT program, and N-cadherin, vimentin, and fibronectin are obtained as mesenchymal markers instead [52]. Several signaling pathways, including as Notch and TGF-, contribute to EMT (transforming growth factor-). Recent research has revealed that Notch signaling and EMT share a number of overlapping roles in the control of cell viability [56, 57]. Snail and ZEB, transcription factors that down regulate E-cadherin and upregulate N-cadherin, respectively, are involved in TGF-mediated EMT [58, 59]. EMT and Notch signaling regulate pancreatic cancer therapy.
Tang et al. wanted to know if miR-34a increases the risk of pancreatic cancer and whether this increase is linked to Notch signaling and EMT [60]. Pancreatic cancer cells’ invasion and migration were inhibited by miR-34a in vitro, but were increased by miR-34a inhibitors (PANC-1 and SW-1990). These effects could be overcome by Snail1 overexpression or shRNA. Notch1 shRNA decreased MiR-34a’s anti-apoptotic effects on pancreatic cancer cells. MiR-34a targets Snail1 and Notch1, according to luciferase reporter assays. MiR-34a inhibited pancreatic cancer growth in vivo by reducing the expression of Snail1 and Notch1. The expression of Snail1 and Notch1 is regulated by MiR-34a, which inhibits the spread of pancreatic cancer [60].
For the X chromosome to become inactive in female mammals, an 18 kb gene-silencing long non-coding RNA called X inactive-specific transcript (XIST) is required [61]. LncRNAs and miRNAs interact in many forms of cancer, according to research [62,63,64,65], and so add additional mysteries to the regulatory networks of miRNAs and lncRNAs.
Sun et al. looked into how pancreatic cancer (PC) growth and progression are impacted by the XIST [66]. They first noticed that PC tissues and PC cell lines both had much higher levels of the lncRNA XIST. PANC-1 cells’ migration, invasion, and proliferation were significantly reduced when XIST was knocked down, and their rate of apoptosis was accelerated. In contrast, BxPC-3 cells’ expression of XIST was significantly boosted. BxPC-3 and PANC-1 cells were subcutaneously injected into nude mice to assess tumor development. The cells had been transfected with various vectors. Studies conducted in vivo proved that XIST promoted tumor growth. Another research revealed that low miR-34a-5p (miR34a-5p) levels in PC tissues were indicative of a poor prognosis. miR-34a-5p was also demonstrated highly negatively linked with XIST, indicating that it might be an XIST target gene. Finally, they discovered that miR-34a-5p counteracted XIST’s ability to promote malignant behavior. XIST and miR-34a-5p were identified as possible therapeutic targets for PC based on these findings [66].
miRNAs may negatively impact critical genes that are involved in stem cell self-renewal and survival, according to mounting evidence [67,68,69,70]. Recent research has shown that the three miR-34 family members are directly regulated by p53, and miR-34’s functional activity has been linked to tumor-suppressive qualities [14, 71,72,73,74,75]. miR-34 has been observed to have direct control on the Bcl-2 protein [76]. The results of He and his colleagues showed that as a result of creating abnormal levels of miR-34, cell cycle progression was halted in cells from both healthy and cancerous tissue, which confirms previous research that miR-34 is capable of reducing the activity of genes that move the cell cycle forward [14]. According to a study, MiR-34 expression was markedly down regulated in p53-mutant gastric cancer cells, and boosting miR-34 expression prevented the cancer cells from proliferating [72]. . MiR-34 loss causes cancer chemoresistance [77]. miR-34a is implicated in colon and pancreatic cancer p53-mediated apoptosis [73, 74].
Ji et al. looked into the connection between miR-34 and pancreatic cancer stem cells by investigating the part miR-34 plays in the human pancreas cancer cell lines MiaPaCa2 and BxPC3 which have mutated p53 [78]. Upon reintroduction of miR-34 into pancreatic cancer cells through either using miR-34 mimics to transfect or through lentiviral MiR-34-MIF infection, a decrease of Bcl-2 and Notch1/2 occurred. This subsequently led to restricted G1 and G2/M cell cycle, increased susceptibility to chemotherapy, and decreased growth and invasion of clonogenic cancer cells. In CD44+/CD133 + MiaPaCa2 cells, a decrease in miR-34 presence was combined with a rise in Notch/Bcl-2 and a higher concentration of tumor stem/progenitor cells, tumor sphere-forming cells, and tumor initiating cells. MiR-34 restoration greatly reduced both the formation of tumorspheres in vitro and the growth of tumors in vivo, as well as the amount of tumor-initiating cells (by 87%). Our findings suggest that miR-34 may partially restore the tumor-suppressive properties of p53-deficient human pancreatic cancer cells. Their findings are consistent with the theory that miR-34 controls the capacity of pancreatic cancer stem cells to self-renew and/or decide their own fate, presumably by directly influencing their targets Bcl-2 and Notch. P53-miR34-deficient human pancreatic cancer may benefit greatly from miR-34 recovery as a new molecular therapeutic, presumably through suppressing pancreatic cancer stem cells [78]. Table 1 presents information detailing various research conducted on the topic of miR-34 and its relation to pancreatic cancer.
MircroRNA-34 and gastric cancer
The TP53 gene on chromosome 17p13.1 encodes the 393 amino - acids long nuclear p53 protein, which is essential for maintaining genome integrity [89]. Research has demonstrated that the transcription factor known as p53 has a role in the apoptotic process by targeting many components [90]. New research has shown that p53 contributes to carcinogenesis by modulating miRNAs in yet another mechanism [91,92,93]. Corney et al. (2007) found miR-34c and miR-34b jointly target p53, purposely decrease cell division and development without adhesion. Expression of miR-34b/c was also upregulated because p53 binds to its promoter In humans, a non-synonymous change at codon 72 of the p53 protein causes a proline to be replaced by arginine (Arg72Pro, rs1042522) [94]. The biological features of the Arg72Pro polymorphism are different, with the 72Pro variation displaying a lower risk for apoptosis than the 72Arg variant [95]. In light of this, research shows looked at the connection among the Arg72Pro polymorphism also different forms of malignancy, including gastric cancer [96,97,98]. The promoter of pri-miR-34b/c contains the polymorphism rs4938723T/C [99]. It has been demonstrated that GATA-X binding is impacted by changing the rs4938723 polymorphism from T to C and, in turn, promoter transcription activity [99,100,101]. Hepatocellular carcinoma is only one of several cancers linked to the polymorphism rs4938723 in miR-34b/c, which has generated a lot of research [99, 102, 103], colorectal cancer [103, 104], nasopharyngeal carcinoma [104], esophageal squamous cell carcinoma [105, 106], osteosarcoma [106] and renal cell cancer [107].
Pan et al. looked at the connection between a polymorphism in TP53 (Arg72Pro) and a variant in miR-34b/c (rs4938723), both of which connected to a higher risk of GC [108]. They examined the frequency of such two polymorphisms in a population of 289 controls and 197 GC patients who shared the same gender, age, geographic location, and ethnicity. They carried out this utilizing DNA direct sequencing, length of restriction fragment polymorphism, and the polymerase chain reaction. miR-34b/c rs4938723 genotype CT or CT/CC individuals had significantly lower odds of developing GC than TT individuals (OR = 0.66; 95% CI, 0.45–0.97; and OR = 0.67; 0.47–0.97). It was discovered that those possessing the miR-34b/c rs4938723 CT/CC and TP53 CG/CC components displayed a 0.62-fold lesser chance to become afflicted by stomach cancer in comparison to people with miR-34b/c rs4938723 TT and TP53 CG/CC genotypes, implying that miR-34b/c rs4938723 could possibly stop the onset of gastric cancer [108].
IGF2BP3, also known as IMP3, a protein that bind mRNA for insulin-like growth factor 2. The elevated levels of IGF2 binding protein 3 in pancreatic cancer led to its discovery [109]. Soon after its discovery, IGF2BP3’s role as an overexpressed family member in tumors like squamous cell carcinoma was clarified [110], lung cancer [111], melanoma [112], colon cancer [113], liver cancer [114]. This may have an oncogenic function in cancer, since its expression was abnormally upregulated. Additionally, growing data suggested that IGF2BP3 was a useful biomarker for several malignancies, including colon cancer [115] and GC [116].
Zhou et al.; studied IGF2BP2’s function in stomach cancer [117]. They employed real-time polymerase chain reaction (qRT-PCR) and Western blotting to analyze IGF2BP3’s expression in both GC cell lines and original tissues, and they utilized a series of in vitro functional experiments to measure IGF2BP3’s biological activity. Using luciferase assays and rescue studies, TargetScan predicted that it was regulated by miRNAs, and those predictions were verified. After doing an expression microarray study on GC cell lines, IGF2BP3 was shown up-regulated gene. IGF2BP3 expressed more in GC tissues than in normal gastric epithelium. Poor disease-specific survival was associated with increased IGF2BP3 expression. Removal of IGF2BP3 dramatically reduced cell invasion and proliferation. IGF2BP3 is known to be suppressed by tumor-suppressive miRNA, particularly miR-34a, in addition to copy number growth. Investigators found that primary gastric cancer (GC) samples had a negative correlation between IGF2BP3 mRNA expression and miR-34a expression. Augmenting expression of IGF2BP3 nullified the inhibitory consequences of miR-34a. They briefly explained its carcinogenic importance and demonstrated with certainty how the silencing of miR-34a leads to the activation of IGF2BP3 in gastric carcinogenesis. Their research uncovered a promising predictive biomarker with use in the clinic [118]. .
Chemotherapy is a key method for treating gastric cancer, however it is undermined by multidrug resistance (MDR) GC [119]. In a wide range of malignancies, miRNA abnormalities are crucial for the emergence and maintenance of MDR. The evolution of GC and MDR has been linked to the histone deacetylase Sirtuin 1 (SIRT1) [120, 121]. SIRT1 may aid in the creation of MDR by regulating the amount of specific proteins, including P-gp and MRP1, responsible for resistance [122, 123]. At SCG-7901 cells, miR-34a was found to decrease SIRT1 expression [124]. Moreover, a variety of malignancies’ start and progression are significantly influenced by hsa-miR-34a-5p [125,126,127], and chemoresistance influence them too [127,128,129].
Deng et al. conducted an examination of the expression levels and actions of hsa-miR-34a-5p and SIRT1 to better understand how they work together to modulate the MDR and reaction to chemotherapy of GC cells [130]. Results of studies on the effects of SGC-7901/5-Fu on multi-drug resistant GC cells showed that the concentration of hsa-miR-34a-5p was less in comparison to normal SGC-7901 cells. When the cells were exposed to chemotherapy, there was an increased expression of hsa-miR-34a-5p which caused apoptosis, and consequently prohibited the migration and invasion of drug-resistant GC cells. The amplified hsa-miR-34a-5p was also found to impede the growth of a drug-resistant tumor in vivo. Speaking to how hsa-miR-34a-5p achieves its purpose, it is suspected that it binds with the target genes contained in the 3’-untranslated region (UTR). In the course of further investigations, target genes involved in the phenomenon, such as MRP1, P-glycoprotein, and SIRT1, were identified. It was inferred that hsa-miR-34a-5p inhibited SIRT1, P-gp, and MRP1, thus making multi-drug resistant GC cells more susceptible to chemotherapy. Additionally, it was noted that the overexpression of hsa-miR-34a-5p may mitigate the resistance of GC cells to multiple drugs by decreasing the expression of SIRT1, P-gp, or MRP1 [130].
Tgif2 has been associated to numerous cancer types. Many ovarian cancers owe their genesis or advancement in part to TGIF2 over-expression [131]. MiR-34a might contribute controlling its expression. MiR-34a has been shown in a study to decrease osteoclastogenesis and Tgif2, hence preventing bone metastases and osteoporosis [132].
Hu et al. conducted a study to evaluate if the expression levels of Tgif2 and miR-34a have an effect on gastric cancer (GC) tissues and adjacent normal tissues. To do this, western blotting and real-time polymerase chain reaction (PCR) methods were applied. On top of this, Tgif2 siRNA or miR-34a mimic was administered to the tissues and the expression levels were tested again using western blotting and PCR. The water-soluble tetrazolium salt and an Annexin-V-FITC Apoptosis Detection Kit I were utilized to observe cell proliferation and apoptosis, respectively. The results showed that the expression of miR-34a was higher in normal tissues compared to malignant tissues, whereas the expression of Tgif2 was the opposite. When transfected with the miR-34a mimic and Tgif2 siRNA, the Tgif2 expression decreased significantly in the GC cells. It was also seen that the cell proliferation was lower and rate of apoptosis was higher in the cells exposed to both the miR-34a mimic and Tgif2 siRNA contrastive to those not exposed to them. Overall, these findings show that miR-34a may be a potential therapeutic tool for controlling tumor invasion and metastasis due to its ability to suppress Tgif2 expression in GC [133].
A protein called survivin, also referred to as baculoviral suppressor of apoptosis repeat-containing 5 (BIRC5), triggers apoptosis to prevent cells from deteriorating (IAP). Upregulation of survivin occurs in almost every human tumor because of its role in and cell cycle control [134, 135]. GC patients who had an abnormally high level of survivin expression were thought to have a better chance of survival [136, 137].
In their research, Yang et al. examined the link between perseverance of survivin and the propagation and infiltration of gastric cancer cells affected by miR-335 and miR-34a and identified a connection between the amount of miR-335 and/or miR-34a expressed and the overall survival of those afflicted with GC [138]. A trial was conducted to observe the following fifty GC patients that had been initially diagnosed between 2011 and 2015. Utilizing miRNA microarray technology, the miRNA expression pattern was investigated in eight tumors and the surrounding healthy cells. Additionally, qRT-PCR, Western blotting, and a luciferase assay were used to validate the assumed link between miR-335 and survivin mRNA. Researchers later conducted studies to understand the role of miR-335 and miR-34a on gastric cancer cell’s proliferation, apoptosis, and invasion. This was done through Transwell assay, flow cytometry, and the CCK-8 test. Furthermore, the latency between decreased miR-335 or miR-34a expression and lymph node metastasis in larger tumors was also investigated. Results showed that if miR-335 or miR-34a was absent, the prognosis was poor; and if both miRs were absent the prognosis was even worse. miR-335 works directly to suppress the synthesis of survivin by connecting to the 3’-UTR region, whereas miR-34a acts more indirectly and functions to restrict cell invasion and proliferation in addition to promoting apoptosis. However, increased expression of survivin that did not have the 3’-untranslated region was able to counteract these functions. Overall, our findings demonstrate the potential influence miR-335 and miR-34a have on restraining GC cell proliferation, apoptosis, and invasion despite previous theories [138]. Table 2 presents details of multiple investigations concerning the role of miR-34 in cases of gastric cancer.
MircroRNA-34 and colon cancer
Integral membrane protein SYT1 (synaptotagmin I) on 12q21.2 had a significant impact in synovial sarcomas. SYT1 is a candidate ring chromosome and has structural rearrangements with three separate genes [170]. Utilizing a 32 K BAC array, Nord et al. identified SYT1 as a new oncogene in glioblastoma [171]. Important fresh visions into the molecular genetic basis of colon cancer were offered by Zhu H et al., who also found that SYT1 might be used as a potential marker in the disease [172].
Hichao et al. utilized qRT-PCR to analyze the miR-34a expression in both colon cancer tissues and associated cell lines [173]. An examination was conducted to evaluate the function of miR-34a and SYT1 in the proliferation, invasion, migration, and apoptosis of cells through in vitro cell functional studies. A fluorescent protein-labeled reporter assay revealed an interaction between miR-34a and SYT1; when the levels of SYT1 were raised, the effects of miR-34a on cell growth and death were reversed, thereby confirming that the two had a causative connection. Further analysis of human colon cancer specimens uncovered that miR-34a was expressed in smaller amounts than in healthy colon mucosal epithelial cells; in addition, miR-34a upregulation hampered cell proliferation, migration, and invasion while down regulation caused cell mortality in vitro. Evidence that miR-34a directly targets the 3’ untranslated region (UTR) of SYT1 suggested that miR-34a affected SYT1 in colon cancer. When SYT1 was deactivated, execution of cell migration, invasion, and proliferation was severely restricted, along with raised cell death, indicating that SYT1 possibly serves as an oncogene in colon cancer. A significant decrease in cell proliferation and an increase in cell death were both observed when SYT1 was increased in the absence of miR-34a. Altogether, these conventions signify that miR-34a and SYT1 are essential elements for the proliferation of colon cancer [173].
The original tumor cells undergo a process known as EMT which is responsible for the further development of metastasis [174]. The production of EMT is stimulated by multiple signaling molecules which are released by tumor cells. These cytokines are able to activate receptor tyrosine kinases (RTKs) [175]. The class of RTKs which is inclusive of the PDGFR and c-Kit receptors extends to include the CSF1R, which is coded by the c-fms gene [176,177,178]. Both c-fms and v-fms have been identified as having the power to transform [179, 180]. The binding of CSF1 to its latest ligand, interleukin 34 (IL34), causes the homodimerization of CSF1R and brings about its activation [181, 182]. Subsequently, associated successive signaling pathways such as PI3K/AKT, JAK/STAT, and Ras/Raf/MAPK are initiated [182,183,184]. Targeting CSF1R in TAMs is a viable method for treating these cells since they rely on this receptor to carry out essential processes [185]. Little is known, however, about the importance of CSF1R expression in tumor cells with an epithelial background. It is noteworthy that breast cancer metastasis and progression are associated with increased expression of CSF1 and CSF1R [186]. Patients with advanced colorectal cancer have considerably increased serum CSF1 levels, which may indicate a function for CSF1R signaling in the growth of CRC [187].
Shi et al. determined that the microRNA miR-34a has a direct impact on the tyrosine kinase receptor CSF1R, but also serves as the initiator of p53 and miR-34a working together to restrict colorectal cancer [188]. The investigators developed a plan to anticipate death and group by molecular patterns in the TCGA-COAD and other three colorectal cancer cohorts using mRNA expression data. They identified and experimentally ratified targets for microRNAs and transcription factors, examining the workings that influence acquired chemoresistance, metastasis, migration, and EMT. They examined how the amount of protein made and the CpG-methylation marks on the primary human CRC tissues affected the cancer. The expression of CSF1R, CSF1, and IL34 in primary CRCs associated with a more mesenchymal-like subtype and poorer patient survival outcomes. It was determined that miR-34a and CSF1R had an inverse correlation, since miR-34a directly inhibits CSF1R. Additionally, p53 lessened the expression of CSF1R by raising miR-34a, whilst SNAIL amplified the expression by decreasing miR-34a both directly and indirectly in what can be seen as a feed-forward cycle. CSF1R is a receptor that was activated by a STAT3-mediated system to cause EMT, migration, colonization, and metastasis in CRC cells. The CpG-methylation of miR-34a increased the expression of CSF1R, which in turn generated a resistance to 5-FU in CRC cells. This process was demonstrated by an elevated expression of CSF1R from the tumor invasion edge to distant metastasis being associated with CpG-methylation of the miR-34a promoter in the primary CRC. This illustrates that the loss of miR-34a in tumor cells and its repression by CSF1R could have an effect on therapeutic and prognostic measures of colorectal cancer [188].
A specialized system of response causes the expression of antizyme; this protein functions to restrict the amount of polyamine found within a cell. When the quantity of cellular polyamine grows, the translation of antizyme mRNA transitions to a frame shift that increases the formation of a fully operational antizyme protein [189]. AZ binds to the ODC monomer, which deactivates ODC and triggers its destruction by the 26 S proteasome when ubiquitin is not present. This reduces the absorption of PA, while also accelerating its excretion. OAZ1 has had its chemical reactions thoroughly investigated. OAZ2 and OAZ3 are shown to be involved in translation when there is PA-induced frame-shifting. All these AZs bind to and inhibit ODC, which results in less PA intake [190, 191]. Li et al. planned a study to determine if miR-34a contributes to the pathophysiology of CCa MDR [192]. This study has found that cancer cells become resistant to chemotherapy when miR-34a expression is decreased. Reduced levels of miR-34a in both clinical cancer samples and in vitro-produced multidrug-resistant cancer cells were found to correlate with increased resistance to chemotherapy. As a result, functional regulation of miR-34a/OAZ2 signaling was essential for the successful treatment of cancer with chemotherapy. Multidrug-resistant cancer cells become more resistant to chemotherapy when miR-34a expression is decreased. Multidrug-resistant cancer cells from patients who are resistant to oxaliplatin treatment and in vitro-produced multidrug-resistant cancer cells were all found to have decreased miR-34a expression. Reduced miR-34a expression in these cancer cells correlated with increased resistance to chemotherapy. Functionally, miR-34a inhibition increased oxaliplatin resistance in chemosensitive HCT-8 cells while ectopic exogenous miR-34a synthesis allowed the reexposure of multidrug-resistant HCT-8/OR cells to oxaliplatin therapy. The functional regulation of miR-34a/OAZ2 signaling was essential for the successful treatment of cancer using chemotherapy. Thus, the inhibition of miR-34a/OAZ2 signaling by chemotherapeutic drugs is key to the development of MDR in cancer cells. Ultimately, the progression of antiapoptotic pathways and ATP-binding cassette (ABC) transporters connected to MDR is greatly increased as a result of the inhibition of miR-34a/OAZ2 signaling by chemotherapeutic drugs. It was shown through this study that the correct cellular response to cancer chemotherapy depends on the miR-34a/OAZ2 cascade [192].
In many cancers, PDGFRA is overexpressed and plays a crucial role in encouraging cell proliferation and distant metastasis [193,194,195,196,197].
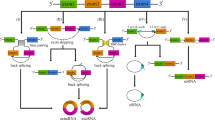
Li et al. explored the impact of miR-34a on the growth and propagation of colon cancer (Figure 3) [198]. Analysis of miR-34a expression began with studies indicating that levels of this microRNA were significantly lower in colon cancer tissues and cell lines than normal. Subsequent research then indicated that miR-34a had the capacity to induce G1 phase arrest, reduce the growth rate of colon cancer cells, restrict metastasis, and block epithelial mesenchymal transition. To further investigate these effects, bioinformatic and luciferase experiments went on to point to Platelet-derived growth factor receptor alpha (PDGFRA) as a direct target for miR-34a. Forwarding tests, involving Western blotting and quantitative reverse-transcription polymerase chain reaction analysis provided further evidence that miR-34a was instrumental in cutting the expression of PDGFRA in colon cancer cells. Functional assays verified that miR-34a exerted an inhibitory action on colon cancer by targeting PDGFRA. Taken together, this evidence implies that miR-34a has a suppressive effect on colon cancer through the regulation of PDGFRA [198]. Table 3 presents an overview of research conducted looking into the relationship between miR-34 and Colorectal Cancer (CRC).
miR-34a has an effect on the genes associated with colorectal cancer, which have been highlighted in orange. This figure adapted from [199]
MircroRNA-34 and esophageal cancer
The E2F transcription factor 5 (E2F5) has an oncogenic function in a number of cancers, suggesting that E2f family transcription factors may be an essential downstream route [221].
Jiang et al.’s research aims to investigate miR-34a and E2F5’s inhibitory impact on ESCC [222]. In order to ascertain its therapeutic value, the researchers examined the miR-34a expression in various embryonic stem cell-related cancer (ESCC) cell lines and patient malignancies. To study its function in proliferation, apoptosis and migration, RNA mimics and inhibitors were employed and gain- and loss-of-function experiments on E2F5 were conducted. It was found that ESCC inherently expressed low levels of miR-34a. Yet, it had the capability to impede cancer cell proliferation, migration and was pro-apoptotic. Furthermore, researchers confirmed that miR-34a specifically attaches to E2F5 resulting in increased proliferation, mobility and survival of tumor cells. This is in accordance with the idea that miR-34a can regulate the cell-cycle regulator E2F5, which may be of great importance in comprehending the anti-cancer properties of miR-34a [222].
FNDC3B is a big region that may be found on chromosome 3 (q26.31) (360 kb). The fibronectin family includes FNDC3B [223] has a biological role that is yet poorly understood. An increasing total of oncogenic drivers have been uncovered in the heightened region of 3q amplicon, with FNDC3B being the most recently identified crucial oncogenic driver gene [224]. MMP-2 and MMP-9 of the MMP family, often referred to as gelatinases, are essential to the process of cancer invasion and progression due to their ability to degrade the type-IV collagen making up the basement membrane. At the onset of tumor invasion, these two particular MMPs become of great significance. As the tumors grow bigger, they generate chemicals that could alter the local microenvironment and consequently, change the way the tumor behaves [225].
Yang et al. investigated how miR-34 controls the growth of ESCC [17]. To validate the presence of miR-34a in ESCC tissue samples, the reverse transcription polymerase chain reaction was employed. Transwell assays and a wound healing test were conducted to evaluate how miR-34a affects ESCC cell line migration and invasion. To further study its influence, Western blotting and luciferase reporter assays were implemented. Quantitative polymerase chain reaction revealed that ESCC tissues featured far less miR-34a than neighboring normal tissues. Additionally, overexpression of miR-34a brought a marked decrease of numerous proteins, including MMP-2, MMP-9, and FNDC3B coding and 3’-untranslated sections, in ESCC cell migration and invasion. These outcomes indicate that microRNA 34a is able to impede cell migration and invasion in ESCC by lessening the expression of MMP-2, MMP-9, and FNDC3B [17].
A novel gene subfamily within the PLC family encodes the enzyme phospholipase C epitope 1 (PLCE1) [226]. To produce 3-phosphoinositide (IP3) and diacylglycerol (DAG), phospholipase C, gamma 1 (PLCE1) hydrolyzes membrane phosphatidyl inositol 4, 5 diphosphate (PIP2), which in turn regulates metabolism, cell growth, and distinctions [227]. The significance of PLCE1 is shown with its role in a variety of important human malignant tumors, including skin cancer [228, 229], bladder [230, 231], colorectal [232, 233], neck and head [234, 235] and ESCC cancers [236, 237]. Nevertheless, PLCE1 acts as an oncogene in endometrial, urothelial, and bladder cancers while acting as a tumour suppressor in colorectal cancers (ESCCs).
Phospholipase C elipson 1 (PLCE1), a gene linked to disease development, was specifically demonstrated by Cui et al. to be expressed at higher levels in esophageal carcinomas [238]. This research demonstrated the relevance of miRNAs in managing the expression of their respective genes. They observed that tissue containing esophageal squamous cell carcinoma demonstrated a dramatic decrease in the amount of miR-34a expressed [239]. In addition, experiments were conducted on cell lines of ESCC where miR-34a was either activated or decreased to find out its role. Results indicated that PLCE1 was a direct, functional target of miR-34a; its action as a tumor suppressor was further validated since it reduced the growth rate, hampered the migration of cells and reversed the EMT characteristics of ESCC cell lines, increasing the cells’ sensitivity to apoptosis. Further research has been conducted on the relationship between PLCE1 and MiR-34a in ESCC tumors, which has increased our understanding of the tumorigenicity in both in vivo and in vitro settings. Their studies have revealed that miR-34a functions to suppress the expression of PLCE1 and as a result, inhibit the development of cancer. Because of these new findings, the miR-34a/PLCE1 axis has become a potential target for the treatment of ESCC, in addition to providing detailed insights into the molecular mechanism of ESCC metastasis [239].
MiR-34a has been shown to target cyclin D1, CDK4, CDK6, E2F3, Bcl-2, SIRT1, cyclin E2, and MYCN, enhancing G0/G1 arrest and apoptosis [14, 240, 241].
Shi et al. utilized qRT-PCR to analyze the performance and significance of miR-34a in esophageal cancer. For their research, they collected samples from both human esophageal cancer tissue and adjacent esophageal tissue [242]. . In order to examine the impact of miR-34a on human esophageal cancer cells, four separate assays were applied: the CCK8 assay, flow cytometry, in vitro migration tests, and MSP assays. It was established that miR-34a was expressed at lower levels in human esophageal cancer tissues compared to other types of cells. After the ectopic expression of miR-34a in esophageal cancer cells, cell growth and migration were suppressed. Western blotting experiments revealed that miR-34a caused levels of cyclin D1 and CDK6 to decrease. Although p53 expression was not related to miR-34a, it was demonstrated that 5-AZA-dC reduced miR-34a’s DNA methylation making it a possible mechanism of miR-34a’s inhibition of human esophageal cancer [242]. Table 4 presents various research projects that have investigated the connection between miR-34 and esophageal cancer.
Exosomal microRNA-34 and gastrointestinal cancers
Exosomes are nanoparticles that range in size from 30 to 150 nm and have been found in a number of physiological fluids [252]. Exosomes have been found to be effective carriers for transporting miRNA into cells in a recent research. Exosomes are easier to enter cells than other conventional gene carriers because their phospholipid structure is similar to that of the cell membrane [253]. Exosomes are endogenous vesicles that often do not provoke an immune response when they reach the human circulatory system [254]. Multiple studies published during the last decade demonstrate that exosomes may efficiently transport siRNA, DNA, and proteins to their intended recipient cells [255].
The study carried out by Zuo et al. explored the production of miR-34a, which was covered with exosomes, in relation to its anti-cancer effects in pancreatic cancer [256]. Using confocal microscopy and flow cytometry, exomiR-34a’s transfection efficiency was evaluated after it was synthesized using an ultrasonic technique. Real-time quantitative PCR was utilized to assess miR-34a and the gene it inhibits, Bcl-2 (qRT-PCR). To confirm the effects of exomiR-34a on pancreatic cancer, the development of the cells was monitored with the MTT assay. In addition, Western blotting and Annexin-V/PI double staining were performed to measure the apoptosis of the cells. To further investigate the anti-cancer properties of exomiR-34a, a xenograft model in naked mice containing human Panc28 pancreatic cancer cells can be utilized. The capacity to cross the cell membrane and modify the expression of genes related to apoptosis make ExomiR-34a a powerful tool in the fight against pancreatic cancer. Inhibition of Bcl-2 expression was found to cause the death of cancerous cells as well as slow the proliferation of Panc28 cells. An in vivo study using xenograft naked mice further showed that tumor creation could be greatly delayed when treated with this exomiR. The results of both in vivo and in vitro studies suggest that exomiR-34a may be an effective means of stopping the spread of pancreatic cancer. Therefore, researchers came to the conclusion that ExomiR-34a may be a highly impactful therapeutic approach for human pancreatic cancer [256].
Exosomes are desirable nano-carriers because of their biocompatibility, lack of immunogenicity, lack of toxicity, permeability through biological barriers, and stability in the bloodstream [257]. It was also shown that the exosomal payloads could differ based on the biological source of the exosomes and the local micro - environment. For instance, texosomes, a kind of tumor-derived exosomes (TEX), may convey various signaling molecules to neighboring cells, which can then impact cellular functions and the pathogenesis of cancer [258]. Hosseini et al. studied the preventive and progressive influences of the freshly devised nano-carrier composite by encapsulating miR-34a into the cancer of the colon cell line CT-26 using exosomes obtained from tumors (TEXs) [259]. Sample cells CT-26 lacking nutrients were incubated in a buffer containing CaCl2, which had been loaded with miR-34a-containing exosomes. The CT-26 cells were then differentially exposed to varying doses of either TEXs or TEX-miR-34a, and their viability was observed. The survival rate and rate of movement of CT-26 cells exposed to TEX-miR-34a and TEX were looked into. Furthermore, the expression levels of IL-6R, STAT3, PD-L1, and VEGF-A, key molecules associated with tumor formation, were examined in the treated CT-26 cells to study the effects on tumor proliferation. The results indicated that with TEX-miR-34a, the rate of apoptosis in CT-26 cells amplified in accordance with the quantity of the treatment. Furthermore, the levels of IL-6R, STAT3, PD-L1, and VEGF-A in the treated CT-26 cells were drastically decreased. The TEX-miR-34a treated cells had a weaker capacity to migrate than the untreated ones. There was a noticeable decrease in the proliferation and migration of tumor cells once they were exposed to TEXs enhanced with miR-34a. miR-34a entry into tumor cells via exosomes released by starved CT-26 cells was effectively possible, which regulates the genes involved in tumorogenesis. Despite there being no direct effect on cancer cells, TEXs have the potential to act as an adjuvant in the treatment of CRC [259]. Fibroblasts associated with cancer are a crucial element of the milieu around a tumour and are necessary for the growth and propagation of the cancer [260,261,262]. It has been seen that CAF-released exosomes are a vital molecule-carrier for intercellular communication [263,264,265]. The goal of Shi et al.‘s study was to determine how GC synthesis and associated operations are influenced by CAFs and associated exosomes [266]. The study showed that miRNA-34 concentrations were lower in GCFs and GC cell lines. When scientists increased miRNA-34’s levels in the GC lines, the ability of the cells to spread, move, and proliferate decreased. What’s more, when GCFs with higher levels of miRNA-34 were incorporated into the GC cells, aggressive growth was inhibited. Furthermore, supporting evidence for the GCFs’ roles as anti-tumor agents was found through both in vivo and in vitro trials; taking up GCF-produced exosomes via the GC cells. The exosomal miRNA-34 diminished tumor growth in vivo and impeded the GC cells’ invasion and proliferation in vitro. Finally, examination found 16 genes that had the possibility of being targeted by miRNA-34 as downstream targets. When taken together, the capacity of exosomal miRNA-34 produced by GCFs to act as a therapeutic agent for gastrointestinal cancers was revealed [266]. Table 5 lists various studies on exosomal miR-34 and GI cancers.
Conclusion
The prognosis of cancer is poor due to the spread of cancer cells. The miR-34 family has been studied for its potential to act as a tumor suppressor, slowing the malignancy’s growth and carcinogenesis. MiR-34a has been the focus of countless investigations and has been found to possess a tumor-suppressing effect. Issues such as the relatively quick degradation of miRNA and an immune response to treatments featuring the miRNA present roadblocks to its utilization. The need for nano-carriers has been established, but even then, their toxicity and the possibility for undesired side effects could prove fatal. Despite these obstacles, miR-34a remains a promising choice for cancer treatment, and the miR-34 family as a whole should continue to be further explored for its potential applications in oncotherapy.
Data availability
Not applicable.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2021;71(3):209–49.
Macha MA, Seshacharyulu P, Krishn SR, Pai P, Rachagani S, Jain M, et al. MicroRNAs (miRNAs) as biomarker(s) for prognosis and diagnosis of gastrointestinal (GI) cancers. Curr Pharm Des. 2014;20(33):5287–97.
Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol. 2019;14(1):26–38.
Di Nicolantonio F, Vitiello PP, Marsoni S, Siena S, Tabernero J, Trusolino L, et al. Precision oncology in metastatic colorectal cancer—from biology to medicine. Nat Reviews Clin Oncol. 2021;18(8):506–25.
Hosein AN, Dougan SK, Aguirre AJ, Maitra A. Translational advances in pancreatic ductal adenocarcinoma therapy. Nat cancer. 2022;3(3):272–86.
Nakamura Y, Kawazoe A, Lordick F, Janjigian YY, Shitara K. Biomarker-targeted therapies for advanced-stage gastric and gastro-oesophageal junction cancers: an emerging paradigm. Nat Reviews Clin Oncol. 2021;18(8):473–87.
Vidigal JA, Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 2015;25(3):137–47.
Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–9.
Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105.
Zadran S, Remacle F, Levine RD. miRNA and mRNA cancer signatures determined by analysis of expression levels in large cohorts of patients. Proc Natl Acad Sci U S A. 2013;110(47):19160–5.
Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17(2):193–9.
Otmani K, Lewalle P. Tumor suppressor miRNA in cancer cells and the tumor microenvironment: mechanism of deregulation and clinical implications. Front Oncol. 2021;11:708765.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97.
He L, He X, Lim LP, De Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–4.
Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7(16):2591–600.
Wu H, Huang M, Liu Y, Shu Y, Liu P. Luteolin induces apoptosis by up-regulating miR-34a in human gastric cancer cells. Technol Cancer Res Treat. 2015;14(6):747–55.
Yang L, Song X, Zhu J, Li M, Ji Y, Wu F, et al. Tumor suppressor microRNA-34a inhibits cell migration and invasion by targeting MMP-2/MMP-9/FNDC3B in esophageal squamous cell carcinoma. Int J Oncol. 2017;51(1):378–88.
Sun T-Y, Xie H-J, Li Z, Kong L-F, Gou X-N, Li D-J, et al. miR-34a regulates HDAC1 expression to affect the proliferation and apoptosis of hepatocellular carcinoma. Am J Translational Res. 2017;9(1):103.
Hiyoshi Y, Schetter AJ, Okayama H, Inamura K, Anami K, Nguyen GH, et al. Increased microRNA-34b and-34c predominantly expressed in stromal tissues is associated with poor prognosis in human colon cancer. PLoS ONE. 2015;10(4):e0124899.
Long L-M, Zhan J-K, Wang H-Q, Li S, Chen Y-Y, Liu Y-S. The clinical significance of miR-34a in pancreatic ductal carcinoma and associated molecular and cellular mechanisms. Pathobiology. 2017;84(1):38–48.
Jin K, Xiang Y, Tang J, Wu G, Li J, Xiao H, et al. miR-34 is associated with poor prognosis of patients with gallbladder cancer through regulating telomere length in tumor stem cells. Tumor Biology. 2014;35(2):1503–10.
Gao H, Zhao H, Xiang W. Expression level of human miR-34a correlates with glioma grade and prognosis. J Neurooncol. 2013;113(2):221–8.
Shi H, Zhou S, Liu J, Zhu J, Xue J, Gu L, et al. miR-34a inhibits the in vitro cell proliferation and migration in human esophageal cancer. Pathology-Research Pract. 2016;212(5):444–9.
Chu J, Li H, Xing Y, Jia J, Sheng J, Yang L, et al. LncRNA MNX1-AS1 promotes progression of esophageal squamous cell carcinoma by regulating miR-34a/SIRT1 axis. Biomed Pharmacother. 2019;116:109029.
Yoon JH, Choi WS, Kim O, Choi BJ, Nam SW, Lee JY, et al. Gastrokine 1 inhibits gastric cancer cell migration and invasion by downregulating RhoA expression. Gastric Cancer. 2017;20(2):274–85.
Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, et al. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275(1):44–53.
Cheng C, Qin Y, Zhi Q, Wang J, Qin C. Knockdown of long non-coding RNA HOTAIR inhibits cisplatin resistance of gastric cancer cells through inhibiting the PI3K/Akt and Wnt/β-catenin signaling pathways by up-regulating miR-34a. Int J Biol Macromol. 2018;107:2620–9.
Peng Y, Guo J-J, Liu Y-M, Wu X-L. MicroRNA-34A inhibits the growth, invasion and metastasis of gastric cancer by targeting PDGFR and MET expression. Biosci Rep. 2014;34(3).
Lai M, Du G, Shi R, Yao J, Yang G, Wei Y, et al. MiR-34a inhibits migration and invasion by regulating the SIRT1/p53 pathway in human SW480 cells. Mol Med Rep. 2015;11(5):3301–7.
Xu X, Chen W, Miao R, Zhou Y, Wang Z, Zhang L, et al. miR-34a induces cellular senescence via modulation of telomerase activity in human hepatocellular carcinoma by targeting FoxM1/c-Myc pathway. Oncotarget. 2015;6(6):3988.
Suárez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res. 2009;104(4):442–54.
Bhardwaj A, Singh S, Singh AP. MicroRNA-based cancer therapeutics: big hope from small RNAs. Mol Cell Pharmacol. 2010;2(5):213–9.
Catela Ivkovic T, Voss G, Cornella H, Ceder Y. microRNAs as cancer therapeutics: a step closer to clinical application. Cancer Lett. 2017;407:113–22.
Luo D, Chen J, Huang S, Xu J, Song X, Yu P. MicroRNA–18b acts as an oncogene in gastric cancer by directly targeting kruppel–like factor 6. Mol Med Rep. 2019;19(3):1926–34.
Amerizadeh F, Khazaei M, Maftouh M, Mardani R, Bahrami A. miRNA Targeting Angiogenesis as a potential Therapeutic Approach in the treatment of colorectal cancers. Curr Pharm Des. 2018;24(39):4668–74.
Herreros-Villanueva M, Duran-Sanchon S, Martín AC, Pérez-Palacios R, Vila-Navarro E, Marcuello M, et al. Plasma MicroRNA signature validation for early detection of Colorectal Cancer. Clin Transl Gastroenterol. 2019;10(1):e00003.
An X, Sarmiento C, Tan T, Zhu H. Regulation of multidrug resistance by microRNAs in anti-cancer therapy. Acta Pharm Sin B. 2017;7(1):38–51.
Gao P, He M, Zhang C, Geng C. Integrated analysis of gene expression signatures associated with colon cancer from three datasets. Gene. 2018;654:95–102.
Jin Z, Selaru FM, Cheng Y, Kan T, Agarwal R, Mori Y, et al. MicroRNA-192 and – 215 are upregulated in human gastric cancer in vivo and suppress ALCAM expression in vitro. Oncogene. 2011;30(13):1577–85.
Waters AM, Der CJ. KRAS: the critical driver and therapeutic target for pancreatic cancer. Cold Spring Harbor Perspect Med. 2018;8(9):a031435.
Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant cK-ras genes. Cell. 1988;53(4):549–54.
Waters AM, Der CJ. KRAS: the critical driver and therapeutic target for pancreatic Cancer. Cold Spring Harb Perspect Med. 2018;8(9).
Collins MA, Bednar F, Zhang Y, Brisset JC, Galbán S, Galbán CJ, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122(2):639–53.
Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin Cancer Res. 2000;6(8):2969–72.
Hidalgo-Sastre A, Lubeseder-Martellato C, Engleitner T, Steiger K, Zhong S, Desztics J, et al. Mir34a constrains pancreatic carcinogenesis. Sci Rep. 2020;10(1):9654.
Wang L, Yu J, Xu J, Zheng C, Li X, Du J. The analysis of microRNA-34 family expression in human cancer studies comparing cancer tissues with corresponding pericarcinous tissues. Gene. 2015;554(1):1–8.
Wan J, Deng D, Wang X, Wang X, Jiang S, Cui R. LINC00491 as a new molecular marker can promote the proliferation, migration and invasion of colon adenocarcinoma cells. OncoTargets Therapy. 2019;12:6471.
Saleh AD, Cheng H, Martin SE, Si H, Ormanoglu P, Carlson S, et al. Integrated genomic and functional microRNA analysis identifies mir-30-5p as a tumor suppressor and potential therapeutic nanomedicine in head and neck CancermiR-30 as tumor suppressor and therapeutic target in cancer. Clin Cancer Res. 2019;25(9):2860–73.
Declerck PJ, De Mol M, Vaughan DE, Collen D. Identification of a conformationally distinct form of plasminogen activator inhibitor-1, acting as a noninhibitory substrate for tissue-type plasminogen activator. J Biol Chem. 1992;267(17):11693–6.
Wang T, Xu H, Liu X, Chen S, Zhou Y, Zhang X. Identification of key genes in colorectal cancer regulated by miR-34a. Med Sci Monitor: Int Med J Experimental Clin Res. 2017;23:5735.
Lin X, Lin B-w, Chen X-l, Zhang B-l, Xiao X-j, Shi J-s, et al. PAI-1/PIAS3/Stat3/miR-34a forms a positive feedback loop to promote EMT-mediated metastasis through Stat3 signaling in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;493(4):1464–70.
Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–90.
Gu Y, Masiero M, Banham AH. Notch signaling: its roles and therapeutic potential in hematological malignancies. Oncotarget. 2016;7(20):29804.
Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci. 2008;105(17):6392–7.
Xin X, Lin F, Wang Q, Yin L, Mahato RI. ROS-responsive polymeric micelles for triggered simultaneous delivery of PLK1 inhibitor/miR-34a and effective synergistic therapy in pancreatic cancer. ACS Appl Mater Interfaces. 2019;11(16):14647–59.
Leong KG, Niessen K, Kulic I, Raouf A, Eaves C, Pollet I, et al. Jagged1-mediated notch activation induces epithelial-to-mesenchymal transition through slug-induced repression of E-cadherin. J Exp Med. 2007;204(12):2935–48.
Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci U S A. 2008;105(17):6392–7.
Gurzu S, Silveanu C, Fetyko A, Butiurca V, Kovacs Z, Jung I. Systematic review of the old and new concepts in the epithelial-mesenchymal transition of colorectal cancer. World J Gastroenterol. 2016;22(30):6764–75.
Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19(11):1438–49.
Tang Y, Tang Y, Cheng YS. miR-34a inhibits pancreatic cancer progression through Snail1-mediated epithelial-mesenchymal transition and the notch signaling pathway. Sci Rep. 2017;7:38232.
Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, et al. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc Natl Acad Sci U S A. 2016;113(37):10322–7.
Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21(41):11680–7.
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6(26):22513–25.
Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–8.
Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088.
Sun Z, Zhang B, Cui T. Long non-coding RNA XIST exerts oncogenic functions in pancreatic cancer via miR-34a-5p. Oncol Rep. 2018;39(4):1591–600.
Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122(1):6–7.
Hatfield S, Ruohola-Baker H. microRNA and stem cell function. Cell Tissue Res. 2008;331(1):57–66.
Papagiannakopoulos T, Kosik KS. MicroRNAs: regulators of oncogenesis and stemness. BMC Med. 2008;6(1):1–4.
Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–23.
Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proceedings of the National Academy of Sciences. 2008;105(36):13421-6.
Ji Q, Hao X, Meng Y, Zhang M, DeSano J, Fan D, et al. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8(1):1–12.
Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26(5):731–43.
Chang T-C, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–52.
Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6(13):1586–93.
Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17(15):1298–307.
Zenz T, Mohr J, Eldering E, Kater AP, Bühler A, Kienle D, et al. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood J Am Soc Hematol. 2009;113(16):3801–8.
Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS ONE. 2009;4(8):e6816.
Adams BD, Parsons C, Slack FJ. The tumor-suppressive and potential therapeutic functions of miR-34a in epithelial carcinomas. Expert Opin Ther Targets. 2016;20(6):737–53.
Tan X, Zhou L, Wang H, Yang Y, Sun Y, Wang Z, et al. Differential expression profiles of microRNAs in highly and weakly invasive/metastatic pancreatic cancer cells. Oncol Lett. 2018;16(5):6026–38.
Ma Y, Chai N, Jiang Q, Chang Z, Chai Y, Li X, et al. DNA methyltransferase mediates the hypermethylation of the microRNA 34a promoter and enhances the resistance of patient-derived pancreatic cancer cells to molecular targeting agents. Pharmacol Res. 2020;160:105071.
Terashima M, Ishimura A, Wanna-Udom S, Suzuki T. MEG8 long noncoding RNA contributes to epigenetic progression of the epithelial-mesenchymal transition of lung and pancreatic cancer cells. J Biol Chem. 2018;293(47):18016–30.
Li CH, Xiao Z, Tong JH, To KF, Fang X, Cheng AS, et al. EZH2 coupled with HOTAIR to silence MicroRNA-34a by the induction of heterochromatin formation in human pancreatic ductal adenocarcinoma. Int J Cancer. 2017;140(1):120–9.
Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS ONE. 2011;6(8):e24099.
Akula SM, Ruvolo PP, McCubrey JA. TP53/miR-34a-associated signaling targets SERPINE1 expression in human pancreatic cancer. Aging. 2020;12(3):2777–97.
Zou L, Chen FR, Xia RP, Wang HW, Xie ZR, Xu Y, et al. Long noncoding RNA XIST regulates the EGF receptor to promote TGF-β1-induced epithelial-mesenchymal transition in pancreatic cancer. Biochem Cell Biol. 2020;98(2):267–76.
Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, Mendell JT, et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10(8):1470–80.
Adamska A, Ferro R, Lattanzio R, Capone E, Domenichini A, Damiani V, et al. ABCC3 is a novel target for the treatment of pancreatic cancer. Adv Biol Regul. 2019;73:100634.
Benchimol S, Lamb P, Crawford LV, Sheer D, Shows TB, Bruns GA, et al. Transformation associated p53 protein is encoded by a gene on human chromosome 17. Somat Cell Mol Genet. 1985;11(5):505–10.
Zhao J, Lu Y, Shen HM. Targeting p53 as a therapeutic strategy in sensitizing TRAIL-induced apoptosis in cancer cells. Cancer Lett. 2012;314(1):8–23.
He X, He L, Hannon GJ. The guardian’s little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67(23):11099–101.
Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, et al. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266.
Soutto M, Chen Z, Saleh MA, Katsha A, Zhu S, Zaika A, et al. TFF1 activates p53 through down-regulation of miR-504 in gastric cancer. Oncotarget. 2014;5(14):5663–73.
Matlashewski GJ, Tuck S, Pim D, Lamb P, Schneider J, Crawford LV. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol. 1987;7(2):961–3.
Dumont P, Leu JI, Della Pietra AC 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33(3):357–65.
Cheng C, Lingyan W, Yi H, Cheng Z, Huadan Y, Xuting X, et al. Association between TLR2, MTR, MTRR, XPC, TP73, TP53 genetic polymorphisms and gastric cancer: a meta-analysis. Clin Res Hepatol Gastroenterol. 2014;38(3):346–59.
Francisco G, Menezes PR, Eluf-Neto J, Chammas R. Arg72Pro TP53 polymorphism and cancer susceptibility: a comprehensive meta-analysis of 302 case-control studies. Int J Cancer. 2011;129(4):920–30.
Tang W, Zhou X, Nie S, Yang Z, Zhu H, Wu X, et al. Association of p53 Arg72Pro polymorphism with gastric cancer: a meta-analysis. Biomarkers. 2012;17(7):597–603.
Xu Y, Liu L, Liu J, Zhang Y, Zhu J, Chen J, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int J Cancer. 2011;128(2):412–7.
Hasegawa M, Nishiyama C, Nishiyama M, Akizawa Y, Mitsuishi K, Ito T, et al. A novel – 66T/C polymorphism in fc epsilon RI alpha-chain promoter affecting the transcription activity: possible relationship to allergic diseases. J Immunol. 2003;171(4):1927–33.
van Rietschoten JG, Westland R, van den Bogaard R, Nieste-Otter MA, van Veen A, Jonkers RE, et al. A novel polymorphic GATA site in the human IL-12Rbeta2 promoter region affects transcriptional activity. Tissue Antigens. 2004;63(6):538–46.
Han Y, Pu R, Han X, Zhao J, Zhang Y, Zhang Q, et al. Associations of pri-miR-34b/c and pre-miR-196a2 polymorphisms and their multiplicative interactions with hepatitis B virus mutations with hepatocellular carcinoma risk. PLoS ONE. 2013;8(3):e58564.
Son MS, Jang MJ, Jeon YJ, Kim WH, Kwon CI, Ko KH, et al. Promoter polymorphisms of pri-miR-34b/c are associated with hepatocellular carcinoma. Gene. 2013;524(2):156–60.
Oh J, Kim JW, Lee BE, Jang MJ, Chong SY, Park PW, et al. Polymorphisms of the pri-miR-34b/c promoter and TP53 codon 72 are associated with risk of colorectal cancer. Oncol Rep. 2014;31(2):995–1002.
Yin J, Wang X, Zheng L, Shi Y, Wang L, Shao A, et al. Hsa-miR-34b/c rs4938723 T > C and hsa-miR-423 rs6505162 C > A polymorphisms are associated with the risk of esophageal cancer in a Chinese population. PLoS ONE. 2013;8(11):e80570.
Zhang J, Huang X, Xiao J, Yang Y, Zhou Y, Wang X, et al. Pri-miR-124 rs531564 and pri-miR-34b/c rs4938723 polymorphisms are associated with decreased risk of esophageal squamous cell carcinoma in Chinese populations. PLoS ONE. 2014;9(6):e100055.
Zhang S, Qian J, Cao Q, Li P, Wang M, Wang J, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with renal cell cancer risk in a Chinese population. Mutagenesis. 2014;29(2):149–54.
Pan XM, Sun RF, Li ZH, Guo XM, Qin HJ, Gao LB. Pri-miR-34b/c rs4938723 polymorphism is associated with a decreased risk of gastric cancer. Genet Test Mol Biomarkers. 2015;19(4):198–202.
Müeller-Pillasch F, Lacher U, Wallrapp C, Micha A, Zimmerhackl F, Hameister H, et al. Cloning of a gene highly overexpressed in cancer coding for a novel KH-domain containing protein. Oncogene. 1997;14(22):2729–33.
Clauditz TS, Wang CJ, Gontarewicz A, Blessmann M, Tennstedt P, Borgmann K, et al. Expression of insulin-like growth factor II mRNA-binding protein 3 in squamous cell carcinomas of the head and neck. J Oral Pathol Med. 2013;42(2):125–32.
Wang T, Fan L, Watanabe Y, McNeill PD, Moulton GG, Bangur C, et al. L523S, an RNA-binding protein as a potential therapeutic target for lung cancer. Br J Cancer. 2003;88(6):887–94.
Pryor JG, Bourne PA, Yang Q, Spaulding BO, Scott GA, Xu H. IMP-3 is a novel progression marker in malignant melanoma. Mod Pathol. 2008;21(4):431–7.
Li D, Yan D, Tang H, Zhou C, Fan J, Li S, et al. IMP3 is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Ann Surg Oncol. 2009;16(12):3499–506.
Jeng YM, Chang CC, Hu FC, Chou HY, Kao HL, Wang TH, et al. RNA-binding protein insulin-like growth factor II mRNA-binding protein 3 expression promotes tumor invasion and predicts early recurrence and poor prognosis in hepatocellular carcinoma. Hepatology. 2008;48(4):1118–27.
Lochhead P, Imamura Y, Morikawa T, Kuchiba A, Yamauchi M, Liao X, et al. Insulin-like growth factor 2 messenger RNA binding protein 3 (IGF2BP3) is a marker of unfavourable prognosis in colorectal cancer. Eur J Cancer. 2012;48(18):3405–13.
Kim HJ, Kim GE, Lee JS, Lee JH, Nam JH, Choi C. Insulin-like growth factor-II mRNA-binding protein 3 expression in effusion cytology: a marker for metastatic adenocarcinoma cells and a potential prognostic indicator in gastric adenocarcinoma. Acta Cytol. 2014;58(2):167–73.
Zhou Y, Huang T, Siu HL, Wong CC, Dong Y, Wu F, et al. IGF2BP3 functions as a potential oncogene and is a crucial target of miR-34a in gastric carcinogenesis. Mol Cancer. 2017;16(1):77.
Peng Y, Fan JY, Xiong J, Lou Y, Zhu Y. miR-34a enhances the susceptibility of gastric cancer to platycodin D by targeting survivin. Pathobiology. 2019;86(5–6):296–305.
Ruan T, Liu W, Tao K, Wu C. A review of research progress in multidrug-resistance mechanisms in gastric cancer. Onco Targets Ther. 2020;13:1797–807.
Dong G, Wang B, An Y, Li J, Wang X, Jia J, et al. SIRT1 suppresses the migration and invasion of gastric cancer by regulating ARHGAP5 expression. Cell Death Dis. 2018;9(10):977.
Qiu G, Li X, Che X, Wei C, He S, Lu J, et al. SIRT1 is a regulator of autophagy: implications in gastric cancer progression and treatment. FEBS Lett. 2015;589(16):2034–42.
Kim HB, Lee SH, Um JH, Kim MJ, Hyun SK, Gong EJ, et al. Sensitization of chemo-resistant human chronic myeloid leukemia stem-like cells to Hsp90 inhibitor by SIRT1 inhibition. Int J Biol Sci. 2015;11(8):923–34.
Ling S, Li J, Shan Q, Dai H, Lu D, Wen X, et al. USP22 mediates the multidrug resistance of hepatocellular carcinoma via the SIRT1/AKT/MRP1 signaling pathway. Mol Oncol. 2017;11(6):682–95.
Deng X, Zheng H, Li D, Xue Y, Wang Q, Yan S, et al. MicroRNA-34a regulates proliferation and apoptosis of gastric cancer cells by targeting silent information regulator 1. Exp Ther Med. 2018;15(4):3705–14.
Gao J, Li N, Dong Y, Li S, Xu L, Li X, et al. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene. 2015;34(31):4142–52.
Xu XP, Peng XQ, Yin XM, Liu Y, Shi ZY. miR-34a-5p suppresses the invasion and metastasis of liver cancer by targeting the transcription factor YY1 to mediate MYCT1 upregulation. Acta Histochem. 2020;122(6):151576.
Zuo Y, Zheng W, Liu J, Tang Q, Wang SS, Yang XS. MiR-34a-5p/PD-L1 axis regulates cisplatin chemoresistance of ovarian cancer cells. Neoplasma. 2020;67(1):93–101.
Pu Y, Zhao F, Li Y, Cui M, Wang H, Meng X, et al. The miR-34a-5p promotes the multi-chemoresistance of osteosarcoma via repression of the AGTR1 gene. BMC Cancer. 2017;17(1):45.
Pu Y, Zhao F, Wang H, Cai S. MiR-34a-5p promotes multi-chemoresistance of osteosarcoma through down-regulation of the DLL1 gene. Sci Rep. 2017;7:44218.
Deng XJ, Zheng HL, Ke XQ, Deng M, Ma ZZ, Zhu Y, et al. Hsa-miR-34a-5p reverses multidrug resistance in gastric cancer cells by targeting the 3’-UTR of SIRT1 and inhibiting its expression. Cell Signal. 2021;84:110016.
Imoto I, Pimkhaokham A, Watanabe T, Saito-Ohara F, Soeda E, Inazawa J. Amplification and overexpression of TGIF2, a novel homeobox gene of the TALE superclass, in ovarian cancer cell lines. Biochem Biophys Res Commun. 2000;276(1):264–70.
Krzeszinski JY, Wei W, Huynh H, Jin Z, Wang X, Chang TC, et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2014;512(7515):431–5.
Hu Y, Pu Q, Cui B, Lin J. MicroRNA-34a inhibits tumor invasion and metastasis in gastric cancer by targeting Tgif2. Int J Clin Exp Pathol. 2015;8(8):8921–8.
Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8(1):61–70.
Sah NK, Khan Z, Khan GJ, Bisen PS. Structural, functional and therapeutic biology of survivin. Cancer Lett. 2006;244(2):164–71.
Zhang Z, Kong Y, Yang W, Ma F, Zhang Y, Ji S, et al. Upregulation of microRNA-34a enhances the DDP sensitivity of gastric cancer cells by modulating proliferation and apoptosis via targeting MET. Oncol Rep. 2016;36(4):2391–7.
Lin L, Jiang H, Huang M, Hou X, Sun X, Jiang X, et al. Depletion of histone deacetylase 1 inhibits metastatic abilities of gastric cancer cells by regulating the miR-34a/CD44 pathway. Oncol Rep. 2015;34(2):663–72.
Yang B, Huang J, Liu H, Guo W, Li G. miR-335 directly, while miR-34a indirectly modulate survivin expression and regulate growth, apoptosis, and invasion of gastric cancer cells. Tumour Biol. 2016;37(2):1771–9.
Tanabe S, Quader S, Ono R, Cabral H, Aoyagi K, Hirose A, et al. Cell cycle regulation and DNA damage response networks in diffuse- and intestinal-type gastric cancer. Cancers (Basel). 2021;13:22.
Wang AM, Huang TT, Hsu KW, Huang KH, Fang WL, Yang MH, et al. Yin Yang 1 is a target of microRNA-34 family and contributes to gastric carcinogenesis. Oncotarget. 2014;5(13):5002–16.
Kim CH, Kim HK, Rettig RL, Kim J, Lee ET, Aprelikova O, et al. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med Genomics. 2011;4:79.
Tahara H, Kay MA, Yasui W, Tahara E. MicroRNAs in cancer: the 22nd Hiroshima cancer seminar/the 4th Japanese association for RNA interference joint international symposium, 30 August 2012, Grand Prince Hotel Hiroshima. Jpn J Clin Oncol. 2013;43(5):579 – 82.
Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47(5):784–91.
Pei LJ, Sun PJ, Ma K, Guo YY, Wang LY, Liu FD. LncRNA-SNHG7 interferes with miR-34a to de-sensitize gastric cancer cells to cisplatin. Cancer Biomark. 2021;30(1):127–37.
Sun C, Zhang S, Liu C, Liu X. Curcumin promoted miR-34a expression and suppressed proliferation of gastric cancer cells. Cancer Biother Radiopharm. 2019;34(10):634–41.
Zhang Y, Yuan Y, Zhang Y, Cheng L, Zhou X, Chen K. SNHG7 accelerates cell migration and invasion through regulating miR-34a-Snail-EMT axis in gastric cancer. Cell Cycle. 2020;19(1):142–52.
Chen H, Zhu XM, Luo ZL, Hu YJ, Cai XC, Gu QH. Sevoflurane induction alleviates the progression of gastric cancer by upregulating the miR-34a/TGIF2 axis. Eur Rev Med Pharmacol Sci. 2020;24(22):11883–90.
Song Z, Liang X, Wang Y, Han H, Yang J, Fang X, et al. Phenylboronic acid-functionalized polyamidoamine-mediated miR-34a delivery for the treatment of gastric cancer. Biomater Sci. 2019;7(4):1632–42.
Zhang S, Chen P, Huang Z, Hu X, Chen M, Hu S, et al. Sirt7 promotes gastric cancer growth and inhibits apoptosis by epigenetically inhibiting miR-34a. Sci Rep. 2015;5:9787.
Zhou Y, Ding BZ, Lin YP, Wang HB. MiR-34a, as a suppressor, enhance the susceptibility of gastric cancer cell to luteolin by directly targeting HK1. Gene. 2018;644:56–65.
Jafari N, Abediankenari S, Hossein-Nataj H. miR-34a mimic or pre-mir-34a, which is the better option for cancer therapy? KatoIII as a model to study miRNA action in human gastric cancer cells. Cancer Cell Int. 2021;21(1):178.
Jafari N, Abediankenari S, Hosseini-Khah Z, Valizadeh SM, Torabizadeh Z, Zaboli E, et al. Expression patterns of seven key genes, including β-catenin, Notch1, GATA6, CDX2, miR-34a, miR-181a and miR-93 in gastric cancer. Sci Rep. 2020;10(1):12342.
Cao W, Yang W, Fan R, Li H, Jiang J, Geng M, et al. miR-34a regulates cisplatin-induce gastric cancer cell death by modulating PI3K/AKT/survivin pathway. Tumour Biol. 2014;35(2):1287–95.
Wei B, Huang QY, Huang SR, Mai W, Zhong XG. MicroRNA–34a attenuates the proliferation, invasion and metastasis of gastric cancer cells via downregulation of MET. Mol Med Rep. 2015;12(4):5255–61.
Hui WT, Ma XB, Zan Y, Wang XJ, Dong L. Prognostic significance of MiR-34a expression in patients with gastric cancer after radical gastrectomy. Chin Med J (Engl). 2015;128(19):2632–7.
Mohammadian F, Abhari A, Dariushnejad H, Zarghami F, Nikanfar A, Pilehvar-Soltanahmadi Y, et al. Upregulation of Mir-34a in AGS gastric cancer cells by a PLGA-PEG-PLGA chrysin nano formulation. Asian Pac J Cancer Prev. 2015;16(18):8259–63.
Cheng C, Qin Y, Zhi Q, Wang J, Qin C. Knockdown of long non-coding RNA HOTAIR inhibits cisplatin resistance of gastric cancer cells through inhibiting the PI3K/Akt and Wnt/β-catenin signaling pathways by up-regulating miR-34a. Int J Biol Macromol. 2018;107(Pt B):2620–9.
Yan LH, Chen ZN, Li L, Chen J, Mo XW, Qin YZ et al. E2F-1 promotes DAPK2-induced anti-tumor immunity of gastric cancer cells by targeting miR-34a. Tumour Biol. 2016.
Cao W, Fan R, Wang L, Cheng S, Li H, Jiang J, et al. Expression and regulatory function of miRNA-34a in targeting survivin in gastric cancer cells. Tumour Biol. 2013;34(2):963–71.
Zhang H, Li S, Yang J, Liu S, Gong X, Yu X. The prognostic value of miR-34a expression in completely resected gastric cancer: tumor recurrence and overall survival. Int J Clin Exp Med. 2015;8(2):2635–41.
Ping W, Senyan H, Li G, Yan C, Long L. Increased lactate in gastric cancer tumor-infiltrating lymphocytes is related to impaired T cell function due to miR-34a deregulated lactate dehydrogenase A. Cell Physiol Biochem. 2018;49(2):828–36.
Azimi M, Totonchi M, Rahimi M, Firouzi J, Sahranavard P, Emami Razavi A, et al. An integrated analysis to predict micro-RNAs targeting both stemness and metastasis in human gastric cancer. J Gastroenterol Hepatol. 2021;36(2):436–45.
Zhang HH, Gu GL, Zhang XY, Li FZ, Ding L, Fan Q, et al. Primary analysis and screening of microRNAs in gastric cancer side population cells. World J Gastroenterol. 2015;21(12):3519–26.
Kipkeeva FM, Muzaffarova Т, Nikulin MP, Apanovich PV, Narimanov MN, Malikhova OA, et al. A group of miRNA as candidates for prognostic biomarkers of gastric cancer metastasis. Bull Exp Biol Med. 2020;169(1):77–80.
Rossi AFT, Contiero JC, Manoel-Caetano FDS, Severino FE, Silva AE. Up-regulation of tumor necrosis factor-α pathway survival genes and of the receptor TNFR2 in gastric cancer. World J Gastrointest Oncol. 2019;11(4):281–94.
Hu CE, Liu YC, Zhang HD, Huang GJ. JMJD2A predicts prognosis and regulates cell growth in human gastric cancer. Biochem Biophys Res Commun. 2014;449(1):1–7.
Wang G, Liu G, Ye Y, Fu Y, Zhang X. Upregulation of miR-34a by diallyl disulfide suppresses invasion and induces apoptosis in SGC-7901 cells through inhibition of the PI3K/Akt signaling pathway. Oncol Lett. 2016;11(4):2661–7.
Jang E, Kim E, Son HY, Lim EK, Lee H, Choi Y, et al. Nanovesicle-mediated systemic delivery of microRNA-34a for CD44 overexpressing gastric cancer stem cell therapy. Biomaterials. 2016;105:12–24.
Ibarrola-Villava M, Llorca-Cardeñosa MJ, Tarazona N, Mongort C, Fleitas T, Perez-Fidalgo JA, et al. Deregulation of ARID1A, CDH1, cMET and PIK3CA and target-related microRNA expression in gastric cancer. Oncotarget. 2015;6(29):26935–45.
Ito T, Ouchida M, Ito S, Jitsumori Y, Morimoto Y, Ozaki T, et al. SYT, a partner of SYT-SSX oncoprotein in synovial sarcomas, interacts with mSin3A, a component of histone deacetylase complex. Lab Invest. 2004;84(11):1484–90.
Nord H, Hartmann C, Andersson R, Menzel U, Pfeifer S, Piotrowski A, et al. Characterization of novel and complex genomic aberrations in glioblastoma using a 32K BAC array. Neurooncology. 2009;11(6):803–18.
Zhu H, Wu TC, Chen WQ, Zhou LJ, Wu Y, Zeng L, et al. Screening for differentially expressed genes between left–and right–sided colon carcinoma by microarray analysis. Oncol Lett. 2013;6(2):353–8.
Lu H, Hao L, Yang H, Chen J, Liu J. miRNA-34a suppresses colon carcinoma proliferation and induces cell apoptosis by targeting SYT1. Int J Clin Exp Pathol. 2019;12(8):2887–97.
Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–34.
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–96.
Roussel M, Sherr C, Barker P, Ruddle F. Molecular cloning of the c-fms locus and its assignment to human chromosome 5. J Virol. 1983;48(3):770–3.
Yarden Y, Ullrich A. Growth factor receptor tyrosine kinases. Annu Rev Biochem. 1988;57(1):443–78.
Heisterkamp N, Groffen J, Stephenson JR. Isolation of v-fms and its human cellular homolog. Virology. 1983;126(1):248–58.
Roussel MF, Dull TJ, Rettenmier CW, Ralph P, Ullrich A, Sherr CJ. Transforming potential of the c-fms proto-oncogene (CSF-1 receptor). Nature. 1987;325(6104):549–52.
Coussens L, Van Beveren C, Smith D, Chen E, Mitchell RL, Isacke CM, et al. Structural alteration of viral homologue of receptor proto-oncogene fms at carboxyl terminus. Nature. 1986;320(6059):277–80.
Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61(2):203–12.
Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, Cella M, et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of langerhans cells and microglia. Nat Immunol. 2012;13(8):753–60.
Novak U, Harpur AG, Paradiso L, Kanagasundaram V, Jaworowski A, Wilks AF et al. Colony-stimulating factor 1-induced STAT1 and STAT3 activation is accompanied by phosphorylation of Tyk2 in macrophages and Tyk2 and JAK1 in fibroblasts. 1995.
Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–34.
Cannarile MA, Weisser M, Jacob W, Jegg A-M, Ries CH, Rüttinger D. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J Immunother Cancer. 2017;5(1):1–13.
Richardsen E, Uglehus RD, Johnsen SH, Busund L-T. Macrophage-colony stimulating factor (CSF1) predicts breast cancer progression and mortality. Anticancer Res. 2015;35(2):865–74.
Mroczko B, Szmitkowski M, Okulczyk B. Hematopoietic growth factors in colorectal cancer patients. 2003.
Shi X, Kaller M, Rokavec M, Kirchner T, Horst D, Hermeking H. Characterization of a p53/miR-34a/CSF1R/STAT3 feedback loop in colorectal cancer. Cell Mol Gastroenterol Hepatol. 2020;10(2):391–418.
Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, et al. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80(1):51–60.
Mangold U. The antizyme family: polyamines and beyond. IUBMB Life. 2005;57(10):671–6.
Coffino P. Regulation of cellular polyamines by antizyme. Nat Rev Mol Cell Biol. 2001;2(3):188–94.
Li Y, Gong P, Hou JX, Huang W, Ma XP, Wang YL, et al. miR-34a regulates multidrug resistance via positively modulating OAZ2 signaling in Colon cancer cells. J Immunol Res. 2018;2018:7498514.
Demoulin J-B, Essaghir A. PDGF receptor signaling networks in normal and cancer cells. Cytokine Growth Factor Rev. 2014;25(3):273–83.
Zhu Y, Wang Y, Guan B, Rao Q, Wang J, Ma H, et al. C-kit and PDGFRA gene mutations in triple negative breast cancer. Int J Clin Exp Pathol. 2014;7(7):4280–5.
Roh J-W, Huang J, Hu W, Yang X, Jennings NB, Sehgal V, et al. Biologic effects of platelet-derived growth factor receptor α blockade in uterine CancerInhibition of PDGFRα in uterine cancer. Clin Cancer Res. 2014;20(10):2740–50.
Stock A-M, Hahn SA, Troost G, Niggemann B, Zänker KS, Entschladen F. Induction of pancreatic cancer cell migration by an autocrine epidermal growth factor receptor activation. Exp Cell Res. 2014;326(2):307–14.
Hayes BJ, Riehle KJ, Shimizu-Albergine M, Bauer RL, Hudkins KL, Johansson F, et al. Activation of platelet-derived growth factor receptor alpha contributes to liver fibrosis. PLoS ONE. 2014;9(3):e92925.
Li C, Wang Y, Lu S, Zhang Z, Meng H, Liang L, et al. MiR-34a inhibits colon cancer proliferation and metastasis by inhibiting platelet-derived growth factor receptor α. Mol Med Rep. 2015;12(5):7072–8.
Fawzy MS, Ibrahiem AT, AlSel BTA, Alghamdi SA, Toraih EA. Analysis of microRNA-34a expression profile and rs2666433 variant in colorectal cancer: a pilot study. Sci Rep. 2020;10(1):16940.
Li Y, Gu F, Lin X. The role of miR-141/ Sirt1 in colon cancer. J buon. 2020;25(6):2665–71.
Roy S, Levi E, Majumdar AP, Sarkar FH. Expression of miR-34 is lost in colon cancer which can be re-expressed by a novel agent CDF. J Hematol Oncol. 2012;5:58.
Li W, Han W, Ma Y, Cui L, Tian Y, Zhou Z, et al. P53-dependent miRNAs mediate nitric oxide-induced apoptosis in colonic carcinogenesis. Free Radic Biol Med. 2015;85:105–13.
Li C, Liu T, Zhang Y, Li Q, Jin LK. LncRNA-ZDHHC8P1 promotes the progression and metastasis of colorectal cancer by targeting miR-34a. Eur Rev Med Pharmacol Sci. 2019;23(4):1476–86.
He J, Zhao H, Liu X, Wang D, Wang Y, Ai Y, et al. Sevoflurane suppresses cell viability and invasion and promotes cell apoptosis in colon cancer by modulating exosome–mediated circ–HMGCS1 via the miR–34a–5p/SGPP1 axis. Oncol Rep. 2020;44(6):2429–42.
Wu J, Wu G, Lv L, Ren YF, Zhang XJ, Xue YF, et al. MicroRNA-34a inhibits migration and invasion of colon cancer cells via targeting to Fra-1. Carcinogenesis. 2012;33(3):519–28.
Orosz E, Kiss I, Gyöngyi Z, Varjas T. Expression of circulating miR-155, miR-21, miR-221, miR-30a, miR-34a and miR-29a: comparison of colonic and rectal cancer. Vivo. 2018;32(6):1333–7.
Cai MH, Xu XG, Yan SL, Sun Z, Ying Y, Wang BK, et al. Regorafenib suppresses colon tumorigenesis and the generation of drug resistant cancer stem-like cells via modulation of miR-34a associated signaling. J Exp Clin Cancer Res. 2018;37(1):151.
Jiang H, Ge F, Hu B, Wu L, Yang H, Wang H. rs35301225 polymorphism in miR-34a promotes development of human colon cancer by deregulation of 3’UTR in E2F1 in Chinese population. Cancer Cell Int. 2017;17:39.
Wang L, Bu P, Ai Y, Srinivasan T, Chen HJ, Xiang K et al. A long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division. Elife. 2016;5.
Bin H, Mei H, Hui W, Bing Z. The correlation between miR – 34a-3p, miR – 31, PLEK2 and the occurrence, development and prognosis of colorectal cancer. Cell Mol Biol (Noisy-le-grand). 2022;68(1):192–200.
Wang L, Wang E, Wang Y, Mines R, Xiang K, Sun Z et al. miR-34a is a microRNA safeguard for citrobacter-induced inflammatory colon oncogenesis. Elife. 2018;7.
Sun N, Zhang G, Liu Y. Long non-coding RNA XIST sponges miR-34a to promotes colon cancer progression via Wnt/β-catenin signaling pathway. Gene. 2018;665:141–8.
Peng CL, Zhao XJ, Wei CC, Wu JW. LncRNA HOTAIR promotes colon cancer development by down-regulating miRNA-34a. Eur Rev Med Pharmacol Sci. 2019;23(13):5752–61.
Bu P, Chen KY, Chen JH, Wang L, Walters J, Shin YJ, et al. A microRNA miR-34a-regulated bimodal switch targets notch in colon cancer stem cells. Cell Stem Cell. 2013;12(5):602–15.
Fang C, Qiu S, Sun F, Li W, Wang Z, Yue B, et al. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017;410:50–62.
Bu P, Wang L, Chen KY, Srinivasan T, Murthy PK, Tung KL, et al. A miR-34a-numb feedforward loop triggered by inflammation regulates asymmetric stem cell division in intestine and colon cancer. Cell Stem Cell. 2016;18(2):189–202.
Eslamizadeh S, Zare AA, Talebi A, Tabaeian SP, Eshkiki ZS, Heydari-Zarnagh H, et al. Differential expression of miR-20a and miR-145 in colorectal tumors as potential location-specific miRNAs. Microrna. 2021;10(1):66–73.
Siemens H, Neumann J, Jackstadt R, Mansmann U, Horst D, Kirchner T, et al. Detection of miR-34a promoter methylation in combination with elevated expression of c-Met and β-catenin predicts distant metastasis of colon cancer. Clin Cancer Res. 2013;19(3):710–20.
Ji G, Zhou W, Li X, Du J, Li X, Hao H. Melatonin inhibits proliferation and viability and promotes apoptosis in colorectal cancer cells via upregulation of the microRNA-34a/449a cluster. Mol Med Rep. 2021;23(3).
Akao Y, Khoo F, Kumazaki M, Shinohara H, Miki K, Yamada N. Extracellular disposal of tumor-suppressor miRs-145 and – 34a via microvesicles and 5-FU resistance of human colon cancer cells. Int J Mol Sci. 2014;15(1):1392–401.
Li L, Wu C, Zhao Y. miRNA-34a enhances the sensitivity of gastric cancer cells to treatment with paclitaxel by targeting E2F5. Oncol Lett. 2017;13(6):4837–42.
Jiang H, Guo Y, Huang K, Lu R, Peng X, Lin S. MicroRNA–34a inhibits esophageal squamous cell carcinoma progression by targeting E2F5. J buon. 2019;24(6):2514–22.
Chen CF, Hsu EC, Lin KT, Tu PH, Chang HW, Lin CH, et al. Overlapping high-resolution copy number alterations in cancer genomes identified putative cancer genes in hepatocellular carcinoma. Hepatology. 2010;52(5):1690–701.
Cai C, Rajaram M, Zhou X, Liu Q, Marchica J, Li J, et al. Activation of multiple cancer pathways and tumor maintenance function of the 3q amplified oncogene FNDC3B. Cell Cycle. 2012;11(9):1773–81.
Duffy MJ, Maguire TM, Hill A, McDermott E, O’Higgins N. Metalloproteinases: role in breast carcinogenesis, invasion and metastasis. Breast Cancer Res. 2000;2(4):1–6.
Shibatohge M, Kariya K-i, Liao Y, Hu C-D, Watari Y, Goshima M, et al. Identification of PLC210, a caenorhabditis elegansphospholipase C, as a putative effector of Ras. J Biol Chem. 1998;273(11):6218–22.
Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem. 2001;70(1):281–312.
Oka M, Edamatsu H, Kunisada M, Hu L, Takenaka N, Dien S, et al. Enhancement of ultraviolet B-induced skin tumor development in phospholipase Cε-knockout mice is associated with decreased cell death. Carcinogenesis. 2010;31(10):1897–902.
Bai Y, Edamatsu H, Maeda S, Saito H, Suzuki N, Satoh T, et al. Crucial role of phospholipase Cε in chemical carcinogen-induced skin tumor development. Cancer Res. 2004;64(24):8808–10.
Ou L, Guo Y, Luo C, Wu X, Zhao Y, Cai X. RNA interference suppressing PLCE1 gene expression decreases invasive power of human bladder cancer T24 cell line. Cancer Genet Cytogenet. 2010;200(2):110–9.
Ling Y, Chunli L, Xiaohou W, Qiaoling Z. Involvement of the PLCε/PKCα pathway in human BIU-87 bladder cancer cell proliferation. Cell Biol Int. 2011;35(10):1031–6.
Wang X, Zbou C, Qiu G, Fan J, Tang H, Peng Z. Screening of new tumor suppressor genes in sporadic colorectal cancer patients. Hepatogastroenterology. 2008;55(88):2039–44.
Hu H, Yang J, Sun Y, Yang Y, Qian J, Jin L, et al. Putatively functional PLCE1 variants and susceptibility to esophageal squamous cell carcinoma (ESCC): a case–control study in eastern Chinese populations. Ann Surg Oncol. 2012;19(7):2403–10.
Ma H, Wang L-E, Liu Z, Sturgis EM, Wei Q. Association between novel PLCE1variants identified in published esophageal cancer genome-wide association studies and risk of squamous cell carcinoma of the head and neck. BMC Cancer. 2011;11(1):1–9.
Bourguignon LY, Gilad E, Brightman A, Diedrich F, Singleton P. Hyaluronan-CD44 interaction with leukemia-associated RhoGEF and epidermal growth factor receptor promotes Rho/Ras co-activation, phospholipase Cϵ-Ca2 + signaling, and cytoskeleton modification in head and neck squamous cell carcinoma cells. J Biol Chem. 2006;281(20):14026–40.
Abnet CC, Freedman ND, Hu N, Wang Z, Yu K, Shu X-O, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet. 2010;42(9):764–7.
Wu C, Hu Z, He Z, Jia W, Wang F, Zhou Y, et al. Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat Genet. 2011;43(7):679–84.
Cui X, Chen Y, Liu L, Li L, Hu J, Yang L, et al. Heterozygote of PLCE1 rs2274223 increases susceptibility to human papillomavirus infection in patients with esophageal carcinoma among the Kazakh populations. J Med Virol. 2014;86(4):608–17.
Cui XB, Peng H, Li RR, Mu JQ, Yang L, Li N, et al. MicroRNA-34a functions as a tumor suppressor by directly targeting oncogenic PLCE1 in Kazakh esophageal squamous cell carcinoma. Oncotarget. 2017;8(54):92454–69.
Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582(10):1564–8.
Wei JS, Song YK, Durinck S, Chen QR, Cheuk AT, Tsang P, et al. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27(39):5204–13.
Shi H, Zhou S, Liu J, Zhu J, Xue J, Gu L, et al. miR-34a inhibits the in vitro cell proliferation and migration in human esophageal cancer. Pathol Res Pract. 2016;212(5):444–9.
Zang HL, Li YH, Huang GM. Long-chain non-coding RNA Linc00888 promotes the proliferation and migration of esophageal cancer cells by downregulating miR-34a expression. Eur Rev Med Pharmacol Sci. 2020;24(21):11081–9.
Asadi M, Shanehbandi D, Mohammadpour H, Hashemzadeh S, Sepehri B. Expression level of miR-34a in tumor tissue from patients with esophageal squamous cell carcinoma. J Gastrointest Cancer. 2019;50(2):304–7.
Lin Z, Chen Y, Lin Y, Lin H, Li H, Su X, et al. Potential miRNA biomarkers for the diagnosis and prognosis of esophageal cancer detected by a novel absolute quantitative RT-qPCR method. Sci Rep. 2020;10(1):20065.
Mahawongkajit P, Tomtitchong P. Expression of miRNA in 5-FU resistant esophageal cancer. Mol Clin Oncol. 2020;13(2):221–7.
Dai S, Ye Z, Wang F, Yan F, Wang L, Fang J, et al. Doxorubicin-loaded poly(ε-caprolactone)-Pluronic micelle for targeted therapy of esophageal cancer. J Cell Biochem. 2018;119(11):9017–27.
Cui X, Zhao Z, Liu D, Guo T, Li S, Hu J, et al. Inactivation of miR-34a by aberrant CpG methylation in Kazakh patients with esophageal carcinoma. J Exp Clin Cancer Res. 2014;33(1):20.
Li J, Wang K, Chen X, Meng H, Song M, Wang Y, et al. Transcriptional activation of microRNA-34a by NF-kappa B in human esophageal cancer cells. BMC Mol Biol. 2012;13:4.
Hu Y, Correa AM, Hoque A, Guan B, Ye F, Huang J, et al. Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int J Cancer. 2011;128(1):132–43.
Hemmatzadeh M, Mohammadi H, Babaie F, Yousefi M, Ebrazeh M, Mansoori B, et al. Snail-1 silencing by siRNA inhibits migration of TE-8 esophageal cancer cells through downregulation of metastasis-related genes. Adv Pharm Bull. 2018;8(3):437–45.
Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, et al. Reassessment Exosome Composition Cell. 2019;177(2):428–e4518.
van der Meel R, Fens MH, Vader P, van Solinge WW, Eniola-Adefeso O, Schiffelers RM. Extracellular vesicles as drug delivery systems: lessons from the liposome field. J Control Release. 2014;195:72–85.
Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9.
Jiang XC, Gao JQ. Exosomes as novel bio-carriers for gene and drug delivery. Int J Pharm. 2017;521(1–2):167–75.
Zuo L, Tao H, Xu H, Li C, Qiao G, Guo M, et al. Exosomes-coated miR-34a displays potent antitumor activity in pancreatic cancer both in vitro and in vivo. Drug Des Devel Ther. 2020;14:3495–507.
Barile L, Vassalli G, Exosomes. Therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63–78.
Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8(19):4083–99.
Hosseini M, Baghaei K, Amani D, Ebtekar M. Tumor-derived exosomes encapsulating miR-34a promote apoptosis and inhibit migration and tumor progression of colorectal cancer cells under in vitro condition. Daru. 2021;29(2):267–78.
Franco OE, Shaw AK, Strand DW, Hayward SW. Cancer associated fibroblasts in cancer pathogenesis. Semin Cell Dev Biol. 2010;21(1):33–9.
Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6(5):392–401.
Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A, et al. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res. 2008;68(3):918–26.
Hu Y, Yan C, Mu L, Huang K, Li X, Tao D, et al. Fibroblast-derived exosomes contribute to chemoresistance through priming cancer stem cells in colorectal cancer. PLoS ONE. 2015;10(5):e0125625.
Luga V, Wrana JL. Tumor-stroma interaction: revealing fibroblast-secreted exosomes as potent regulators of wnt-planar cell polarity signaling in cancer metastasis. Cancer Res. 2013;73(23):6843–7.
Richards KE, Zeleniak AE, Fishel ML, Wu J, Littlepage LE, Hill R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene. 2017;36(13):1770–8.
Shi L, Wang Z, Geng X, Zhang Y, Xue Z. Exosomal miRNA-34 from cancer-associated fibroblasts inhibits growth and invasion of gastric cancer cells in vitro and in vivo. Aging. 2020;12(9):8549–64.
Chen S, Mao Y, Chen W, Liu C, Wu H, Zhang J, et al. Serum exosomal miR-34a as a potential biomarker for the diagnosis and prognostic of hepatocellular carcinoma. J Cancer. 2022;13(5):1410–7.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
WG, and JZ contributed to manuscript drafting and data collection. All authors approved the final paper. MM and JZ oversaw the study.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Gao, W., Zhou, J. & Morshedi, M. MicroRNA-34 and gastrointestinal cancers: a player with big functions. Cancer Cell Int 24, 163 (2024). https://doi.org/10.1186/s12935-024-03338-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-024-03338-w