Abstract
Background
Thyroid carcinoma (THCA) is a malignant endocrine tumor all around the world, which is influenced by genetic and environmental factors.
Objective
To explore the association between TPO rs2048722, PTCSC2 rs925489, SEMA4G rs4919510 polymorphisms and THCA susceptibility in Chinese population.
Methods
We recruited 365 THCA patients and 498 normal controls for the study. Logistic regression analysis was used to evaluate the association between TPO rs2048722, PTCSC2 rs925489, SEMA4G rs4919510 polymorphisms and THCA susceptibility. MDR was used to assess the genetic interactions among the three SNPs.
Results
Overall analysis demonstrated that rs925489 of PTCSC2 was evidently associated with increased risk of THCA in multiple genetic models (OR = 1.59, 95%CI = 1.12–2.24, p = 0.009). The results of stratified analysis illustrated that rs2048722 of TPO can significantly increase the THCA susceptibility of participants less than or equal to 44 years old and smokers. Similarly, rs925489 of PTCSC2 obviously improved the risk of THCA among participants older than 44 years, males, smokers and drinkers. However, rs4919510 of SEMA4G has a protective effect on the development of THCA among participants with less than or equal to 44 years old and non-drinkers. Interestingly, there was a strong genetic interaction among the three SNPs in the occurrence of THCA risk.
Conclusion
TPO rs2048722, PTCSC2 rs925489 and SEMA4G rs4919510 polymorphisms were evidently associated with the risk of THCA in the Chinese population, which was affected by age, gender, smoking and drinking consumption.
Similar content being viewed by others
Introduction
Thyroid carcinoma (THCA) is a common malignant endocrine tumor with rapid growth, accounting for 1–2% of all cancers [1]. According to the world health organization (WHO, Global cancer statistics 2018) reported in 2018, more than 576,233 new THCA patients were diagnosed, of which 41,071 appeared in death [2]. Based on the pathological characteristics of the tumor, THCA could be further divided into five types including papillary (PTC), follicular (differentiated), poorly differentiated, anaplastic and medullary [3]. Pervious epidemiology studies have reported that environmental and hereditary parameters might affect the onset of a pathology of THCA [4]. The risk of differentiated thyroid carcinoma (DTC) was observably increased among the participants older than 55 years (HR 1.78) [5]. Smoking males had an evident reduced risk of THCA [6], and the risk of THCA was lower among the recruiters who smoked and drinked at the same time (HR 0.80) [7]. Up to now, the detail of THCA molecular mechanism was still unknown, just as many other human cancers.
For the past years, an increasing evidence suggested that hereditary parameters played a crucial role in the development of THCA [8, 9]. Various susceptibility genes and single nucleotide polymorphisms (SNPs) locus of THCA were identified by genome-wide association studies (GWAS) [10, 11]. The study found that the rs2048722 CT + TT genotype of thyroid peroxidase (TPO) in the Japanese population had markedly higher serum anti-thyroid peroxidase antibody (TPOAb) levels compared with CC genotype autoimmune thyroid disease patients [12]. Another study found that rs965513 of papillary thyroid cancer susceptibility candidate gene 2 (PTCSC2) in the Kazakh population was apparently associated with an increased risk of PTC [13]. Furthermore, a meta-analysis, which including 12,517 cases and 15,624 controls belonged to 18 case–control researches were conducted, and the statistical analysis results confirmed that miR-608 rs4919510 polymorphism was connected with THCA susceptibility among Chinese population, and miR-608 rs4919510 targeted Semaphorin-4G (SEMA4G) [14]. Up to now, the relationship between TPO rs2048722, PTCSC2 rs925489 and SEMA4G rs4919510 polymorphism and THCA sensibility and the interaction among the three SNPs in Chinese persons were not reported.
Hence, in our current research based on Chinese population, we designed a case–control study to inquire the interaction between the three SNPs (rs2048722, rs925489 and rs4919510) polymorphism and THCA risk among Chinese persons, and the interaction between the three SNPs in THCA development.
Material and methods
Study population
The study was approved by the ethics committee of the First Affiliated Hospital of Xi'an Jiaotong University. Meanwhile, this study was conducted in accordance with the Declaration of Helsinki. The informed documents were written by all participants prior to entering the study. Of this cohort, 365 THCA patients including 97 males and 268 females were recruited. THCA patients who was newly diagnosed by clinical factors and histopathological examination, meanwhile who has family cancer history and other diseases were excluded. In addition, a total of 498 unrelated healthy controls including 137 males and 361 females without any thyroid pathology and other cancers were recruited from the same hospital during the same time.
DNA extraction and genotyping
In this study, 5 ml peripheral blood samples from each participants were collected by specialized technicians and then stored into test tubes containing EDTA [15]. Next, genomic DNA were isolated from blood samples following standard GoldMag whole blood genomic DNA purification kit (GoldMag Co. Ltd. Xi’an city, China) extraction procedures. DNA quality was checked utilizingNanoDrop 2000 platform (Thermo Fisher Scientific, Waltham, MA, USA). The single-nucleotide polymorphisms (SNPs) including TPO rs2048722, PTCSC2 rs925489 and SEMA4G rs4919510 with the minor allele frequency more than 0.05 were selected from the 1000 Genomes Project (http://www.internationalgenome.org/). The corresponding amplification primers of each SNP were designed by Agena Bioscience Assay Design Suite V2.0 software (https://agenacx.com/online–tools/). The SNPs genotype were performed by MassARRAY Nanodispenser and MassARRAY iPLEX platform (both from Agena Bios 95% CIence, San Diego, CA, USA),with standard recommended instructions. Subsequently, Agena Bioscience TYPER version 4.0 software was used to manage all data, as our pervious describe [16, 17].
Statistical analysis
The statistical analyses were conducted by SPSS 20.0 (SPSS, USA) software. Goodness-of-fit χ2 test was applied to evaluate if the selected SNPs deviated from Hardy–Weinberg equilibrium (HWE) among controls. Difference in the distribution of demographic factors and frequencies of were calculated by χ2 test among patients and controls. In addition, the risk of THCA associated with the candidate SNPs polymorphism was estimated using the odds ratio (OR) and 95% confidence interval (95% CI) after adjusting age, sex, smoking and drinking. We used MDR software (version 4.0.2) to assess the interaction of candidate SNPs for THCA. PancanQTL (http://gong-lab.hzau.edu.cn/PancanQTL/) database was applied to analyze SNPs genotype expression. In this study, all statistical tests, p < 0.05 was considered statistically significant.
Results
Characteristics of study individuals
A total of 365 THCA patients and 498 unrelated healthy controls were recruited into the current study. The demographic parameters of the study participants were shown in Table 1, the mean age was 43.98 ± 15.12 years old for THCA patients and 44.16 ± 12.37 years old for healthy controls, there was no obvious difference in age between the two groups (p = 0.744). Statistical analysis results showed that there were not significant difference between patients and controls in terms of sex (p = 0.760), smoking (p = 0.492), and drinking consumption (p = 0.638), respectively. In addition, we also made statistics on the lymph node metastasis and THCA stage of the case group. In conclusion, the cases and controls were not evidently different in terms of sex, age, smoking, and drinking consumption, thus excluding confounding factors from interfering with the study results.
Associations between SNPs polymorphism and THCA risk
The SNP ID, chromosome, MAF and HWE p value of each candidate SNP were presented in Table 2. Our results showed that the distribution of genotypes in the healthy controls was consistent with HWE (all p > 0.05). Multiple genetic models and allele frequencies were used to assess the relationships between the SNPs and THCA risk. Our results suggested that the variant C allele in PTCSC2 rs925489 presented a significantly increasing THCA risk (OR = 1.51, 95% CI = 1.10- 2.07, p = 0.011). However, no significant difference between other SNPs (TPO rs2048722 and SEMA4G rs4919510) and TC risk were observed (p = 0.245 and p = 0.385).
Subsequently, we evaluated the influence of TPO rs2048722, PTCSC2 rs925489 and SEMA4G rs4919510 polymorphisms with THCA risk under four different genetic models. The results of the genetic models were listed in Table 3. In total, the polymorphism of PTCSC2 rs925489 were observed enhancing THCA risk under the co-dominant genetic model (OR = 1.59, 95% CI = 1.12–2.24, p = 0.009), the dominant genetic model (OR = 1.58, 95% CI = 1.12–2.23, p = 0.009) and the additive model (OR = 1.54, 95% CI = 1.10–2.15, p = 0.010). In addition, there was no significant difference between TPO rs2048722 and SEMA4G rs4919510 polymorphisms and the risk of THCA under four genetic models (p > 0.05).
Stratified analysis of the effect of SNPs polymorphism in demographic parameters
Furthermore, we carried out the stratification analysis to improve a more comprehensive insight into the effect of 3 SNPs (TPO rs2048722, PTCSC2 rs925489, and SEMA4G rs4919510) in THCA. The results of statistical analysis of age, sex, smoking and drinking were shown in Table 4, Table 5, Table 6 and Table 7, respectively, and the results of lymph node stratification were shown in Additional file 1: Table S2.
Age
Stratified results (Table 4) demonstrated that TPO rs2048722 was evidently increase the risk of THCA among participants less than or equal to 44 years old in multiple genetic models [allelic model: OR (95% CI) = 1.38 (1.04–1.83), p = 0.026; co-dominant model: OR (95% CI) = 1.86 (1.05–3.28), p = 0.033; recessive model: OR (95% CI) = 1.67 (1.02–2.73), p = 0.041; additive model: OR (95% CI) = 1.35 (1.02–1.79), p = 0.039]. PTCSC2 rs925489 was significantly associated with an increased risk of THCA in people older than 44 years in the allelic model [OR (95% CI) = 2.29 (1.44–3.64), p < 0.001], co-dominant model [OR (95% CI) = 2.22 (1.34–3.69), p = 0.002], dominant model [OR (95% CI) = 2.30 (1.39–3.81), P = 0.001] and additive model [OR (95% CI) = 2.32 (1.42- 3.79), p < 0.001]. However, SEMA4G rs4919510 had a protective effect on the risk of developing THCA among participants less than or equal to 44 years old in co-dominant [OR (95% CI) = 0.52 (0.33–0.83), p = 0.006] and dominant model [OR (95% CI) = 0.59 (0.38–0.91), p = 0.017].
Sex
Table 5 illustrated that PTCSC2 rs925489 was associated with increased THCA risk among males in alleles [OR (95% CI) = 2.77 (1.48–5.17), P = 0.001], co-dominance [OR (95% CI) = 3.59 (1.74–7.41), p < 0.001], dominant [OR (95% CI) = 3.42 (1.67–6.98), p < 0.001] and additive model [OR (95% CI) = 3.03 (1.52–6.02)), p = 0.001]. However, rs2048722 in TPO and rs4919510 in SEMA4G were not significantly associated with THCA risk in both male and female populations.
Smoking
Stratified results indicated (Table 6) that rs2048722 in TPO obviously increased susceptibility to THCA among smoking populations in multiple genetic models [allelic model: OR (95% CI) = 1.48 (1.10–1.99), p = 0.009; co-dominant model: OR (95% CI) = 2.14 (1.13–4.06), p = 0.019; dominant model: OR (95% CI) = 1.81 (1.07–3.06), p = 0.026; and additive model: OR (95% CI) = 1.47 (1.07–2.02), p = 0.019]. Rs925489 in PTCSC2 was significantly associated with increased risk of THCA in smokers only in the allelic model [OR (95% CI) = 1.84 (1.11–3.05), p = 0.017]. However, SEMA4G rs4919510 was not found to be evidently associated with the risk of THCA in smoking stratification.
Drinking
Table 7 indicated that rs2048722 in TPO is not significantly associated with the risk of THCA in drinking stratification, while PTCSC2 rs925489 can evidently increase the risk of THCA in drinking populations, with allele [OR (95% CI) = 1.90 (1.14–3.15), p = 0.012], dominant [OR (95% CI) = 1.82 (1.04–3.19), p = 0.036], and additive model [OR (95% CI) = 1.88 (1.10–3.21), p = 0.021]. Interestingly, rs4919510 in SEMA4G was significantly associated with reduced THCA risk among non-drinkers in multiple genetic models [allelic: OR (95% CI) = 0.77 (0.59–1.00), p = 0.049; co-dominant: OR (95% CI) = 0.56 (0.35–0.89), P = 0.014; dominant: OR (95% CI) = 0.57 (0.37–0.88), p = 0.012; and additive: OR (95% CI) = 0.75 (0.56–1.00), p = 0.049].
Lymph node metastasis
In the case group, rs2048722 in TPO, rs925489 in PTCSC2 and rs4919510 in SEMA4G were not found to be notably correlated with lymph node metastasis.
In general, stratified analysis results demonstrated that TPO rs2048722 could significantly increase THCA susceptibility among participants less than or equal to 44 years old and smokers. Similarly, PTCSC2 rs925489 evidently increased the risk of THCA in people older than 44 years, males, smokers and drinkers. However, rs4919510 in SEMA4G notably reduced the risk of THCA among people less than or equal to 44 years old and non-drinkers.
Analysis of MDR
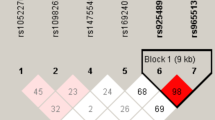
The MDR software was used to evaluate three SNPs with high-order interactions in THCA. Regarding the THCA risk model, the single-locus model rs925489, the two-locus model rs925489, rs4919510 and the three-locus model rs2048722, rs925489 and rs4919510 all have higher accuracy and testability, among which the three-locus model has the highest concordance of 10/10, and p = 0.001 (Table 8). Figure 1A and 1B indicated the interaction between the three SNPs, where the color closer to red indicates stronger synergy, and closer to blue indicates stronger redundancy. Taken together, TOP rs2048722, PTCSC2 rs925489 and SEMA4G rs4919510 may have strong genetic interactions in the occurrence of THCA.
Analysis of MDR and SNP genotype expression. A SNP-SNP interaction dendrogram of MDR analysis. B Fruchterman-reingold of MDR analysis. (The closer to red the stronger the synergy, the closer to the blue the more redundancy.) C Rs925489 genotype expression of THCA. D Rs4919510 genotype expression of THCA
Analysis of SNP genotype expression
The analysis of SNP genotype expression in THCA declared that rs925489 had significant differences among different genotypes in cis-eQTL and trans-eQTL (CC < CT < TT, Fig. 1C), indicating that the genotype change of rs925489 of THCA may directly or indirectly affect the expression of related genes. Different genotypes of rs4919510 have obvious differences in cis-eQTL (CC > CG > GG, Fig. 1D), which indicates that the genotype change of rs4919510 of THCA directly affects the expression of related genes. Unfortunately, the expression of rs2048722 different genotypes in THCA were not found.
Discussion
As we all know, THCA is most frequent head and neck tumors, and is reported that THCA has a highly morbidity all over the world [18]. More and more researchers have given evidences that genetic factors play an important role in the pathogenesis of THCA [19]. As a membrane-bound glycoprotein, TPO catalyzes thyroid hormone enzymes and regulates thyroid function [20]. Various studies have been confirmed that multiple TPO gene mutations may give rise to dysfunction of the TPO enzyme and varieties human disease [21]. Aleksander et.al suggested that TPO rs11675434 polymorphism was related with autoimmune thyroid disease among Polish Caucasian population [22]. In addition, the study found that the rs2048722 CT + TT genotype of TPO had evidently higher serum anti-thyroid peroxidase antibody (TPOAb) levels compared with CC genotype autoimmune thyroid disease patients in the Japanese population [12]. In this study, rs2048722 in TPO was also found to be a significant risk gene for THCA among the Chinese population aged less than or equal to 44 years old and smoking in the stratified analysis.
As long noncoding RNAs (lncRNAs), the SNP (rs965513) in PTCSC2 was evidently associated with PTC risk, and similar to TPO, PTCSC2 also regulates thyroid hormone levels and thyroid function [23]. Similarly, PTCSC2 is a susceptibility gene in familial non-medullary thyroid cancer [24]. Furthermore, PTCSC2 rs965513 was obviously associated with an increased risk of PTC in the Kazakh population [13]. This study is the first to confirm that PTCSC2 rs925489 was notably associated with increased susceptibility to THCA risk in different genetic models. Interestingly, PTCSC2 rs925489 all evidently increased the risk of THCA in Chinese populations older than 44, males, smokers and drinkers. Taken together, genetic variation in PTCSC2 affects the risk of developing THCA.
SEMA4G is known to the semaphorin family and involved over 20 genes classified into 7 difference subfamilies. It was reported that the SEMA4G gene has a DNA damage-binding and repair function [25]. The rs4919510 is located on 10q24.31 in the SEMA4G gene intron region. Furthermore, Wu et al. performed a meta-analysis to report that rs4919510 was significantly related with improved PTC sensibility, and rs4919510 regulated SEMA4G [14]. In this study, stratified analysis also demonstrated that SEMA4G rs4919510 was evidently associated with a reduced risk of THCA among Chinese participants less than or equal to 44 years old and non-drinkers, indicating that rs4919510 significantly reduced the risk of THCA.
Genetic variations affecting THCA susceptibility are related to age, sex, smoking and alcohol consumption. Previous studies have shown that PCNXL2 SNPs can increase THCA risk in population older than 45 and reduce the risk of THCA among females or participants with less than or equal to 45 years old [26]. Furthermore, IL1A SNPs were identified as biomarkers of THCA risk in males or individuals age ≤ 48 years, while IL1B SNPs detected strong correlations with THCA susceptibility among women and population aged > 48 years [27]. Similar to this findings, our study revealed that TPO rs2048722 had higher THCA risk in participants age ≤ 44 years or smokers; PTCSC2 rs925489 was also a risk factor for THCA susceptibility among population age > 44 years, men, smokers or drinker; and SEMA4G rs4919510 reduced THCA risk in recruiter age ≤ 44 years or non-drinkers. In a word, genetic variations to THCA susceptibility may be due to the involvement of age, sex, smoking, and drinking.
In this study, the association between TPO rs2048722, PTCSC2 rs925489, SEMA4G rs4919510 polymorphisms and THCA susceptibility was explored in the Chinese population, but limitations remained. The study only studied the THCA susceptibility gene in the Chinese population, and further studies on other populations still need to be explored. In addition, it is still necessary to explore the effects of TPO, PTCSC2 and SEMA4G expression on the biological functions and regulatory pathways related to the pathogenesis and treatment of THCA at the animal and cellular levels in the later stage of the study.
Conclusions
In summary, by investigation of Chinese population of THCA patients and unrelated healthy controls, the association of TPO rs2048722, SEMA4G rs4919510, PTCSC2 rs925489 polymorphism and TC susceptibility was demonstrated. Our study shown that PTCSC2 rs925489 were observed with an increasing risk factor of THCA in the overall analysis. Stratified analysis results found that PTCSC2 rs925489 increased the risk of THCA in the Chinese population older than 44 years, males, smokers and drinkers. TPO rs2048722 was an obvious risk locus of THCA in Chinese population with less than or equal to 44 years old and smokers. Nevertheless, SEMA4G rs4919510 was evidently associated with a reduced risk of THCA in Chinese population with less than or equal to 44 years old and non-drinkers. The purpose of this study was to find the key markers of the occurrence and treatment of THCA, in order to achieve personalized treatment.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the zenodo repository (https://zenodo.org/record/6668025#.Yq_MvPkaWUk).
References
Wen J, Gao Q, Wang N, Zhang W, Cao K, Zhang Q, et al. Association of microRNA-related gene XPO5 rs11077 polymorphism with susceptibility to thyroid cancer. Medicine. 2017;96(14): e6351.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Chmielik E, Rusinek D, Oczko-Wojciechowska M, Jarzab M, Krajewska J, Czarniecka A, et al. Heterogeneity of thyroid cancer. Pathobiology J Immunopathol Mol Cell Biol. 2018;85(1–2):117–29.
Hińcza K, Kowalik A, Kowalska AJG. Current knowledge of germline genetic risk factors for the development of non-medullary thyroid cancer. Genes. 2019;10(7):482.
Trimboli P, Piccardo A, Signore A, Valabrega S, Barnabei A, Santolamazza G, et al. Patient age is an independent risk factor of relapse of differentiated thyroid carcinoma and improves the performance of the American thyroid association stratification system. Thyroid. 2020;30(5):713–9.
Cho A, Chang Y, Ahn J, Shin H, Ryu S. Cigarette smoking and thyroid cancer risk: a cohort study. Br J Cancer. 2018;119(5):638–45.
Yeo Y, Han K, Shin DW, Kim D, Jeong SM, Chun S, et al. Changes in smoking, alcohol consumption, and the risk of thyroid cancer: a population-based Korean cohort study. Cancers. 2021;13(10).
Pozdeyev N, Gay LM, Sokol ES, Hartmaier R, Deaver KE, Davis S, et al. Genetic analysis of 779 advanced differentiated and anaplastic thyroid cancers. Clin Cancer Res. 2018;24(13):3059–68.
Jin M, Song DE, Ahn J, Song E, Lee YM, Sung TY, et al. Genetic profiles of aggressive variants of papillary thyroid carcinomas. Cancers. 2021;13(4):892.
Zhou W, Brumpton B, Kabil O, Gudmundsson J, Thorleifsson G, Weinstock J, et al. GWAS of thyroid stimulating hormone highlights pleiotropic effects and inverse association with thyroid cancer. Nat Commun. 2020;11(1):3981.
Jendrzejewski JP, Sworczak K, Comiskey DF, de la Chapelle A. Clinical implications of GWAS variants associated with differentiated thyroid cancer. Endokrynol Pol. 2019;70(5):423–9.
Tomari S, Watanabe M, Inoue N, Mizuma T, Yamanaka C, Hidaka Y, et al. The polymorphisms in the thyroid peroxidase gene were associated with the development of autoimmune thyroid disease and the serum levels of anti-thyroid peroxidase antibody. Endocr J. 2017;64(10):1025–32.
Mussazhanova Z, Rogounovitch TI, Saenko VA, Krykpayeva A, Espenbetova M, Azizov B, et al. The contribution of genetic variants to the risk of papillary thyroid carcinoma in the Kazakh population: study of common single nucleotide polymorphisms and their clinicopathological correlations. Front Endocrinol. 2020;11: 543500.
Wu S, Yuan W, Shen Y, Lu X, Li Y, Tian T, et al. The miR-608 rs4919510 polymorphism may modify cancer susceptibility based on type. 2017;39(6):1010428317703819.
Wang B, Wang Y, Wang L, He X, He Y, Bai M, et al. The role of FOXO3 polymorphisms in susceptibility to tuberculosis in a Chinese population. Mol Genet Genom Med. 2019:e770.
Li Y, Sun Y, Yang Q, Wu J, Xiong Z, Li S, et al. Variants in COL6A3 gene influence susceptibility to esophageal cancer in the Chinese population. Cancer Genet. 2019;238:23–30.
Feng Y, Su Y, Ma C, Jing Z, Yang X, Zhang D, et al. 3′ UTR variants of TNS3, PHLDB1, NTN4, and GNG2 genes are associated with IgA nephropathy risk in Chinese Han population. Int Immunopharmacol. 2019;71:295–300.
Accardo G, Conzo G, Esposito D, Gambardella C, Mazzella M, Castaldo F, et al. Genetics of medullary thyroid cancer: an overview. Int J Surg. 2017;41:S2–6.
Santos LS, Gomes BC, Bastos HN, Gil OM, Azevedo AP, Ferreira TC, et al. Thyroid cancer: the quest for genetic susceptibility involving DNA repair genes. Genes. 2019;10(8):586.
Godlewska M, Banga PJ. Thyroid peroxidase as a dual active site enzyme: focus on biosynthesis, hormonogenesis and thyroid disorders of autoimmunity and cancer. Biochimie. 2019;160:34–45.
Yu Y, Wang S, Zhang X, Xu S, Li Y, Liu Q, et al. Clinical implications of TPO and AOX1 in pediatric papillary thyroid carcinoma. Transl Pediat. 2021;10(4):723–32.
Jendrzejewski J, Liyanarachchi S, Nagy R, Senter L, Wakely PE, Thomas A, et al. Papillary thyroid carcinoma: association between germline DNA variant markers and clinical parameters. Thyroid. 2016;26(9):1276–84.
Wang Y, He H, Li W, Phay J, Shen R, Yu L, et al. MYH9 binds to lncRNA gene PTCSC2 and regulates FOXE1 in the 9q22 thyroid cancer risk locus. Proc Natl Acad Sci USA. 2017;114(3):474–9.
Hińcza K, Kowalik A, Kowalska A. Current knowledge of germline genetic risk factors for the development of non-medullary thyroid cancer. Genes. 2019;10(7):482.
Yong KJ, Milenic DE, Baidoo KE, Brechbiel MWJPO. Cell killing mechanisms and impact on gene expression by gemcitabine and 212Pb-trastuzumab treatment in a disseminated ip tumor model. PLoS One 2016; 11(7):e0159904.
Hao R, Han P, Zhang L, Bi Y, Yan J, Li H, et al. Genetic polymorphisms in the PCNXL2 gene are risk factors for thyroid cancer in the Chinese population. Fut Oncol (Lond Engl). 2021;17(34):4677–86.
Li H, Duan N, Zhang Q, Shao Y. IL1A & IL1B genetic polymorphisms are risk factors for thyroid cancer in a Chinese Han population. Int Immunopharmacol. 2019;76: 105869.
Acknowledgements
We are grateful to all participants for providing blood samples. We also appreciate the reviewers and editors for their efforts and patience.
Funding
No applicable.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. GN designed the research study. ZS wrote the first draft of the manuscript. LZ collected the samples needed for this study. YS performed material preparation, data collection and analysis and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the ethics committee of the First Affiliated Hospital of Xi'an Jiaotong University. Meanwhile, this study was conducted in accordance with the Declaration of Helsinki. Informed consent was obtained from all individual participants included in the study.
Consent for publication
The authors affirm that human research participants provided informed consent for publication of information in the First Affiliated Hospital of Xi'an Jiaotong University.
Competing interests
The authors have declared that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
SNPs primers and stratification of lymph node metastasis with THCA risk.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Shen, Z., Sun, Y. & Niu, G. Variants in TPO rs2048722, PTCSC2 rs925489 and SEMA4G rs4919510 affect thyroid carcinoma susceptibility risk. BMC Med Genomics 16, 19 (2023). https://doi.org/10.1186/s12920-023-01447-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-023-01447-5