Abstract
Background
Some patients infected with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) go on to experience post-COVID-19 condition or long COVID. Preliminary findings have given rise to the theory that long COVID may be due in part to a deranged immune response. In this study, we assess whether there is an association between SARS-CoV-2 infection and the incidence of immune-mediated inflammatory diseases (IMIDs).
Methods
Matched cohort study using primary care electronic health record data from the Clinical Practice Research Datalink Aurum database. The exposed cohort included 458,147 adults aged 18 years and older with a confirmed SARS-CoV-2 infection and no prior diagnosis of IMIDs. They were matched on age, sex, and general practice to 1,818,929 adults with no diagnosis of confirmed or suspected SARS-CoV-2 infection. The primary outcome was a composite of any of the following IMIDs: autoimmune thyroiditis, coeliac disease, inflammatory bowel disease (IBD), myasthenia gravis, pernicious anaemia, psoriasis, rheumatoid arthritis (RA), Sjogren’s syndrome, systemic lupus erythematosus (SLE), type 1 diabetes mellitus (T1DM), and vitiligo. The secondary outcomes were each of these conditions separately. Cox proportional hazard models were used to estimate adjusted hazard ratios (aHR) and 95% confidence intervals (CI) for the primary and secondary outcomes, adjusting for age, sex, ethnic group, smoking status, body mass index, relevant infections, and medications.
Results
Six hundred and nighty six (0.15%) and 2230 (0.12%) patients in the exposed and unexposed cohort developed an IMID during the follow-up period over 0.29 person-years, giving a crude incidence rate of 4.59 and 3.65 per 1000 person-years, respectively. Patients in the exposed cohort had a 22% increased risk of developing an IMID, compared to the unexposed cohort (aHR 1.22, 95% CI 1.12 to 1.33). The incidence of three IMIDs was significantly associated with SARS-CoV-2 infection. These were T1DM (aHR 1.56, 1.09 to 2.23), IBD (aHR 1.36, 1.18 to 1.56), and psoriasis (1.23, 1.05 to 1.42).
Conclusions
SARS-CoV-2 was associated with an increased incidence of IMIDs including T1DM, IBD and psoriasis. However, these findings could be potentially due to ascertainment bias. Further research is needed to replicate these findings in other populations and to measure autoantibody profiles in cohorts of individuals with COVID-19.
Similar content being viewed by others
Summary box
What is already known on this topic
-
A subsection of the population infected with SARS-CoV-2 go on to experience post-COVID-19 condition or long COVID.
-
Preliminary findings, such as case reports of post-COVID-19 immune-mediated inflammatory diseases, increased autoantibodies in COVID-19 patients, and molecular mimicry of the SARS-CoV-2 virus have given rise to the theory that long COVID may be due in part to a deranged immune response.
What this study adds
-
SARS-CoV-2 infection was associated with a 22% relative increase in the risk of developing certain immune-mediated inflammatory diseases, including type 1 diabetes mellitus, inflammatory bowel disease, and psoriasis.
-
These findings support the hypothesis that a subgroup of long COVID may be caused by immune-mediated inflammatory mechanisms.
Background
Emerging in late 2019, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the virus causing the coronavirus disease-2019 (COVID-19) pandemic, as of March 2023, resulted in over 6 million deaths worldwide [1, 2]. The acute presentation can range from being completely asymptomatic to sepsis, organ failure and death [3]. The effects of COVID-19 are not limited solely to acute infection but have also manifested in a series of post-acute sequelae commonly referred to as long COVID or post-COVID-19 condition [4, 5].
The World Health Organisation define this as symptoms occurring in people with a history of probable or confirmed SARS-CoV-2 infection three months after the onset of COVID-19 that cannot be explained by an alternative diagnosis [5, 6]. With over a third of people with COVID-19 reporting persistent symptoms and over 1.7 million UK residents self-reporting the condition, long COVID is emerging as one of the major public health challenges of the modern era [7, 8]. Despite this, the pathogenesis behind the condition remains unclear [4, 9].
One theory is that SARS-CoV-2 infection causes an inappropriate immune response that leads to the varied symptoms of long COVID. This arose from evidence of a marked and persistent increase in autoantibodies in patients with COVID-19 compared to uninfected controls and high rates of patients hospitalised with COVID-19 being transiently positive for anti-phospholipid (aPL) antibodies [10, 11].
Some of these autoantibodies were also deemed as potential risk factors for long COVID [12]. Several systematic reviews have collated case reports of patients with a history of COVID-19 who have experienced deranged immune manifestations. Tang et al. found 187 reports and Novelli et al. found 382 reports of autoimmune-like phenomena following COVID-19 [13, 14]. Among those with a history of COVID-19, one review reported thyroid dysfunction in up to 20% of patients, which is linked with B and T-cell autoimmunity [15].
Autoimmunity may be due to the degree of homology existing between some human self-proteins and components of SARS-CoV-2, a phenomenon termed molecular mimicry [16]. Molecular mimicry combined with the immune system dysregulation that occurs during SARS-CoV-2 infection may be the mechanism driving the development of immune-mediated inflammatory diseases. Alternatively, the reaction could arise from tissue damage and the release of autoantigens as a result of SARS-CoV-2 infection.
This preliminary evidence has been derived largely from case series, case reports, small cohort studies, or systematic reviews of these study types, which are weak study designs for ascertaining causal inference. Stronger study designs are needed that include appropriate control groups and large sample sizes. Furthermore, the data were drawn largely from patients with moderate or severe COVID-19, which underrepresents the mild or asymptomatic cases that make up most SARS-CoV-2 infections and that can also go on to develop long COVID [17]. To address these limitations, we conducted a retrospective matched cohort study using data from a large primary care database to assess the incidence of immune-mediated inflammatory diseases (IMIDs) in patients with SARS-CoV-2 infection compared to matched individuals with no record of SARS-CoV-2 infection.
Methods
Study design and data source
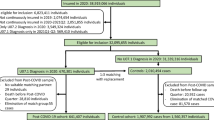
A retrospective cohort study was undertaken using data extracted from the Clinical Practice Research Datalink (CPRD) Aurum database between the 31st of January 2020 and the 30th of June 2021. The CPRD Aurum database consists of routinely collected, pseudo-anonymised data from general practices across England [18]. The data were extracted using the data extraction for epidemiological research (DExtER) tool, which facilitates extraction based on predefined parameters [19].
Study population
Patients were eligible to enter the study if they were at least 18 years old at the study start date, had no prior history of the IMIDs included in the primary outcome (see below), had an acceptable patient flag indicating provision of good quality data, and if they were registered with an eligible general practice for at least 12 months to allow sufficient time for recording baseline information.
Exposure
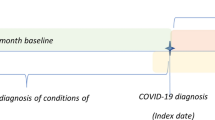
All patients with a SNOMED-CT coded diagnosis of either a positive reverse transcriptase polymerase chain reaction (RT-PCR) or lateral flow antigen test for SARS-CoV-2 were included in the exposed cohort, and the date of coded diagnosis was assigned as the index date. Patients with a suspected COVID-19 diagnosis were not included to increase the specificity of the exposure definition. For each exposed patient, up to four patients were selected who did not have a coded record of a positive RT-PCR or lateral flow antigen test, or a diagnosis of suspected or confirmed diagnosis of COVID-19, and were matched on age, sex and registered general practice. This made up the unexposed cohort. The same index date of the exposed patients was assigned to the corresponding matched unexposed patients to avoid immortal time bias [20]. Data from the COVID-19 Second Generation Surveillance System was not used for this study as it comprised of data from swab testing in Public Health England (PHE) labs and NHS hospitals primarily for hospitalised patients and healthcare workers as opposed to data from the wider population which was required for this study.
Outcomes
The primary outcome was a composite of the incidence of any of the following IMIDS: autoimmune thyroiditis, coeliac disease, inflammatory bowel disease (IBD), myasthenia gravis, pernicious anaemia, psoriasis, rheumatoid arthritis (RA), Sjogren’s syndrome, systemic lupus erythematosus (SLE), type 1 diabetes mellitus (T1DM), and vitiligo. These conditions were selected as they cover a range of different systems and constitute many of the most prevalent IMIDs in the UK. The secondary outcomes were the individual diseases included in the primary outcome, to discern which of these IMIDs, if any, had the strongest association with SARS-CoV-2 infection. SNOMED-CT code lists used for the ascertainment of each IMID, as well as the exposure codes, are given at https://github.com/Umer-Syed/COVIDAutoimmune. In light of the CPRD policy on data governance, we have not reported outcomes that had below five events due to disclosure risk.
Follow-up period
Participants were followed up from the index date to the end of the follow-up. The end of follow-up was defined as the earliest of any of the following: a coded diagnosis of an IMID, date of death, study end date (30 June 2021), date of practice de-registration, and date of the last practice contribution to the CPRD Aurum database.
Covariates
Age, sex, body mass index (BMI), smoking status, ethnicity, previous exposure to relevant viral infections (Epstein-Barr virus (EBV), human cytomegalovirus (CMV), human herpesvirus 6 (HHV-6), human T lymphotropic virus type 1 (HTLV-1), hepatitis C virus (HCV), influenza A virus, and parvovirus B19), and previous prescriptions of selected medications (procainamide, hydralazine, quinidine, and isoniazid) were included as potential confounders. Previous studies found these variables to be associated with at least one of the outcome IMIDs and were thus adjusted for in the analysis [21,22,23,24,25,26,27,28,29,30,31,32,33].
Age was divided into the following bands: 18 to 29, 30 to 39, 40 to 49, 50 to 59, 60 to 69, and ≥ 70 years. Ethnicity was identified through SNOMED CT codes and was classified into the following groups: white, South Asian, black, mixed ethnicity and other. BMI was divided in accordance with the WHO classification: underweight (body mass index (BMI) < 18.5 kg/m2), normal weight (18.5–24.9 kg/m2), overweight (25–29.9 kg/m2) and obese (≥ 30 kg/m2) [34]. Smoking status was categorised as current smoker, ex-smoker and never smoked. A separate ‘data missing’ category was used where data were missing for ethnicity, smoking status, and BMI.
Statistical analysis
Baseline characteristics of patients stratified by their exposure status were summarised using simple descriptive statistics. The number and percentage of each of the outcome events for the unexposed and exposed cohorts were reported and the crude incidence rates per 1000 person-years were calculated. Cox proportional hazards regression models were used to estimate the unadjusted and adjusted hazard ratios (HRs) with 95% confidence intervals (CI), for each of the outcomes among patients in the exposed and unexposed cohorts. P-values below 0.05 were considered statistically significant. In order to ensure our analysis was valid, a calculation to determine the Schoenfeld residual was undertaken. If this test yielded a value of < 0.05, then the data was not normally distributed and thus the proportional hazard assumption would not be met. All analyses were conducted using Stata Version 17, the do-file for this is given at https://github.com/Umer-Syed/COVIDAutoimmune.
Results
Study population
We identified 458,147 patients with confirmed SARS-CoV-2 infection and matched them to 1,818,929 patients who lacked a confirmed or suspected diagnosis of COVID-19. Table 1 shows the baseline characteristics of patients in both cohorts. The mean age was 43.6 years (SD 17.1) in the exposed cohort and 42.8 (SD 18.0) in the unexposed cohort. Both groups had slightly more females than males (54.7% versus 45.3%, respectively). A slightly larger proportion of the exposed cohort were of white and South Asian ethnicity compared to the unexposed group (64.4% versus 59.4%, and 12.2% versus 10.6%, respectively). However, the unexposed cohort had a slightly higher amount of missing ethnicity data (21.6% versus 16.2%, respectively). The mean BMI was similar between groups but there were slightly more current smokers in the unexposed cohort (26.5% versus 22.1%, respectively). Exposure to the selected infections and medications was similar between both groups.
Primary analysis
Six hundred ninety-six (0.15%) patients in the exposed cohort developed the primary outcome compared to 2230 (0.12%) within the unexposed cohort. The median (interquartile range [IQR]) follow-up was 0.29 years (0.24–0.42) for both groups. The results of the primary analysis are reported in Table 2 and Fig. 1. The crude incidence rate (IR) per 1000 person-years was higher for the exposed cohort than the unexposed cohort (4.59 versus 3.65 per 1000 person-years, respectively). This yielded a crude hazard ratio of 1.26 (95% CI 1.16–1.37) for the composite primary outcome. When adjusted for pre-selected covariates, the HR slightly reduced to 1.22 (1.12–1.33) but remained statistically significant. The proportional hazard assumption was met based on Schoenfeld residuals for the composite outcome. Furthermore, a matched analysis yielded a hazard ratio of 1.25 (95% CI 1.15–1.36). Characteristics of patients stratified by their primary outcome status have been tabulated in Additional file 1: Supplementary Table 2.
Secondary analysis
Table 3 and Fig. 1 report the results for each individual IMID as separate outcomes. Of the eleven conditions, SARS-CoV-2 infection was significantly associated with an increased incidence of T1DM, IBD and psoriasis. T1DM was 56% more likely to occur in the exposed cohort compared to the unexposed cohort (aHR 1.56, 95% CI 1.09 to 2.23). IBD was 36% more likely to occur in the exposed cohort compared to the unexposed cohort (aHR 1.36, 95% CI 1.18 to 1.56). This was the most common IMID to be diagnosed during the study period (39.6% of all IMIDs diagnosed in the exposed cohort and 36.6% in the unexposed cohort). Psoriasis was 23% more likely to occur in the exposed cohort compared to the unexposed cohort (1.23, 1.05 to 1.42) and was the second most diagnosed IMID, representing more than 30% of all new diagnoses of IMIDs in both cohorts.
Discussion
Main findings
Exposure to SARS-CoV-2 infection was associated with a 22% relative increase in the incidence of any of the eleven IMIDs considered in our study compared to a matched unexposed group during the same period. This was after adjustment for several important confounding factors and during a relatively short period of follow-up. We also found that this association was specific to an increased incidence of T1DM, inflammatory bowel disease, and psoriasis in the SARS-CoV-2 infected cohort.
Comparison with existing literature
The relatively high incidence of psoriasis in the SARS-CoV-2 infected cohort is supported by other reports from the literature which found increased cases of psoriasis, and flares of existing disease, following COVID-19 [13]. Evidence on the incidence of IBD following COVID-19 is scarcer, although ulcerative colitis has been reported to develop post-infection [13]. A systematic review on T1DM and COVID-19 noted that between 1.77 and 15.6% of newly diagnosed patients, depending on the study, had preceding COVID-19 [35].
SARS-CoV-2 may be associated with IMIDs due to several putative mechanisms that result in the release of autoantibodies following infection. All three conditions that were found to have a significantly increased incidence following SARS-CoV-2 infection in our study have at least a limited association with autoantibodies. T1DM is associated with islet cells and other autoantibodies, psoriasis is linked with anti-nuclear antibodies (ANAs) and inflammatory bowel disease has a limited association with pancreatic autoantibodies (PAB) [36,37,38]. The reason for the increased incidence of these conditions following SARS-CoV-2 infection is unclear as they are not typically the most strongly associated with the presence of autoantibodies. This requires further exploration in future mechanistic studies.
Strengths and limitations
A large sample size was included, which provided sufficient statistical power to assess for differences in the incidence of IMIDs between the exposed and unexposed cohorts over a relatively short follow-up period. This also allowed us to assess the relative incidence of eleven of the more common IMIDs across the two comparison groups. We included IMIDs in our outcome such as T1DM, that are likely to be well-recorded in primary care records. The use of primary care data meant that we were able to adjust for important demographic and clinical risk factors that are known to be associated with the incidence of IMIDs. The use of data from practices across a national database also improved the generalisability of our findings.
The study had several limitations. We had missing data for ethnicity (22% missing), BMI (18%), and smoking status (7%), which we accounted for in our analyses using a missing category variable. However, these missing data could lead to biased effect estimates. We also did not have access to data on socioeconomic status but partially accounted for this by matching patients in the unexposed and exposed cohorts on general practice, which would result in patients from both groups sharing their approximate residential geography, which is associated with socioeconomic status.
There is likely to be a degree of misclassification bias between the exposed and unexposed cohorts. There was little community testing for SARS-CoV-2 infection in the first wave of the pandemic, so some members of the unexposed cohort may have been infected but not diagnosed. IMIDs may also have been underdiagnosed during the study period due to the relative inaccessibility of healthcare services during the early phase of the pandemic. It is possible that only more severely affected patients with IMIDs presented to healthcare services during this period.
The study period was restricted as data availability only covered from 31 January 2020 to 30 June 2021. This encompassed three national lockdowns where reduced healthcare appointments led to a backlog of up to 300,000 patients waiting over a year for treatment [39, 40]. Beyond this period, there was reduced availability of community testing for SARS-CoV-2 infection in the UK at a time when an increasing proportion of the population had experienced at least one episode of COVID-19, thus diminishing future comparator populations.
The short follow-up period may have diluted the effect size and power of the study as IMIDs tend to have a clinical latency period and thus the full scope of the potential impact of SARS-CoV-2 infections is likely to have been underrepresented [41]. It also cannot be confirmed whether the true onset of these conditions preceded SARS-CoV-2 infection or the matched index dates. However, we would expect these issues to equally bias our estimates of disease incidence in both the exposed and unexposed cohorts and would therefore not anticipate it affecting the hazard ratios. There also exists the possibility that patients experiencing COVID-19 may have accessed healthcare services more than those with no prior infection and thus had more opportunities to be diagnosed with IMIDs. Likewise, patients with underlying IMIDs may have had their symptoms exacerbated by COVID-19 which resulted in seeking healthcare services and subsequent diagnosis.
Implications for practice, policy, and research
Our findings provide epidemiological evidence that SARS-CoV-2 infection is associated with an increased risk of a range of IMIDs, including T1DM, IBD, and psoriasis. This provides evidence that autoimmunity may be a potential mechanism that accounts for some of the longer-term symptoms and health impacts of a subgroup of those with long COVID. This is particularly of interest given the finding that women are generally at increased risk of both IMIDs as well as Long COVID, that symptoms of long COVID are diverse and often overlap with those of IMIDs, and that the symptoms of both IMIDs and long COVID characteristically follow a relapsing–remitting pattern over time [42].
Further epidemiological studies with a longer follow-up period are needed to confirm our findings and to test for relevant autoantibodies in the serum of participants to correlate with symptoms and clinical findings. These studies could also include other rarer IMIDs potentially associated with COVID-19 such as Guillain-Barré syndrome [14]. Evidence suggests that those who have been vaccinated against COVID-19 are approximately half as likely to develop symptoms lasting over 28 days than unvaccinated individuals [43]. It would be valuable to know if these differences in long COVID incidence rates are also associated with differences in the incidence of IMIDs.
Conclusions
SARS-CoV-2 infection was associated with an increased incidence of several IMIDs, including type 1 diabetes mellitus, inflammatory bowel disease, and psoriasis. This lends support to the hypothesis that the long-term effects of COVID-19 or long COVID may in part be related to autoimmune mechanisms. Further research is needed to replicate these findings in other populations, over a longer time period and to sample autoantibody profiles in people with long COVID and matched control groups.
Availability of data and materials
Access to anonymized patient data from CPRD is subject to a data-sharing agreement containing detailed terms and conditions of use following protocol approval from the MHRA Independent Scientific Advisory Committee. The dataset for this study is not publicly available but may be requested from the corresponding author at a.subramanian@bham.ac.uk if in accordance with data governance approvals. Details about Independent Scientific Advisory Committee applications and data costs are available on the CPRD website (cprd.com).
References
Hu B, Guo H, Zhou P, Shi Z-L. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19(3):141–54.
WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int. Accessed 19 Apr 2022.
Liu Y, Sawalha AH, Lu Q. COVID-19 and autoimmune diseases. Curr Opin Rheumatol. 2021;33(2):155–62.
Burke MJ, del Rio C. Long COVID has exposed medicine’s blind-spot. Lancet Infect Dis. 2021;21(8):1062–4.
Soriano JB, Murthy S, Marshall JC, Relan P, Diaz JV. A clinical case definition of post-COVID-19 condition by a Delphi consensus. Lancet Infect Dis. 2022;22(4):e102–7.
National Institute for Health and Care Excellence (NICE). COVID-19 rapid guideline: managing the longterm effects of COVID-19 NICE; 2022.
Taquet M, Dercon Q, Luciano S, Geddes JR, Husain M, Harrison PJ. Incidence, co-occurrence, and evolution of long-COVID features: a 6-month retrospective cohort study of 273,618 survivors of COVID-19. PLoS Med. 2021;18(9):e1003773.
Ayoubkhani D, Gaughan C. Updated estimates of the prevalence of post-acute symptoms among people with coronavirus (COVID-19) in the UK: 26 April 2020 to 1 August 2021. Newport: Office for National Statistics; 2021.
The L. Understanding long COVID: a modern medical challenge. Lancet. 2021;398(10302):725.
Wang EY, Mao T, Klein J, Dai Y, Huck JD, Jaycox JR, et al. Diverse functional autoantibodies in patients with COVID-19. Nature. 2021;595(7866):283–8.
Zuo Y, Estes SK, Ali RA, Gandhi AA, Yalavarthi S, Shi H, et al. Prothrombotic autoantibodies in serum from patients hospitalized with COVID-19. Sci Transl Med. 2020;12(570):eabd3876.
Su Y, Yuan D, Chen DG, Ng RH, Wang K, Choi J, et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell. 2022;185(5):881-95.e20.
Tang K-T, Hsu B-C, Chen D-Y. Autoimmune and rheumatic manifestations associated with COVID-19 in adults: an updated systematic review. Front Immunol. 2021;12:645013.
Novelli L, Motta F, De Santis M, Ansari AA, Gershwin ME, Selmi C. The JANUS of chronic inflammatory and autoimmune diseases onset during COVID-19 – a systematic review of the literature. J Autoimmun. 2021;117:102592.
Yong SJ. Long COVID or post-COVID-19 syndrome: putative pathophysiology, risk factors, and treatments. Infect Dis. 2021;53(10):737–54.
Kanduc D, Shoenfeld Y. Molecular mimicry between SARS-CoV-2 spike glycoprotein and mammalian proteomes: implications for the vaccine. Immunol Res. 2020;68(5):310–3.
Burns PB, Rohrich RJ, Chung KC. The levels of evidence and their role in evidence-based medicine. Plast Reconstr Surg. 2011;128(1):305–10.
Wolf A, Dedman D, Campbell J, Booth H, Lunn D, Chapman J, et al. Data resource profile: Clinical Practice Research Datalink (CPRD) Aurum. Int J Epidemiol. 2019;48(6):1740-g.
Gokhale KM, Chandan JS, Toulis K, Gkoutos G, Tino P, Nirantharakumar K. Data extraction for epidemiological research (DExtER): a novel tool for automated clinical epidemiology studies. Eur J Epidemiol. 2021;36(2):165–78.
Lévesque LE, Hanley JA, Kezouh A, Suissa S. Problem of immortal time bias in cohort studies: example using statins for preventing progression of diabetes. BMJ. 2010;340:b5087.
Agrawal A, Sridharan A, Prakash S, Agrawal H. Dendritic cells and aging: consequences for autoimmunity. Expert Rev Clin Immunol. 2012;8(1):73–80.
Amador-Patarroyo MJ, Rodriguez-Rodriguez A, Montoya-Ortiz G. How does age at onset influence the outcome of autoimmune diseases? Autoimmune Dis. 2012;2012:251730.
Beeson PB. Age and sex associations of 40 autoimmune diseases. Am J Med. 1994;96(5):457–62.
Fairweather D, Frisancho-Kiss S, Rose NR. Sex differences in autoimmune disease from a pathological perspective. Am J Pathol. 2008;173(3):600–9.
Calixto OJ, Anaya JM. Socioeconomic status. The relationship with health and autoimmune diseases. Autoimmun Rev. 2014;13(6):641–54.
Versini M, Jeandel P-Y, Rosenthal E, Shoenfeld Y. Obesity in autoimmune diseases: not a passive bystander. Autoimmun Rev. 2014;13(9):981–1000.
Costenbader KH, Karlson EW. Cigarette smoking and autoimmune disease: what can we learn from epidemiology? Lupus. 2006;15(11):737–45.
Cooper GS, Stroehla BC. The epidemiology of autoimmune diseases. Autoimmun Rev. 2003;2(3):119–25.
Moroni L, Bianchi I, Lleo A. Geoepidemiology, gender and autoimmune disease. Autoimmun Rev. 2012;11(6):A386–92.
Smatti MK, Cyprian FS, Nasrallah GK, Al Thani AA, Almishal RO, Yassine HM. Viruses and autoimmunity: a review on the potential interaction and molecular mechanisms. Viruses. 2019;11(8):762.
Hussein HM, Rahal EA. The role of viral infections in the development of autoimmune diseases. Crit Rev Microbiol. 2019;45(4):394–412.
Araújo-Fernández S, Ahijón-Lana M, Isenberg DA. Drug-induced lupus: including anti-tumour necrosis factor and interferon induced. Lupus. 2014;23(6):545–53.
Chang C, Gershwin ME. Drug-induced lupus erythematosus: incidence, management and prevention. Drug Saf. 2011;34(5):357–74.
National Institute for Health and Care Excellence (NICE). Obesity: identifying, assessing and managing obesity in adults, young people and children. 2014. https://www.nice.org.uk/guidance/cg189. Accessed 25 Apr 2022
Nassar M, Nso N, Baraka B, Alfishawy M, Mohamed M, Nyabera A, et al. The association between COVID-19 and type 1 diabetes mellitus: a systematic review. Diabetes Metab Syndr. 2021;15(1):447–54.
Taplin CE, Barker JM. Autoantibodies in type 1 diabetes. Autoimmunity. 2008;41(1):11–8.
ten Bergen LL, Petrovic A, Aarebrot AK, Appel S. Current knowledge on autoantigens and autoantibodies in psoriasis. Scand J Immunol. 2020;92(4):e12945.
Joossens S, Vermeire S, Van Steen K, Godefridis G, Claessens G, Pierik M, et al. Pancreatic autoantibodies in inflamm bowel dis. Inflamm Bowel Dis. 2004;10(6):771–7.
Brown J, Kirke-Wade E. Coronavirus: a history of English lockdown laws. London: The Commons Library. 2021. https://commonslibrary.parliament.uk/research-briefings/cbp-9068/. Accessed 17 Aug 2023.
House of Commons Health and Social Care Committee. Clearing the backlog caused by the pandemic. London: UK Parliament; 2021. https://committees.parliament.uk/publications/8352/documents/85020/default. Accessed 18 Aug 2023
Rosenblum MD, Remedios KA, Abbas AK. Mechanisms of human autoimmunity. J Clin Invest. 2015;125(6):2228–33.
Subramanian A, Nirantharakumar K, Hughes S, Myles P, Williams T, Gokhale KM, et al. Symptoms and risk factors for long COVID in non-hospitalized adults. Nat Med. 2022;28:1706–14.
UK Health Security Agency. The effectiveness of vaccination against long COVID A rapid evidence briefing. London: UK HSA; 2022. https://www.icpcovid.com/sites/default/files/2022-02/Ep%20241-9%20UK%20Health%20Security%20Agency%20The%20effectiveness%20of%20vaccination%20against%20long%20COVID%20Feb%202022.pdf. Accessed 18 Aug 2023
Acknowledgements
Not applicable.
Declarations
The lead author (SH) affirms that this manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
Funding
This study was not externally funded.
Author information
Authors and Affiliations
Contributions
SH and AS conceptualised the research question. US conducted the study under the supervision of AS and SH. DW, JL, and KM provided expert input on immunology and autoimmunity. KN and KG provided expertise on data science and data management. All authors reviewed and approved the submitted form of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study consists of secondary data analysis of pseudo-anonymised data from the CPRD Aurum database and thus approval was only required to access the database. The project was reviewed and approved by the CPRD Independent Scientific Advisory Committee (study #21_000712) on the 24th of February 2022.
Consent for publication
Not applicable.
Competing interests
SH reports receiving funding from NIHR and UKRI. US report no competing interests. AS has no declarations during the study period; after the study was completed, she has taken a post in AstraZeneca. JL receives grant funding from NIHR, UKRI, Versus Arthritis, The Scar Free Foundation, FOREUM, and Bayer Healthcare for which also she acts as a consultant.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Table 1.
Adjusted hazard ratios for other risk factors included in the Cox proportional hazards model. Supplementary Table 2. Population characteristics by outcome cohort. The RECORD statement: A checklist of items, extended from the STROBE statement, that should be reported in observational studies using routinely collected health data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Syed, U., Subramanian, A., Wraith, D.C. et al. Incidence of immune-mediated inflammatory diseases following COVID-19: a matched cohort study in UK primary care. BMC Med 21, 363 (2023). https://doi.org/10.1186/s12916-023-03049-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12916-023-03049-5