Abstract
Background
In filamentous fungi, transport of organic acids across the mitochondrial membrane is facilitated by active transport via shuttle proteins. These transporters may transfer different organic acids across the membrane while taking others the opposite direction. In Aspergillus niger, accumulation of malate in the cytosol can trigger production of citric acid via the exchange of malate and citrate across the mitochondrial membrane. Several mitochondrial organic acid transporters were recently studied in A. niger showing their effects on organic acid production.
Results
In this work, we studied another citric acid producing fungus, Aspergillus carbonarius, and identified by genome-mining a putative mitochondrial transporter MtpA, which was not previously studied, that might be involved in production of citric acid. This gene named mtpA encoding a putative oxaloacetate transport protein was expressed constitutively in A. carbonarius based on transcription analysis. To study its role in organic acid production, we disrupted the gene and analyzed its effects on production of citric acid and other organic acids, such as malic acid. In total, 6 transformants with gene mtpA disrupted were obtained and they showed secretion of malic acid at the expense of citric acid production.
Conclusion
A putative oxaloacetate transporter gene which is potentially involved in organic acid production by A. carbonarius was identified and further investigated on its effects on production of citric acid and malic acid. The mtpA knockout strains obtained produced less citric acid and more malic acid than the wild type, in agreement with our original hypothesis. More extensive studies should be conducted in order to further reveal the mechanism of organic acid transport as mediated by the MtpA transporter.
Similar content being viewed by others
Background
Bio-based production of organic acids by microorganisms as e.g. filamentous fungi in a biorefinery has a high potential as a substitution of chemicals produced from crude oil [1]. The organic acids (e.g. malic acid and succinic acid) can easily be converted and used as building blocks for deriving different commodity and specialty chemicals and in the past decades, filamentous fungi as Aspergillus niger and Aspergillus oryzae have been used in industrial production of organic acids. Recently, Aspergillus carbonarius was reported to be an efficient organic acid producer and as such may have a potential for bio-based production of C4-dicarboxylic acids. When several genetic modifications were made to improve carbon flux towards dicarboxylic acid production (fumaric, succinic and malic acid), an increased production of citric acid was often observed [2,3,4]. This phenomenon might be a result of the transport of organic acids across the mitochondrial membrane where e.g. malic acid is transported to the mitochondria in exchange with citric acid. Recently, we identified a plasma membrane C4-dicarboxylate transporter, which was highly involved in the dicarboxylic acid production [5]. Overexpression of the transporter led to a significant increase of C4-dicarboxylic acid production and decreased citric acid production.
In the well-known organic acid producer, A. niger, which is phylogenetically related to A. carbonarius, the roles of mitochondrial transport of organic acid have been investigated in more details. A. niger is able to produce high amounts of organic acids (e.g. citric acid) from a broad range of substrates [6,7,8]. Considerable research efforts have been taken to reveal the mechanism of citric acid accumulation by the fungus and to redirect carbon flux from the production of citric acid towards other types of organic acids, e.g. itaconic acid production by genetically altered A. niger strains [9, 10]. In A. niger, citrate is produced in the mitochondria through the Tricarboxylic acid cycle (TCA cycle). Citrate is then transported out of the mitochondria into the cytosol by membrane bound citrate transporters [11]. A common belief is that these transporters are integral membrane proteins and function as antiporters where they exchange citrate from the mitochondria with other organic acids from the cytosol (e.g. cytosolic malate) [8, 12, 13]. It has been suggested that the onset of citric acid production is initiated in relation to the concentration of malate in the cytosol, which is then transported into the mitochondria in exchange for citrate. It was observed that malate was accumulated in the cytosol directly preceding start of production of citrate [12], and an A. niger strain transformed with malate dehydrogenase which, in theory, should produce elevated amounts of malate instead produced significantly higher amounts of citrate [8]. This supports the hypothesis that concentration of malate (or possible fumarate or succinate) in the cytosol will not result in secretion of these organic acids but instead produce an increased amount of citric acid due to a possible organic acid transport mechanism where C4-dicarboxylic acids are transported to the mitochondria in exchange with citric acid.
In Saccharomyces cerevisiae, it has been observed that the transport of citrate across the mitochondrial membrane is influenced not only by the concentration of malate, but also by the concentration of iso-citrate, succinate, and phosphoenolpyruvate [14, 15]. It is strongly suspected that the mitochondrial citrate transporter (CTP) is the primary transporter of citrate across the mitochondrial membrane in exchange of malate [16, 17]. In another Aspergillus species from the section Nigri, Aspergillus luchuensis the two putative CTP homologues ctpA and ctpB have been studied in some detail [18, 19]. The ctpB gene was shown to be non-expressed under citric acid production conditions, whereas ctpA was constitutively expressed in both A. luchuensis and A. niger [18, 20]. Although deletion of the ctpA gene resulted in serious growth effects of the resulting mutant strains, the effects on citric acid production were marginally [18, 19]. In addition to the mitochondrial citrate/malate antiporter, the role of two putative citrate/oxoglutarate antiporters was also studied in A. luchuensis [19] and one in A. niger [21]. In S. cerevisiae this antiporter is encoded by the yhm2 gene responsible for exporting citrate from mitochondria to cytosol in exchange of oxoglutarate [22]. Also in this case one of the homologues, yhmB, was shown to be hardly expressed, while the closest homologue to yhm2 (cocA), yhmA was also expressed constitutively in A. luchuensis and A.niger, [18, 20]. Deletion of yhmB showed no effect on citrate production, whereas yhmA and cocA deletion had a clear effect on citrate production, which was further aggravated in a conditional ctpA/yhmA double mutant, where malate and oxo-glutarate production was increased [19].
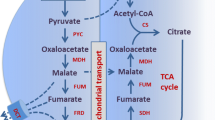
The hypothesis for the present study is based on the idea that when metabolic carbon-flux increases towards organic acids (e.g. dicarboxylic acids) other than citric acid in the cytosol in A. carbonarius, the accumulation of organic acids of interests will not happen due to transport of these acids (or their biosynthetic precursors) into the mitochondria by mitochondrial organic acid transporters in exchange of citrate (Fig. 1). If a transporter that is involved in exchange of organic acids is disrupted, the organic acid, which should be transported to the mitochondria in exchange with citrate, will remain in the cytosol, and in turn may be secreted to the exterior. The ability to reroute organic acid producing biocatalysts into production of more high-value acids would have great economic benefits since several organic acids are considered to be interesting building block chemicals [23]. In the present study, a putative mitochondrial oxaloacetate transporter gene was identified in A. carbonarius ITEM5010 based on a bioinformatic approach using characterized fungal organic acid transporters. Oxaloacetate is the key intermediate in the reductive tricarboxylic acid branch (rTCA branch) and the TCA cycle, which are highly involved in production of a number of organic acids e.g. citric acid, malic acid and succinic acid. In our work, the role of the oxaloacetate transport in organic acid production was investigated by disrupting the transporter gene and examining the changes in organic acid production by the knockout strain.
Results
Identification and expression of mtpA gene
Mitochondrial transporters of carboxylic acids are not well-characterized proteins in Aspergillus sp. To date only a few mitochondrial transporters of carboxylic acids have been studied in filamentous fungi [19, 24]. The number of candidate transporter proteins in a single fungal species is high, in yeast for example at least 34 different mitochondrial transporter proteins are known [25], and 39 are annotated in the A. niger genome [26]. The putative mitochondrial transporter gene investigated in the present study was identified using genome mining based on characterized mitochondrial proteins of fungal origin. For this purpose, homologues of the three genes identified in S. cerevisiae as being involved in citrate or citrate-intermediate transport, CTP1, YHM2 and OAC1 were identified by BLAST analysis in the genome of the most well characterized black Aspergillus genome, A. niger NRRL3 [27]. This resulted in the identification of 5 putative homologues, two for CTP1, two for YHM2 and one for OAC1. In Table 1, an overview of the expression data in A. niger available for these five genes in various publically available RNAseq data sets is given. From this analysis it is clear that the CTP1 and YHM2 homologues, ctpA and yhmA, are expressed to much higher and constitutive levels compared to ctpB or yhmB, respectively, confirming what was also already described by Kirimura et al. (2016) and Kadooka et al. (2018) [18, 19] . The OAC1 homologue was expressed to a similar level in all the studies represented in Table 1 (A. niger An14g06860 [20]. The putative A. carbonarius OAC1 orthologue, which we refer as mtpA was selected for further analysis. In order to confirm the expression of the mtpA gene in A. carbonarius, transcriptional analysis was carried out using reverse transcription polymerase chain reaction (RT-PCR). As shown in Additional file 1: Figure S1, a shorter fragment (~ 200 bp) was obtained by polymerase chain reaction (PCR) amplification of cDNA compared with genomic DNA, since the primers spanned the terminal part of the gene, where the cDNA in contrast to the genomic DNA contained no intron, confirming mtpA expression under these conditions.
Generation of knockout strains and southern blotting
The putative organic acid transporter gene mtpA in A. carbonarius ITEM 5010 was knocked out using a bipartite approach to create knockout strains. Disruption of the target gene in the selected transformants was first verified by PCR (data not shown). This was done with a forward primer binding upstream to the gene in the genome paired with a reverse primer binding to the inserted hygromycin gene, so generation of the predicted PCR product indicated that the transporter gene was replaced by the hygromycin resistant gene (Fig. 2a). In total, 32 transformants were obtained, and of these, 6 transformants were verified as positive knockout giving a gene targeting efficiency of 20%, Southern blot hybridization was then performed on the selected transformant mtpA-1, and the hybridization of probe only occurred with the expected DNA fragment as shown in Fig. 2b, confirming successful gene disruption via homologous recombination without any ectopic integration in other sites of the genome.
Verification of mtpA gene disruption in the transformant a) Disruption of mtpA gene with bipartite substrates in A. carbonarius. b Southern blotting analysis. Lane 1 hydridization of the probe to the genomic DNA fragment (from the wild-type) digested by EcoRI and HindIII, Lane 2, DNA ladder, Lane 3, hydridization of the probe to the genomic DNA fragment (from mtpA-1 transformant) digested by EcoRI and HindIII
Organic acid production
All 6 putative mtpA disruption strains were grown in shake flask cultures and analyzed for organic acid production by High-performance liquid chromatography (HPLC) analysis. As shown in Fig. 3, citric acid and malic acid were analyzed during the cultivation. All the transformants produced less citric acid than the wild type after day 5, meanwhile, a low but significant amount of malic acid was also detected from the culture with transformants but not from the wild type. No other major organic acid peaks were detected in the HPLC profile. The transformant mtpA-1 that was verified via southern blotting was also analyzed in a pH controlled fermentation. As shown in Table 2, it consumed 29 g/l glucose and produced 1.6 g/l citric acid and 0.35 g/l malic acid while the wild type consumed 31 g/l glucose and produced 2.6 g/l citric acid and no malic acid.
Discussion
A putative mitochondrial oxaloacetate transporter gene mtpA was identified in A. carbonarius and knocked out. The mtpA mutant was investigated for its impact on production of citric acid and malic acid. Disruption of the mtpA gene resulted in a reduction in citric acid production and secretion of malic acid in the culture medium. The hypothesis of this research study was that the disruption of this organic acid transporter could reduce the production of citric acid and simultaneously increase accumulation of malic acid or other dicarboxylic acids. Oxaloacetate is used in both cytosolic rTCA branch and mitochondrial TCA cycle respectively to produce malic acid and citric acid (Fig. 1). The deletion of mtpA may stop the transport of oxaloacetate into TCA cycle in mitochondria and save more carbon flux towards downstream rTCA branch in cytosol, which could be the reason for the measured increase in malic acid. However, only mild effects on production of citric acid and malic acid have been obtained. Apart from the fact that these effects may be larger in high citric acid production strains like A. niger H915–1 [28], a reason for this could also be that the MtpA membrane protein is part of a more complex network of transmembrane proteins involved in antiport and transport of different organic acids over the mitochondrial membrane. Similarly modest effects on organic acid production were also reported in the case of knocking out mitochondrial citrate transporters in A. niger and A. luchuensis. Citric acid production was also only slightly affected in ctpA knockout strain suggesting that other transporters also play important roles in the export of citric acid during the production phase [18]. On the other hand, deletion of the citrate/oxoglutarate antiporter yhmA had a more serious effect on citric acid and malic acid production, but also showed serious pleiotropic effects, making conclusion about its role in citric acid production somewhat ambiguous [18, 19]. It should also be noted that the expression of the genes encoding each of the citric acid related mitochondrial transporters is similar in high and low citric acid production strains and conditions (Table 2).
In the study presented here, the effect of deletion of the putative oxaloacetate carrier MtpA may be somewhat moderate due to a rerouting of the organic acid fluxes by other mitochondrial dicarboxylate transporters that can transport malate from the cytosol to mitochondria, where malate is the precursor for oxaloacetate synthesis in the TCA cycle and thus for citric acid production. Since shaking cultures, as performed in this study, are prone to variation, we carried out replicate cultures to determine the statistical relevance of the results obtained. A further more definitive conclusion would require further investigations, including controlled fermentation experiments in different scales and under different optimized conditions or knockouts of mtpA in combination with other putative transporter genes. Also more detailed in vitro transporter specificity studies as performed for A. luchuensis CtpA and YhmA [19], could be performed for MtpA to determine kinetic transport parameters and substrate specificity. In these studies, the function of a transporter was defined in the presence or absence of substrates or even radioactive labeled substrates as was recently performed for the first time for a fungal mitochondrial transporter [30].
Conclusions
A putative oxaloacetate transporter gene was identified in A. carbonarius and further investigated on its effects on citric and malic acid production. The mtpA knockout mutant obtained secreted malic acid at the expense of citric acid which was the only secreted organic acid in the wild type, in agreement with our original hypothesis for the effect of gene disruption on organic acid production.
Methods
Strains and growth conditions
A. carbonarius ITEM 5010 was used as the parental strain to construct transformants. All the fungal strains were cultivated as previously described in [31] . Escherichia coli strain DH5α was used as host for plasmid propagation.
Genomic DNA extraction and plasmid construction
Genomic DNA extraction from freshly grown mycelia and measurement of DNA concentration were carried out as described in [31]. The knockout plasmid pSB4.1.1 containing RP27-hph-βT for hygromycin resistance, origin of replication and ampicillin resistance for growth in E.coli and a specific cassette facilitating simpleUSER cloning was constructed as described in [2]. All primers used in the study were designed with the primer software Primer3 [32] (Table 3). In the knock out plasmid pSB4.1.1-Antiporter upstream and downstream regions to the target gene were identified in the A. carbonarius genome provided by JGI. The upstream and downstream flanking regions with the size of 1 kb were selected for efficient homologous recombination [33] and amplified with primers containing uracil tails (primers 1–4, Table 3). Genomic DNA from A. carbonarius was used as template. The flanking regions of gene were amplified by PCR using Pfu turbo cx polymerase (Agilent) and 65 °C as annealing temperature. The obtained PCR products were then ligated to the simpleUSER cassette in pSB4.1.1 that was already digested with the restriction enzyme PacI and the nicking enzyme Nb.BbvCI to create the complementary overhangs as previously described [2]. Through self-assembly the PCR fragments were cloned into the plasmid followed by transformation of E. coli with the plasmid for further propagation. Plasmid extraction with the GeneJET plasmid miniprep kit (ThermoFisher Scientific) was carried out according to the manufacturer’s protocol. All plasmids were verified by Sanger sequencing using the sequencing service from StarSEQ (Mainz, Germany).
Protoplast transformation
Protoplasts were prepared from wild type A. carbonarius following the procedure described in [4]. Bipartite PCR fragments were made from the knockout plasmids pSB4.1.1-Antiporter and primer 5–8 (Table 3) and prepared for transformation as described in [31]. Transformation was done in 100 μl aliquots of protoplasts according to the method described by [4]. Transformants carrying hygromycin resistance were isolated and transferred to PDA plates containing hygromycin. All the transformants were preserved by mixing spores suspension with 20% glycerol (final concentration in cryostock) and stored at − 80 °C. Validation of positive knockouts was carried out by PCR using primers 9–10 (Table 3).
Transcriptional analysis of mtpA gene
Transcription of the mtpA gene was analyzed by reverse-transcription PCR (RT-PCR). Total RNA purification and cDNA synthesis were prepared as previously described [5]. The cDNA was then used as template in PCR with mtpA gene specific primers no. 13–14 for transcription analysis.
Southern blotting of mtpA transformant
Southern blotting was carried out on a single mtpA transformant for confirmation of mtpA gene disruption. As shown in Fig. 2a, the hybridization probe was prepared by amplifying a part of the hph marker gene with primers 11–12 (Table 3) and labeled using a North2South chemiluminescent detection kit (Thermo Fisher Scientific). Genomic DNA of the transformant was digested by EcoRI and HindIII and hybridized with the probe. Southern blotting was then performed using Whatman Turboblotter transfer system (GE healthcare life sciences) and detected by using a Pierce Chemiluminescent Nucleic acid detection module Kit (Thermo scientific). The blot was imaged in Thermo ECL imager.
Fermentation and shake flask cultivation condition
Fermentation was performed in 50 ml Erlenmeyer flasks in 10 ml media in triplicates. Fresh spore-suspension was added to 10 ml pre-culture medium (Yeast extract 3.6 g/l and peptone 10 g/l) to a final concentration of 2 × 105 spores/ml. Pre-culture was incubated at 25 °C, 200 rpm and 2.5 cm amplitude for 48 h (KS 4000 I control, IKA). For the pH controlled fermentation, the mycelia pellets from the pre-culture were transferred to Erlenmeyer flasks with cotton stoppers and 10 ml production media [34] containing 100 g/l glucose, 2 g/l (NH4)2SO4, 0.15 g/l KH2PO4, 0.15 g/l K2HPO4, 0.1 g/l MgSO4∙7H2O, 0.1 g/l CaCl2∙2H2O, 0.005 g/l NaCl, 0.1 g/l ZnSO4, 0.005 g/l FeSO4·7H2O and 30 g/l CaCO3(for pH maintenance). Flasks were incubated at 25 °C, 200 rpm. The supernatant obtained from each fermentation culture was prepared as described by [35]. 1 ml sample from each flask was taken at day 5. 50 μl 50% H2SO4 was added and the mixture was heated to 80 °C and incubated for 15 min. The samples were then cooled down to the room temperature followed by centrifugation at 8000 rpm for 1 min. The supernatant was filtered through 0.45 μM HPLC-grade regenerated cellulose membrane filters for HPLC analysis. 250 μl of the filtrate were analyzed for the content of sugars and organic acids by HPLC (Dionex Ultimate 3000-LC system) with the conditions described in [31].
Statistical analysis
Comparison of results from triplicates were analyzed by t-test with a significance level of p < 0.05. Error bars on figures are standard error of the mean.
Availability of data and materials
All the data and material presented in the article are available from the corresponding author upon reasonable request.
Abbreviations
- CTP:
-
Citrate transporter
- HPLC:
-
High-performance liquid chromatography
- PCR:
-
Polymerase chain reaction
- rTCA branch:
-
Reductive tricarboxylic acid branch
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- TCA cycle:
-
Tricarboxylic acid cycle
References
Gallezot P. Conversion of biomass to selected chemical products. Chem Soc Rev. 2012;41(4):1538–58.
Hansen NB, Lübeck M, Lübeck PS. Advancing USER cloning into simpleUSER and nicking cloning. J Microbiol Methods. 2014;96(1):42–9.
Yang L, Lübeck M, Lübeck PS. Effects of heterologous expression of phosphoenolpyruvate carboxykinase and phosphoenolpyruvate carboxylase on organic acid production in Aspergillus carbonarius. J Ind Microbiol Biotechnol. 2015;42(11):1533–45.
Yang L, Lübeck M, Lübeck PS. Deletion of glucose oxidase changes the pattern of organic acid production in Aspergillus carbonarius. AMB Express. 2014;4(1):54.
Yang L, Christakou E, Vang J, Lübeck M, Lübeck PS. Overexpression of a C4-dicarboxylate transporter is the key for rerouting citric acid to C4-dicarboxylic acid production in Aspergillus carbonarius. Microb Cell Factories. 2017;16(1):43.
Goldberg I, Rokem JS, Pines O. Organic acids: old metabolites, new themes. J Chem Technol Biotechnol. 2006;81(10):1601–11.
Papagianni M. Advances in citric acid fermentation by Aspergillus niger: biochemical aspects, membrane transport and modeling. Biotechnol Adv. 2007;25(3):244–63.
de Jongh WA, Nielsen J. Enhanced citrate production through gene insertion in Aspergillus niger. Metab Eng. 2008;10(2):87–96.
Blumhoff ML, Steiger MG, Mattanovich D, Sauer M. Targeting enzymes to the right compartment: metabolic engineering for itaconic acid production by Aspergillus niger. Metab Eng. 2013;19:26–32.
Li A, Pfelzer N, Zuijderwijk R, Punt P. Enhanced itaconic acid production in Aspergillus Niger using genetic modification and medium optimization. BMC Biotechnol. 2012;12.
Ruijter GJG, Panneman H, Xu D-, Visser J: Properties of Aspergillus niger citrate synthase and effects of citA overexpression on citric acid production. FEMS Microbiol Lett 2000, 184(1):35–40.
Rohr M, Kubicek CP. Regulatory aspects of citric acid fermentation by Aspergillus niger. Process Biochem. 1981;16:34.
Ruijter G, Kubicek C, Visser J: Production of organic acids by fungi. In Industrial Applications. Edited by Anonymous Springer; 2002:213–230.
Palmieri F, Stipani I, Quagliariello E, Klingenberg M. Kinetic study of the Tricarboxylate carrier in rat liver mitochondria. Eur J Biochem. 1972;26(4):587–94.
Conover TE. Does citrate transport supply both acetyl groups and NADPH for cytoplasmic fatty acid synthesis? Trends Biochem Sci. 1987;12(C):88–9.
Kaplan RS, Mayor JA, Wood DO. The mitochondrial tricarboxylate transport protein. cDNA cloning, primary structure, and comparison with other mitochondrial transport proteins. J Biol Chem. 1993;268(18):13682–90.
Bisaccia F, De Palma A, Dierks T, Krämer R, Palmieri F. Reaction mechanism of the reconstituted tricarboxylate carrier from rat liver mitochondria. Biochim Biophys Acta Bioenerg. 1993;1142(1–2):139–45.
Kirimura K, Kobayashi K, Ueda Y, Hattori T. Phenotypes of gene disruptants in relation to a putative mitochondrial malate–citrate shuttle protein in citric acid-producing Aspergillus niger. Biosci Biotechnol Biochem. 2016:1–10.
Kadooka C, Izumitsu K, Onoue M, Okutsu K, Yoshizaki Y, Takamine K, Goto M, Tamaki H, Futagami T. Mitochondrial citrate transporters CtpA and YhmA are involved in lysine biosynthesis in the white koji fungus, Aspergillus luchuensis mut. kawachii. bioRxiv. 2018:341370.
Hossain AH, Ter Beek A, Punt PJ. Itaconic acid degradation in Aspergillus niger: the role of unexpected bioconversion pathways. Fungal Biol Biotechnol. 2019;6(1):1.
Kirimura K, Kobayashi K, Yoshioka I. Decrease of citric acid produced by Aspergihllus niger through disruption of the gene encoding a putative mitochondrial citrate-oxoglutarate shuttle protein. Biosci Biotechnol Biochem. 2019:1–9.
Castegna A, Scarcia P, Agrimi G, Palmieri L, Rottensteiner H, Spera I, Germinario L, Palmieri F. Identification and functional characterization of a novel mitochondrial carrier for citrate and oxoglutarate in Saccharomyces cerevisiae. J Biol Chem. 2010;285(23):17359–70.
Werpy T, Petersen G: Top value added chemicals from biomass. Results of Screening for Potential Candidates from Sugars and Synthesis Gas 2004, 1.
Yang L, Lübeck M, Lübeck PS. Aspergillus as a versatile cell factory for organic acid production. Fungal Biol Rev. 2017;31(1):33–49.
El Moualij B, Duyckaerts C, Lamotte-Brasseur J, Sluse FE. Phylogenetic classification of the mitochondrial carrier family of Saccharomyces cerevisiae. Yeast. 1997;13(6):573–81.
Pel HJ, De Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, De Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JAE, Van Den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EGJ, Debets AJM, Dekker P, Van Dijck PWM, Van Dijk A, Dijkhuizen L, Driessen AJM, D’Enfert C, Geysens S, Goosen C, Groot GSP, De Groot PWJ, Guillemette T, Henrissat B, Herweijer M, Van Den Hombergh JPTW, Van Den Hondel CAMJJ, Van Der Heijden RTJM, Van Der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, Van Kuyk PA, Lauber J, Lu X, Van Der Maarel MJEC, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, Van Peij NNME, Ram AFJ, Rinas U, Roubos JA, Sagt CMJ, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, Van De Vondervoort PJJ, Wedler H, Wösten HAB, Zeng A-, Van Ooyen AJJ, Visser J, Stam H: Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 2007, 25(2):221–231.
Aguilar-Pontes M, Brandl J, McDonnell E, Strasser K, Nguyen T, Riley R, Mondo S, Salamov A, Nybo JL, Vesth TC. The gold-standard genome of Aspergillus niger NRRL 3 enables a detailed view of the diversity of sugar catabolism in fungi. Stud Mycol. 2018;91:61–78.
Yin X, Shin H, Li J, Du G, Liu L, Chen J. Comparative genomics and transcriptome analysis of Aspergillus niger and metabolic engineering for citrate production. Sci Rep. 2017;7:41040.
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621.
Yuzbasheva EY, Agrimi G, Yuzbashev TV, Scarcia P, Vinogradova EB, Palmieri L, Shutov AV, Kosikhina IM, Palmieri F, Sineoky SP. The mitochondrial citrate carrier in Yarrowia lipolytica: its identification, characterization and functional significance for the production of citric acid. Metab Eng. 2019;54:264–74.
Linde T, Zoglowek M, Lübeck M, Frisvad JC, Lübeck PS. The global regulator LaeA controls production of citric acid and endoglucanases in Aspergillus carbonarius. J Ind Microbiol Biotechnol. 2016;43:1139–47.
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115.
Hynes MJ. Genetic transformation of filamentous fungi. J Genet. 1996;75(3):297–311.
Peleg Y, Stieglitz B, Goldberg I. Malic acid accumulation by Aspergillus flavus - I. biochemical aspects of acid biosynthesis. Appl Microbiol Biotechnol. 1988;28(1):69–75.
Battat E, Peleg Y, Bercovitz A, Rokem JS, Goldberg I. Optimization of L-malic acid production by Aspergillus flavus in a stirred fermentor. Biotechnol Bioeng. 1991;37(11):1108–16.
Acknowledgements
Gitte Hinz-Berg and Sabine Malene Bjerregaard are thanked for excellent laboratory assistance.
Statement on methodology
In this study, description of methods on fungal cultivation, genomic DNA extraction, plasmid construction, protoplast transformation and HPLC analysis was simplified to avoid repetitive information that has been provided by our previous research paper. The detailed descriptions on these methods can be obtained from the reference [31].
Funding
The project was financed by Novozymes A/S and the Danish Strategic Research program MycoFuelChem (DSF grant no. 11–116803). The funders had no role in the design of this study, the collection, analysis, and interpretation of data or in preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
LY, TL, ML, PP and PSL conceived the project and designed experiments. LY and TL performed the experiments and drafted the manuscript. AHH acquired and interpreted data for transcription analysis. AHH, ML, PP and PSL revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study does not contain any experiment with human participants or animals performed by any of the authors.
Consent for publication
Not applicable.
Competing interests
The co-author Peter J. Punt is a member of the editorial board (Section Editor) of this journal, and other authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1: Figure S1.
Transcriptional analysis of the mtpA gene.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yang, L., Linde, T., Hossain, A.H. et al. Disruption of a putative mitochondrial oxaloacetate shuttle protein in Aspergillus carbonarius results in secretion of malic acid at the expense of citric acid production. BMC Biotechnol 19, 72 (2019). https://doi.org/10.1186/s12896-019-0572-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12896-019-0572-0