Abstract
Background
Bacterial pneumonia can affect all age groups, but people with weakened immune systems, young children, and the elderly are at a higher risk. Streptococcus pneumoniae, Klebsiella pneumoniae, Haemophilus influenzae, and Pseudomonas aeruginosa are the most common causative agents of pneumonia, and they have developed high MDR in recent decades in Ethiopia. This systematic review and meta-analysis aimed to determine the pooled prevalence of bacterial pneumonia and multidrug resistance in Ethiopia.
Methods
The articles were searched extensively in the electronic databases and grey literature using entry terms or phrases. Studies meeting the eligibility criteria were extracted in MS Excel and exported for statistical analysis into STATA version 14 software. The pooled prevalence of bacterial pneumonia and multidrug resistance were calculated using a random-effects model. Heterogeneity was assessed by using the I2 value. Publication bias was assessed using a funnel plot and Egger’s test. A sensitivity analysis was done to assess the impact of a single study on the pooled effect size.
Result
Of the 651 studies identified, 87 were eligible for qualitative analysis, of which 11 were included in the meta-analysis consisting of 1154 isolates. The individual studies reported prevalence of bacterial pneumonia ranging from 6.19 to 46.3%. In this systematic review and metanalysis, the pooled prevalence of bacterial pneumonia in Ethiopia was 37.17% (95% CI 25.72–46.62), with substantial heterogeneity (I2 = 98.4%, p < 0.001) across the studies. The pooled prevalence of multidrug resistance in bacteria isolated from patients with pneumonia in Ethiopia was 67.73% (95% CI: 57.05–78.40). The most commonly isolated bacteria was Klebsiella pneumoniae, with pooled prevalence of 21.97% (95% CI 16.11–27.83), followed by Streptococcus pneumoniae, with pooled prevalence of 17.02% (95% CI 9.19–24.86), respectively.
Conclusion
The pooled prevalence of bacterial isolates from bacterial pneumonia and their multidrug resistance were high among Ethiopian population. The initial empirical treatment of these patients remains challenging because of the strikingly high prevalence of antimicrobial resistance.
Similar content being viewed by others
Introduction
Pneumonia is an infection-induced inflammation of the lung tissue due to infectious caused by bacteria and other agents [1]. There is a very wide variety of pneumonia-responsible pathogens with the largest agents are bacteria [2] and resulting in approximately 7 million deaths annually [3].The most common causative agents are Streptococcus pneumoniae (S. pneumoniae), Haemophilus influenzae (H. influenzae), Klebsiella pneumoniae (K. pneumoniae), Pseudomonas aeruginosa (P. aeruginosa), Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus) [4]. In Spain, S. pneumoniae was the leading species in causing bacterial pneumonia which accounts for 31.7% [5]. Varying prevalence of bacterial pneumonia were reported in different parts of Ethiopia; 42.9% in southern Ethiopia and 32.1% in central Ethiopia, with S. pneumoniae and K. pneumoniae were predominant isolates, respectively [6, 7].
Bacterial pneumonia causes complications for everyone, but individuals with weakened immune systems, children, and the elderly are at higher risk [8]. Community and hospital-acquired pneumonia are the two main types of pneumonia. In the community, with a high prevalence, and it causes significant morbidity and mortality [9]. Patients living with HIV/AIDS especially those who had co-infection with one or more microorganisms, and older age individuals are more susceptible for infections with bacterial pneumonia [10]. The positive culture rate was slightly higher in women than in men and higher prevalence rates of lower respiratory tract infections were observed in age groups greater than or equal to 45 years [11].
The main problem concerning about treatment of bacteria causing pneumonia is multidrug resistance (MDR) (antibiotic resistance to at least three or more than three classes), extensively drug resistant (XDR) (resistance to all antibiotics classes except one), and pan-drug resistant (PDR) (resistance to all groups of antibiotics) [12, 13]. There are different mechanisms in which bacteria can escape from the effect of antibiotics. Resistance to one or more groups of antimicrobial agents may be innate or acquired by bacteria. The antibiotic resistance crisis is due to emerging and dissemination antibiotic resistance pathogen in the hospital and environments, inappropriate drug use, over use and consumption of drug resistant pathogens from animal sources and crops [14].
In China most frequently prescribed antibiotics including penicillin, erythromycin, tetracycline and clindamycin resistance were pertained by S. aureus, and S. pneumoniae was highly resistant to erythromycin, azithromycin and clindamycin. E. coli, was resistant to ampicillin, gentamicin, and ciprofloxacin. K. pneumoniae, has the highest resistance to gentamicin and ampicillin [15]. Similarly, cotrimoxazole was 100% resistant to S. aureus and S. pneumoniae. K. pneumoniae was resistant to most of the antibiotics showing more than 50% resistance to ceftriaxone and cefotaxime drugs respectively [16].
Nigeria’s Analysis of pneumonia-associated bacteria among HIV/AIDS patients in Nigeria showed that P. aeruginosa were highly resistant to all antibiotics including ciprofloxacin and ceftazidime whereas E. coli, S. aureus and K. pneumoniae were resistance to commonly prescribing drugs [17]. Gram-negative bacilli were highly resistant to ampicillin tetracycline, ciprofloxacin, and trimethoprim-sulfamethoxazole [7]. On the other hand, most of the isolates were less resistant to amikacin. Methicillin resistance was observed in isolates of S. aureus [18]. This study is the first systematic review and meta-analysis to report the national burden of bacterial pneumonia and MDR in Ethiopia; and it aimed to summarize the findings of local studies and estimate the pooled prevalence of bacterial pneumonia and MDR in Ethiopia.
Methods
Design and protocol registration
This systematic review and meta-analysis were designed to estimate the pooled prevalence of bacterial pneumonia and their multi-drug resistance pattern in Ethiopia based on the preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) [19]. The review protocol was registered in the international prospective register of systematic review (PROSPERO) under registration number CRD42023414098.
Data source and search strategy
A comprehensive search of databases was performed to identify all relevant articles published on bacterial isolates with MDR of bacterial isolates from patients with pneumonia in Ethiopia from January 1, 2000 to April 2023. Articles published in English language were searched in PubMed, google scholar, scopus, science direct, African index medicos, African journal online (AJOL), Ethiopian journals, WHO afro library databases from April 6 to April 16, 2023. In addition to accounting for the studies’ omission during electronic database searches, a direct google search was carried out using listed references in included articles. The comprehensive and extensive searching strategy has been employed using condition, context, population, and outcome of interest (CoCoPop) formulating questions and searching terms were (‘‘prevalence”), (“epidemiology”) (“magnitude”), and (“bacterial pneumonia”) and (“antimicrobial resistance”), (“antibiotic resistance”) and (“antibiotic susceptibility”), (“hospital-acquired pneumonia”), (“community-acquired pneumonia”) and (“Ethiopia”). The search terms were combined using the Boolean operators “OR” and “AND” to fit the advanced searching of articles.
Eligibility criteria
The authors developed a selection criteria checklist for study eligibility before identifying appropriately published, relevant full-text articles either in local or international journals. We included published and preprint (study done at Bahirdar University) of original articles that reported bacterial pneumonia and their antimicrobial resistance pattern in all age populations of Ethiopia, studies written in English, and laboratory-based observational (e.g., cross-sectional) studies. We excluded studies with no confirmation of bacterial isolates using phenotypic and/or genotypic methods, qualitative studies, review papers, commentaries, case series, case reports, conference proceedings, and abstracts.
Data extraction
Data extraction was performed by four independent reviewers (HD, MT, OM, and HE) using a standard extraction format adapted from the Joanna Briggs Institute (JBI) data extraction format [20] and recorded them in a Microsoft Excel spreadsheet.
The extracted data includes, the first author’s name and year of publication, the study period, the study design, the study region, the total sample size, the number of isolates, the criteria for diagnosing bacterial isolates causing pneumonia, the number and percentage of Gram-positive and Gram-negative bacteria, and the prevalence MDR of commonly identified bacteria.
Quality assessment
Four authors (MT, AG, HD, and MAB) carefully assessed the quality of the articles using JBI quality appraisal tool. The full texts of the articles were used to determine whether the study met the selection criteria or whether the eligibility of the article was called into question [30]. By using the critical appraisal checklists, studies with an average score of 50–75% were considered of good quality, while scores greater than 75% were considered of high quality. As a result, articles of both good and high quality were included for the analysis [31] (Supplementary Table 1).
Outcome variables
Two findings were drawn from this systematic review and meta-analysis. The first goal was to determine the pooled estimates of bacterial pneumonia among pneumonia suspected Ethiopian patients. The second goal was to calculate the pooled prevalence of MDR of common pathogens.
Data processing and analysis
The data were analyzed by using STATA version 14.0 statistical software. A random effect model was applied to estimate the pooled estimate and MDR of the isolates. A potential source of heterogeneity was investigated by subgroup and meta-regression analysis. The Cochran’s Q test and I2 statistics were used to quantify and assess the presence of heterogeneity between studies. The p-value of < 0.05 for I2 statistics was used to determine the presence of heterogeneity [21] and Der Simonian-Laired random effects model was employed [22]. Subgroup analysis was done based on the patient’s region, city, study design, and HIV sero-status. The results were presented using table and forest plot. Publication bias was evaluated using inspection of funnel plot symmetry and Egger’s test statistics. The Trim-and-Fill was used in asymmetrical funnel plots to integrate missing studies and estimate adjusted effect size. Meta-regression was also used to further assess the cause of heterogeneity.
Result
Selection and identification of studies
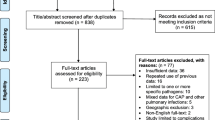
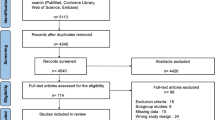
A total of 651 articles were retrieved from databases. About 565 articles remained after removing 86 duplicate articles. From the remaining, 239 articles were excluded after reviewing the title, abstract, and objective of the study. Finally, 87 full-length articles were thoroughly reviewed by predetermined eligibility criteria, and 11 studies were included in the meta-analysis [18, 23,24,25,26,27,28,29,30,31,32] (Fig. 1).
Characteristics of included studies
Table 1 Summarizes the characteristics of 11 studies included in our final meta-analysis. These studies were conducted between 2000 and 2023 in different regions of Ethiopia and were used to estimate the pooled prevalence and MDR of bacterial isolates suspected of pneumonia. From the total studies, 10 of the studies were cross-sectional and 1 study was prospective observational. All of the studies were published on peer review journals except one unpublished data that was obtained from preprint data [18, 23,24,25,26,27,28,29,30,31,32]. About 45.5% (5/11) of the studies were conducted in Amhara region [27,28,29,30, 32], followed by two studies 18.2% (2/11) in Addis Ababa city [23, 24], two in SNNPR [25, 26], one in Oromia region [31] and one in Tigray region [18]. The sample size of the studies included the least from Jimma 133 [31] to the highest from Addis Ababa 549 [23]. The reported prevalence of bacterial pneumonia ranged from 6.19% to in Addis Ababa [23] to 46.3% in Dessie [28], while the prevalence of MDR bacterial pneumonia ranged from 32.7% in Hawassa [26] to 84.6% in Dessie [28] (Table 1).
The pooled prevalence of bacterial pneumonia
A total of 11 studies reported that bacterial pneumonia infections were detected in 1151 samples out of a total of 3423 samples taken from bacterial pneumonia suspected patients who visit the health care system. In this systematic review and meta-analysis, the overall pooled prevalence of bacterial pneumonia in Ethiopia was 37.17% (95% CI 25.72–48.62%) with substantial heterogeneity (I2 = 98.4%, p < 0.001) across the studies. (Fig. 2).
Subgroup analysis
Subgroup analyses revealed 38.71% (95% CI: 33.37–44.05, I2 = 72%, p < 0.001) and 42.45% (95% CI 35.00–49.90, I2 = 78.5%, p = 0.031) pooled prevalence of bacterial pneumonia in the age group > 18 years and studies involving all age groups, respectively, with no statistically significant difference (p = 0.14). Another subgroup analysis was performed on HIV sero-status and indicated that the prevalence of bacterial pneumonia among HIV-negative patients was 34.07% (95% CI 19.32–48.83, I2 = 98.6%, p < 0.001) while it was 44.51% (95% CI 44.06–47.97, I2 = 0%, p = 0.582) in HIV positive patients. Lastly, subgroup by study area showed that the prevalence of bacterial pneumonia was 18.99% in Addis Ababa city, 37.73% SNNP region, and 41.21% in Amhara region with no significant difference across the regions (p = 0.46). The prevalence of bacterial pneumonia pooled from studies showed increment from the period ≤ 2020 (35.49%, 95% CI: 14.51, 56.48) to > 2020 with prevalence of 38.55% (95% CI: 34.23, 42.86). On the other hand, the prevalence of bacterial pneumonia in terms of sample size ≤ 384 was 41.45% (95% CI: 37.62, 45.29) (Table 2).
The pooled prevalence of MDR
In individual study, the magnitude of MDR in Ethiopia was varying from 32.4 to 84.6%. The overall pooled prevalence of MDR of bacteria isolated from patients with pneumonia in Ethiopia was 67.73% (95% CI: 57.05–78.40) with high level of heterogeneity (I2 = 97.2%, p < 0.001) across the studies (Fig. 3). Subgroup analysis performed on HIV serostatus indicated that the prevalence of MDR among HIV positive patients was 76.88% (95% CI: 66.48–87.28%, I2 = 88.3%, p < 0.001) while it was 63.97% (95% CI: 47.52–80.42%, I2 = 98.0%, p < 0.001) in HIV negative patients (Fig. 4).
Pooled prevalence of pneumonia causing bacterial isolates
Summary of Table 3 indicated eleven different types of bacterial isolates. Gram-negative bacteria were the predominant isolates with 61.5% pooled prevalence. The most common bacterial isolate was K. pneumoniae with an overall prevalence of 21.97% (95% CI 16.11–27.83%), followed by S. pneumoniae 17.02 (95% CI 9.19–24.86%), S.aureus 14.47% (95% CI 8.95–19.99%), P. aeruginosa 9.98% (95% CI 6.57–14.13%), E. coli 9.75% (95% CI 7.01–12.50), Citrobacter species 4.12% (95% CI 1.84–6.39%), Enterobacter species 5.10% (95% CI 0.57–9.63%), H. influenzae 3.89%(95% CI 2.45–5.32%), P. mirabilis 2.73% (95% CI 1.20–4.27%), P. vulgaris 1.71% 95% CI -0.92–4.33%) and Acinetobacter species 3% 4.70% (95% CI -0.87–8.54% ) (Table 3).
Sensitivity analysis
According to our sensitivity analysis finding, each study did not affect the pooled estimate of the proportion indicating the precise aggregate result. When individual studies were omitted, the pooled effect size lay within the 95% confidence interval of the overall pooled effect size. This demonstrated that no single study had an impact on the overall pooled prevalence of bacterial pneumonia infection in Ethiopia (Table 4).
Publication bias
The funnel plot was used to assess the impact of the small-studies effect or publication bias on estimated pooled prevalence. In this study, the asymmetry of the funnel plot illustrated the presence of publication bias with over 63.6% of the studies skewed to the right side of the triangular zone (Fig. 5). Furthermore, Egger’s test statistics also confirmed the presence of significant publication bias at a P-value < 0.001 (Table 5) (Fig. 6).
Trim and fill analysis of the pooled prevalence of bacterial pneumonia in Ethiopia
Due to the presence of slightly significant publication bias, we performed a trim and fill analysis. After incorporating 6 studies, the trim and fill analysis revealed a pooled prevalence of bacterial pneumonia in Ethiopia was 21.33% (95% CI:10.86–31.798) (Table 6).
Meta-regression
Meta-regression was carried out to further explore the potential sources of heterogeneity or variability among studies included in the meta-analysis. We included continuous study characteristics as covariates including published year, sample size, and number of isolates. In this study, total isolates and sample size were the responsible variables for the existence of heterogeneity between studies (P < 0.001) (Table 7).
Discussions
Bacterial pneumonia is one of the most serious public health issues due to the high medical and economic costs that result in increased morbidity and mortality in people of all ages worldwide [33]. Bacterial pneumonia is characterized by a productive cough, fever with shaking chills, shortness of breath, sharp chest pain during deep breaths, increasing rate of breathing and confusion may be the most noticeable symptom in the elderly [34]. The main aim of this study was to determine the pooled prevalence of bacterial pneumonia and MDR of bacterial isolates causing pneumonia in Ethiopia, involving about 3428 study participants.
The overall pooled prevalence of bacterial pneumonia in Ethiopia was found to be, 37.17% (95% CI 25.72–48.62%) with a high level of heterogeneity (I2 = 98.4%, p < 0.001). This finding is comparable with a previous review reporting the pooled prevalence of bacterial pneumonia in Sudan (33.33%), and a systematic review and meta-analysis of pneumonia in east African children (34%) [35], Sudan (42%) [36], Asian countries (44.8%) [37], Iran (44%) [38] and India (46.3%) [39]. On the contrary, the finding of the present systematic review and meta-analysis is massively higher than systematic review and meta-analysis on the pooled magnitude of pneumonia among under-five children in Ethiopia which accounted 18.03 [40] and lower than the study in Ghana 84.5% [41], Nigeria 69.6% and 45.2% [42, 43], Zambia 59% [44], Egypt 50.4% [45], Pakistan 75% (37), in different regions of India 52.83% and 58.8%, 83% [46, 47], Bangladesh 61.83% [48], multicenter study in China 74.4% [49], Spain 50.7% [50], and Vietnam 61.8% [51]. This could be due to differences in the study setting, genetic background of the study population, and sample size. Another reason for this discrepancy could be methodological differences as some studies use molecular and serological detection methods for both typical and atypical pneumonia [52].
In this meta-analysis, Gram-negative bacteria accounted for 61.5% of culture-positive samples. Similarly, another review article reported 76.13 to 95.3% of Gram-negative bacteria as the cause of bacterial pneumonia infections [53]. The high prevalence of Gram-negative bacteria in various research is due to differences in sample size, geographic location, study period, study population, and respiratory flora specimen contamination.
Regarding the specific bacteria identified, the most common bacterial isolate causing pneumonia was K. pneumoniae with an overall prevalence of 21.97% (95% CI 16.11–27.83), followed by S. pneumoniae 17.02 (95% CI 9.19–24.86). Similar findings from Cambodia reported K. pneumoniae as the leading cause [54]. According to a study in Nepal, the most common bacterial isolate were K. pneumoniae (27.0%), S. aureus 20.8%, S. pneumoniae 18.8%, E. coli 8.3%, H. influenzae, K. oxytoca, P. aeruginosa, 4.2% each, Enterobacter spp 2.1% and unidentified Gram negative bacteria 10.4% [11]. In Nigeria, K. pneumoniae (23%) was the predominant, followed by S. aureus (21%), S. pneumoniae (13%), P. aeruginosa (9%) and E. coli (3%) [55]. Another comparative cross-sectional also indicated, K. pneumoniae was the predominant bacteria isolated 16 (13.3%) followed by E. Coli 10 (8.3%), S. pneumoniae 10 (8.3%), S. aureus 9(7.5% ), P. aeruginosa 5 (4.1% ), M. catarrhalis 4 (3.3% ) and H. influenzae 2 (1.6%) [17]. Different reported S. pneumoniae was the most frequent bacteria isolated from the sputum culture of community-acquired pneumonia and K. pneumoniae were the second frequent pneumonia causing bacteria [56]. A similar finding was reported from a community-based study depicting S. pneumoniae, H. influenzae, S. aureus as predominantly isolated bacterial [3]. In contrast in the UK, S. pneumoniae was the most commonly isolated species (30%) followed by H. influenzae (19%) and M. catarrhalis (2%) [57]. Another descriptive cross-sectional study was conducted in Malawi, the predominant isolate were S. aureus followed by P. aeruginosa, E. cloacae, and K. pneumonia [58].
Furthermore, the pooled prevalence of multi-drug resistant bacterial pneumonia isolates was 67.73% (95% CI: 57.05–78.40). This finding was in line with studies conducted in Nigeria (67.2%) [59] and systematic review report of 59.7% overall MDR prevalence in Ethiopia [60], and lower than the study conducted in Cameron 79.4% [61]. This finding also alarms the need for integrated efforts of antimicrobial surveillance systems and poses for the development of new antibiotics.
In the current review, substantial heterogeneity with an (I2 = 97.2%, p < 0.001) was found. This study’s substantial heterogeneity is most likely not attributable to publication bias, but rather to variances in methodological concerns such as sample size, target population categories, and patient underpinning circumstances. The other difference could be attributed to the target group from which samples were collected and the antibiotic resistance crisis, primarily because antibiotics lose their efficacy over time due to the emergence and spread of resistance among bacterial pathogens, which is primarily caused by the overuse and inappropriate use of antibiotics, as well as the widespread use of antibiotics in agriculture and the food industry. Antibiotic resistance is a natural phenomenon in bacteria that cannot be stopped; however, various measures can be taken to reduce the rate of its development and devise more effective strategies to control its spread.
Sensitivity analysis, sub-group analysis, and meta-regression have been carried-out to rule out the most possible causes of heterogeneity. The results of sensitivity analysis proved that there is no single study that impacted the pooled effect size. The pooled prevalence of bacterial pneumonia infections in Ethiopia was calculated by omitting each study sequentially and the computed pooled prevalence was within 95% CI of the overall pooled prevalence. Meta-regression has confirmed that a number of total pathogens isolates and sample size were a significant cause of heterogeneity in prevalence of bacterial pneumonia while publication year was not found to be a significant cause. In addition to this, publication bias was assessed using funnel plot and Egger’s test statistics, and trim and fill analysis was performed to fill the bias.
One of the notable strengths of this study is its comprehensive nature, being the first of its kind to conduct a thorough analysis of bacterial pneumonia and MDR within Ethiopia. It encompasses a wide range of studies conducted across multiple regions and cities of the country, providing a robust overview.
Furthermore, the study included various studies done in different target populations using clinical specimens to show a clear picture of bacterial pneumonia and MDR in the country. However, the results should be interpreted with caution, as the reviewed studies were highly heterogeneous in terms of prevalence, aetiology, study setups, study participants, disease conditions, clinical specimens, sample sizes, and AST methods. Therefore, to account for this heterogeneity, the random-effects model of Der Simonian and Laird was implemented in the meta-analyses. Moreover, subgroup analyses, sensitivity analysis, and meta-regression were conducted to further address and mitigate the impact of heterogeneity on the findings.
Conclusion
According to this systematic review and meta-analysis, the pooled prevalence of bacterial pneumonia infection and MDR have alarmingly increased and become a public health threat. The most common etiology identified was K. pneumoniae followed by S. pneumoniae. This indicates an urgent need of routine screening and appropriate treatment for better management of pneumonia suspected patients as well as effective controlling of the emergence of drug resistance. Furthermore, it serves as a wake-up call to international, continental, and national health bureaus, as well as other stakeholders, to develop targeted prevention and control strategies, and strengthen antibiotics stewardship programs for better management of hospital-acquired as well as community-acquired infections. Moreover, the data could be used for future complementary research and evidence-based decision-making both in clinical and public health approaches.
Data availability
All relevant data are included in the manuscript and its supplementary data.
Abbreviations
- AMR:
-
Antimicrobial resistance
- CI:
-
Confidence interval
- CLSI:
-
Clinical Laboratory Standards Institute
- MDR:
-
Multidrug resistance
- PRISMA:
-
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- STATA:
-
statistics and data
- WHO:
-
World Health Organization
References
Mackenzie G. The definition and classification of pneumonia. Pneumonia. 2016;8(1):14.
Shilpa A, Anuradha K, Venkatesha D. Drug susceptibility pattern of aerobic bacterial isolates from pulmonary infection in HIV seropositives and their correlation with CD4 count. IOSR J Dent Med Sci, 2014: p. 37–41.
Benito N, et al. Pulmonary infections in HIV-infected patients: an update in the 21st century. Eur Respir J. 2012;39(3):730–45.
López-Palomo C, et al. Pneumonia in HIV‐infected patients in the HAART era: incidence, risk, and impact of the pneumococcal vaccination. J Med Virol. 2004;72(4):517–24.
Curran A, et al. Bacterial pneumonia in HIV-infected patients: use of the pneumonia severity index and impact of current management on incidence, aetiology and outcome. HIV Med. 2008;9(8):609–15.
Regasa B. Drug resistance patterns of bacterial pathogens from adult patients with pneumonia in Arba Minch hospital, South Ethiopia. Glob J Med Res, 2014. 14(5).
Nurahmed N, et al. Bacterial profile and antimicrobial susceptibility patterns of lower respiratory tract infection among patients attending selected health centers of Addis Ababa, Ethiopia. Egypt J Chest Dis Tuberculosis. 2020;69(2):399.
Sampson S, De Pietro M. What to know about bacterial pneumonia. 2022.
Alshammari MK, et al. Prevalence and etiology of community-and hospital-acquired pneumonia in Saudi Arabia and their Antimicrobial susceptibility patterns: a systematic review. Medicina. 2023;59(4):760.
Ojha CR, et al. Lower respiratory tract infections among HIV positive and control group in Nepal. VirusDisease. 2015;26(1):77–81.
Khushbu Y, Satyam P. Bacteriological Profile of Lower respiratory tract infection (LRTI) among HIV seropositive cases in Central Terai of Nepal. Int J Curr Microbiol App Sci. 2015;4(11):431–42.
Magiorakos A-P, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81.
Sweeney MT, et al. Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J Antimicrob Chemother. 2018;73(6):1460–3.
Giedraitienė A, et al. Antibiotic resistance mechanisms of clinically important bacteria. Medicina. 2011;47(3):19.
Jinghua M, Gaizhuang L, Qiaoli C. Pathogens and antibiotic resistance of children with community-acquired pneumonia 2017.
Shrestha S, et al. Lower respiratory tract pathogens and their antimicrobial susceptibility pattern in a medical hospital of central Nepal. Int J Biomedical Adv Res. 2013;4(5):335–40.
Ojo-Bola O, Oluyege A. Antibiotics resistance of bacteria associated with pneumonia in HIV/AIDS patients in Nigeria. Am J Infect Dis Microbiol. 2014;2(6):138–44.
Adhanom G et al. Species, risk factors, and antimicrobial susceptibility profiles of bacterial isolates from HIV-infected patients suspected to have pneumonia in Mekelle zone, Tigray, northern Ethiopia BioMed research international, 2019. 2019.
Moher D, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Reviews. 2015;4(1):1–9.
Moola S, et al. Conducting systematic reviews of association (etiology): the Joanna Briggs Institute’s approach. JBI Evid Implement. 2015;13(3):163–9.
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58.
Higgins JP, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60.
Negash AA, et al. Bacteremic community-acquired pneumonia in Ethiopian children: etiology, antibiotic resistance, risk factors, and clinical outcome. In Open forum infectious diseases. Oxford University Press US; 2019.
Nurahmed N, et al. Bacterial profile and antimicrobial susceptibility patterns of lower respiratory tract infection among patients attending selected health centers of Addis Ababa, Ethiopia. Egypt J Chest Dis Tuberculosis. 2020;69(2):399–406.
Regasa B. Drug resistance patterns of bacterial pathogens from adult patients with pneumonia in Arba Minch hospital, South Ethiopia. J Med Microbiol Diagnosis. 2014;3(4):1.
Gebre AB, Begashaw TA, Ormago MD. Bacterial profile and drug susceptibility among adult patients with community acquired lower respiratory tract infection at tertiary hospital, Southern Ethiopia. BMC Infect Dis. 2021;21(1):440.
Temesgen D, et al. Bacteriology of community acquired pneumonia in adult patients at Felege Hiwot Referral Hospital, Northwest Ethiopia: a cross-sectional study. Antimicrob Resist Infect Control. 2019;8(1):1–8.
Tilahun M, et al. Bacteriology of community-acquired pneumonia, antimicrobial susceptibility pattern and associated risk factors among HIV patients, Northeast Ethiopia: cross-sectional study. SAGE Open Med. 2023;11:20503121221145569.
Dessie T, et al. Multiresistant bacterial pathogens causing bacterial pneumonia and analyses of potential risk factors from Northeast Ethiopia. Int J Microbiol. 2021;2021:1–9.
Assefa M, et al. Bacterial profile, antimicrobial susceptibility patterns, and associated factors of community-acquired pneumonia among adult patients in Gondar, Northwest Ethiopia: a cross-sectional study. PLoS ONE. 2022;17(2):e0262956.
Regasa B, et al. Antimicrobial susceptibility pattern of bacterial isolates from community-acquired pneumonia patients in Jimma University specialized hospital, Jimma, Ethiopia. Saudi J Health Sci. 2015;4(1):59.
Genetu DE, Zenebe Y. Bacterial Profile and Their Antibiotic Resistance Pattern Among HIV Patients Diagnosed with Pneumonia in Felege-Hiwot Referral Hospital, Bahir Dar, Northwest Ethiopia 2020.
Peyrani P, et al. The burden of community-acquired bacterial pneumonia in the era of antibiotic resistance. Expert Rev Respir Med. 2019;13(2):139–52.
Teja GRA et al. Detection and classification of pneumonia in chest X-ray images using deep learning techniques. J Eng Sci, 2023. 14(03).
Abate BB et al. Prevalence of pneumonia and its associated factors among under five children in East Africa. Meta-analysis and systemic review 2020.
Ibrahim A. Bacterial etiology of community acquired pneumonia and their antimicrobial susceptibility in patients admitted to alshaab teaching hospital. Sudan Med Lab J, 2018. 6(1).
Song J-H, et al. Epidemiology and clinical outcomes of community-acquired pneumonia in adult patients in Asian countries: a prospective study by the Asian network for surveillance of resistant pathogens. Int J Antimicrob Agents. 2008;31(2):107–14.
Kashef N, Djavid GE, Shahbazi S. Antimicrobial susceptibility patterns of community-acquired uropathogens in Tehran, Iran. J Infect Developing Ctries. 2010;4(04):202–6.
Yadav P, et al. Clinico–Bacteriological Profile of community–acquired Pneumonia patients at Tertiary Care Center of North India. Indian J Respiratory Care¦ Volume. 2022;11(2):118.
Abebaw B, Damtie D. Meta-analysis and systematic review of under-five pneumonia in Ethiopia
Pappoe F, Amissah I. Prevalence of bacterial pathogens isolated from sputum cultures of hospitalized adult patients with community-acquired pneumonia at the cape coast teaching hospital, Ghana. J Med Res. 2014;E3(5):58–61.
Ojuawo OB, et al. Clinical and microbiological profile of adult inpatients with community acquired pneumonia in Ilorin, North Central, Nigeria. Afr Health Sci. 2020;20(4):1655–68.
Seid AM et al. Prevalence of community-acquired pneumonia among adult population in Ethiopia: A Systematic Review and Meta-analysis 2023.
Ziko L, et al. Aetiology and prognosis of community-acquired pneumonia at the Adult University Teaching Hospital in Zambia. PLoS ONE. 2022;17(7):e0271449.
El-Sokkary RH, et al. Community acquired pneumonia among adult patients at an Egyptian university hospital: bacterial etiology, susceptibility profile and evaluation of the response to initial empiric antibiotic therapy. Infect Drug Resist. 2018;11:2141.
Panda S, Nandini BP, Ramani T. Lower respiratory tract infection-bacteriological profile and antibiogram pattern. Int J Curr Res Rev. 2012;4:149–55.
Kalita D, et al. High proportion of drug-resistant isolates in adult community-acquired pneumonia from Northeast India: a hospital-based study. Lung India: Official Organ Indian Chest Soc. 2021;38(5):460.
Akter S, Shamsuzzaman S, Jahan F. Community acquired bacterial pneumonia: aetiology, laboratory detection and antibiotic susceptibility pattern. Malays J Pathol. 2014;36(2):97–103.
Qu J, et al. Aetiology of severe community acquired pneumonia in adults identified by combined detection methods: a multi-centre prospective study in China. Volume 11. Emerging Microbes & Infections; 2022. pp. 556–66. 1.
Gutierrez F, et al. Epidemiology of community-acquired pneumonia in adult patients at the dawn of the 21st century: a prospective study on the Mediterranean coast of Spain. Clin Microbiol Infect. 2005;11(10):788–800.
Do Tran H, et al. Community-acquired pneumonia-causing bacteria and antibiotic resistance rate among Vietnamese patients: a cross-sectional study. Medicine. 2022;101(36):e30458.
Herrera M, et al. Comparison of serological methods with PCR-based methods for the diagnosis of community-acquired pneumonia caused by atypical bacteria. J Negat Results Biomed. 2016;15:1–11.
Gupta R, et al. Epidemiology of multidrug-resistant Gram-negative pathogens isolated from ventilator-associated pneumonia in ICU patients. J Global Antimicrob Resist. 2017;9:47–50.
Rammaert B, et al. Klebsiella pneumoniaerelated community-acquired acute lower respiratory infections in Cambodia: clinical characteristics and treatment. BMC Infect Dis. 2012;12(1):1–7.
Urama EU, et al. Microbiological Profile of respiratory tract infections among HIV Sero-positive subjects attending Nnamdi Azikiwe University Teaching Hospital Nnewi, Nigeria. Amer J Med Med Sci. 2018;8:37–42.
Musher DM, et al. Can an etiologic agent be identified in adults who are hospitalized for community-acquired pneumonia: results of a one-year study. J Infect. 2013;67(1):11–8.
Nimmo C, et al. Airway bacteria and respiratory symptoms are common in ambulatory HIV-positive UK adults. Eur Respir J. 2015;46(4):1208–11.
Hartung TK, et al. Etiology of suspected pneumonia in adults admitted to a high-dependency unit in Blantyre, Malawi. Am J Trop Med Hyg. 2011;85(1):105–12.
Adeyemi FM, Adejuyigbe A-nK, Ebhodaghe E, Osho BI, Oyeniyi PO. Molecular characterizationand antibiotic resistance pro. Arch Clin Microbiol. 2018;6(1):269–76.
Muhie OA. Antibiotic use and resistance pattern in Ethiopia: systematic review and meta-analysis International journal of microbiology, 2019. 2019.
Marbou WJ. Bacterial resistance and immunological pro. J Infect Public Heal. 2017;10(3):269–76.
Acknowledgements
Not applicable.
Funding
This study did not receive any specific grant from funding agencies.
Author information
Authors and Affiliations
Contributions
MIhret Tilahun conceived and designed the study. Melaku Ashagrie Belete, Alemu Gedefei, Ermiyas Alemayehu, and Habtu Debash participated in the article searches and data extraction. Melaku Ashagrie Belete, Ousman Mohhamed, Habtu Debash, and Alemu Gedefie conduct a quality assessment of the included studies and perform the statistical analysis and interpretation of the data. Melaku Ashagrie Belete drafts the manuscript. Melaku Ashagrie Belete, Alemu Gedefei, Ermiyas Alemayehu, and Habtu Debash check the validity and monitor the overall process.MIhret Tilahun, Daniel G/tsadik, Hussein Ebrahim, Ousman Mohhamed, and Habtu Debash critically reviewed the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participant
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tilahun, M., Belete, M.A., Gedefie, A. et al. Etiology of bacterial pneumonia and multi-drug resistance pattern among pneumonia suspected patients in Ethiopia: a systematic review and meta-analysis. BMC Pulm Med 24, 182 (2024). https://doi.org/10.1186/s12890-024-03000-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12890-024-03000-1