Abstract
Background
The published data on the association between β2-adrenergic receptor gene polymorphisms and asthma susceptibility are inconclusive. To derive a more precise estimation of this association, a meta-analysis was performed.
Methods
A literature search was conducted in PubMed, Web of Science, EMBASE, Wanfang, and the China National Knowledge Infrastructure (CNKI) databases to identify eligible studies. The pooled odds ratios (ORs) with corresponding 95% confidence intervals (CIs) were used to calculate the strength of the association. A sensitivity analysis was performed to evaluate the influence of individual studies on the overall effect estimates, and funnel plots and Egger’s tests were used for indications of publication bias.
Results
Seventy three studies with three single nucleotide polymorphisms (SNP) (rs1042713, c.G46A, p.Gly16Arg; rs1042714, c.G79C, p.Gln27Glu; rs1042711, c.T-47C, p.Cys19Arg) were finally identified. For the rs1042713 polymorphism, no significant association with asthma risk was found in the overall population. However, a significant protective association was found in the Indian population in the dominant model comparison (OR = 0.72, 95% CI = 0.59–0.87, I2 = 25%, studies = 5, cases = 1190, controls = 1241). A significant risk association was found in the Arab population in the dominant model comparison (OR = 1.75, 95% CI = 1.14–2.70, I2 = 0%, studies = 2, cases = 307, controls = 361) and the homozygote model comparison (OR = 1.88, 95% CI = 1.17–3.02, I2 = 0%, studies = 2, cases = 307, controls = 361), and in the Hispanic-Latino population in the dominant model comparison (OR = 1.68, 95% CI = 1.10–2.55, I2 = 77%, studies = 5, cases = 1026, controls = 1412). For the rs1042714 polymorphism, we found a significant association in the recessive model comparison (OR = 0.83, 95% CI = 0.70–0.98, I2 = 44%, studies = 52, cases = 8242, controls = 16,832), the homozygote genotype comparison (OR = 0.84, 95% CI = 0.72–0.98, I2 = 25%, studies = 52, cases = 8242, controls = 16,832) and the allelic genetic model (OR = 0.91, 95% CI = 0.83–0.99, I2 = 59%, studies = 52, cases = 8242, controls = 16,832) in the overall population. When stratified by age, a significant association was also found in children in the recessive model comparison (OR = 0.59, 95% CI = 0.39–0.88, I2 = 58%, studies = 18, cases = 2498, controls = 2510) and the homozygote genotype comparison (OR = 0.63, 95% CI = 0.43–0.92, I2 = 46%, studies = 18, cases = 2498, controls = 2510), but not in adult. For the rs1042711 polymorphism, no significant associations were found in the any genetic model.
Conclusion
The meta-analysis suggests that the ADRB2 rs1042714 polymorphism has a protective association with asthma in the overall population and the pediatric subgroup.
Similar content being viewed by others
Background
Asthma is a chronic respiratory inflammation disease characterized by airway hyperresponsiveness, reversible airway obstruction and airway wall remodeling [1]. It is believed to be a multifactorial disorder with a strong genetic component in its pathogenesis [2, 3]. So far, many studies have evaluated the association between genetic variants and asthma susceptibility. Numerous genes have been identified as asthma-susceptible genes, from which the β-2 adrenergic receptor (ADRB2) is the most widely studied [4,5,6].
ADRB2 is encoded by an intronless gene on chromosome 5q31, which is abundantly expressed on many airway cells including smooth muscle cells [7, 8]. ADRB2 transcript has a 5′ leader cistron (5′ LC) harboring a short open reading frame (ORF) that encodes a 19-amino acid peptide, which regulates mRNA translation and controls the cellular expression of ADRB2. A variation at position 19 that causes a change from cysteine (Cys) to arginine (Arg) was reported in the 5′ LC, and this variation plays a role in regulating ADRB2 gene expression [9,10,11]. However, little is known regarding the possible role of this polymorphism in asthma. In addition, two missense variations (rs1042713, c.G46A, p.Gly16Arg and rs1042714, c.G79C, p.Gln27Glu) that occur in high allelic frequency in the general population have been identified, corresponding to a change from glycine (Gly) to arginine (Arg) at amino acid position 16 and glutamate (Gln) to glutamine (Glu) at amino acid position 27 [12]. Studies in vitro [13] and primary cultures of cells expressing these endogenous variants [14] illustrated the different phenotypes between the polymorphic receptors. The Gly16 receptor could enhance agonist-promoted downregulation of receptor expression compared with the Arg16 receptor. In contrast, the Glu27 receptor is relatively resistant to agonist-promoted downregulation compared with the Gln27 receptor [13, 14]. Genetic studies have indicated that these variations not only affect the risk of asthma, but also affect the therapeutic outcomes of inhaled β2-adrenergic receptor agonists [15,16,17,18,19,20].
Considering the impact of the asthma risk potentially resulting from ADRB2 gene variations, a number of case-control studies have explored the association between the ADRB2 gene polymorphisms and asthma risk in different ethnicities [21,22,23]. However, these results are conflicting and inconclusive, which are possibly due to the limitations associated with an individual studies and small sample size. To shed light on these contradictory results and to more precisely evaluate the relationship between the ADRB2 gene polymorphisms and asthma risk, several meta-analyses concerning the association between ADRB2 gene polymorphisms and asthma have been reported [15, 24,25,26,27,28,29]. However, these meta-analyses have also shown inconsistent results. After publication of these meta-analyses, many additional case-control studies about the ADRB2 polymorphisms on asthma risk were carried out [30,31,32,33,34]. Therefore, we present the results of a comprehensively updated meta-analysis of all relevant published data to investigate the associations between ADRB2 gene polymorphisms and asthma risk with a focus on rs1042713, rs1042714 and rs1042711 polymorphisms.
Methods
This systematic review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Statement guidelines.
Publication search
Publications were obtained from the PubMed, EMBASE, Web of Science, the Chinese National Knowledge Infrastructure, and Wanfang databases (the last search was conducted on September 1, 2018). The keywords searched in our investigation were (asthma or asthmatic) and (β2-adrenergic receptor or ADRB2 or β2-AR or beta2-adrenoreceptor or β2-adrenoceptor) and (polymorphism or mutation or variant or rs1042713 or G46A or Gly16Arg or rs1042714 or G79C or Gln27Glu or rs1042711 or T-47C or Cys19Arg). The search was performed in duplicate by two independent reviewers (Songlin Zhao and Wei Zhang).
Inclusion and exclusion criteria
The inclusion criteria of our study were as follows: (1) any human studies that estimated the prevalence of the β2-adrenergic receptor polymorphisms and asthma risk were included, which were published in English and Chinese. (2) They were case-control studies. (3) The genotype distributions or allele frequency of each study should be available for estimating an odds ratio with a 95% confidence interval. (4) When eligible papers had insufficient information, we contacted the authors for additional information via email. Studies were excluded from our meta-analysis if their authors did not provide us with the related data.
Data extraction
The basic information extracted from each study was as follows: name of the first author, publication year, country, ethnicity, age of cases and controls, sample size, and genotype frequencies in cases and controls. The data were extracted independently and in duplicate by two reviewers (Songlin Zhao and Wei Zhang) who used a standardized data extraction form. Any disagreement was adjudicated by a third author (Xiuhong Nie).
Study quality assessment and meta-analysis quality assessment
The Newcastle-Ottawa Scale (NOS) was used to assess the quality of the included studies. The items assessed included selection, comparability of case/controls, exposure/outcome, age and gender. The quality scores ranged from 0 to 9. We divided the NOS scores into three levels (higher quality, score ≥ 7; moderate quality, 4 ≤ score < 7; low quality, score < 4).
A Measurement Tool to Assess Systematic Reviews 2 (AMSTAR 2) was used to assess the quality of the systematic reviews [35]. The AMSTAR 2 calculator queries 16 items of relevance that provide insight into the quality of the systematic review methodology.
Statistical analysis
A Hardy-Weinberg equilibrium (HWE) was assessed for each study by use of the Pearson’s chi-square test in control groups, and significance was set at P < 0.05. The pooled ORs for the ADRB2 polymorphisms and asthma risk were calculated for the dominant genetic model, the recessive genetic model, the homozygote genetic model and the allele genetic model. The heterogeneity was assessed by using the Q-test and I2 test. A P-value> 0.10 of Q-test and I2 < 50% indicated a lack of heterogeneity among the studies; then, the fixed-effect model was used. Otherwise, the random effect model was used. Subgroup analyses were performed regarding ethnicity, case age and HWE P-value. The ethnicity subgroups were designed as Caucasian, Hispanic-Latinos, non-Hispanic Blacks, East-Asian, Indian and Arab. Age subgroups were designed as Adult and pediatric subgroups. HWE P-value subgroups were designed as P-value> 0.05 and P-value< 0.05 subgroups. Sensitivity analysis was conducted by sequentially excluding one study at a time to examine the effect of each study on the combined result. The funnel plot and Egger’s test was used to assess the potential publication bias. All the statistical analyses of this meta-analysis were performed using the STATA 11.0 software (State Corporation, College Station, TX, USA).
Results
Characteristics of the studies included in the meta-analysis.
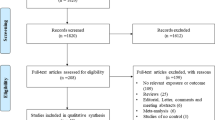
The flow chart in Fig. 1 outlines the study selection process. After a comprehensive search of the PubMed, Web of Science, EMBASE, CNKI and Wanfang databases, a total of 1557 articles were identified. First, we excluded 882 duplicated studies. After reading the abstracts and titles, 530 studies were excluded. The remaining 145 studies were then assessed for inclusion. Of these, 72 studies were excluded because 48 studies were not irrelevance, 16 studies lacked detailed genotypes, and 8 studies lacked a case-control design. Finally, a total of 73 studies met the inclusion criteria and were included in the meta-analysis [8, 12, 17, 21,22,23, 30,31,32,33,34, 36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97]. The characteristics of each eligible study are shown in Additional file 1. Of these 73 studies, each contains two independent studies, and the data is extracted accordingly. During the process, if the sum of genotype distribution of one study is over than 1, the study is excluded from the meta-analysis. The genotype, allele distribution and P-value of HWE for the rs1042713, rs1042714 and rs1042711 polymorphisms are respectively shown in Additional files 2, 3 and 4.
Meta-analysis of ADRB2 rs1042713 polymorphism and asthma risk
Among the 73 publications included in the meta-analysis, 71 studies investigated the association between the ADRB2 rs1042713 polymorphism and asthma risk (Additional file 2). Overall, there was no significant association in any of the genetic model comparisons in the total population (Table 1, Additional file 5: Figure S1, Additional file 6: Figure S2, Additional file 7: Figure S3 and Fig. 2). In the stratified analysis of ethnicity, a significant protective association was found in the Indian population in the dominant model comparison (OR = 0.72, 95% CI = 0.59–0.87, I2 = 25%, studies = 5, cases = 1190, controls = 1241), and a significant risk association was found in the Arab population in the dominant model comparison (OR = 1.75, 95% CI = 1.14–2.70, I2 = 0%, studies = 2, cases = 307, controls = 361) and the homozygote model comparison (OR = 1.88, 95% CI = 1.17–3.02, I2 = 0%, studies = 2, cases = 307, controls = 361) (Table 1, Fig. 2). In addition, we found a significant association in the Hispanic-Latino population in the dominant model comparison (OR = 1.68, 95% CI = 1.10–2.55, I2 = 77%, studies = 5, cases = 1026, controls = 1412) (Table 1, Fig. 2), consistent with a previous study [25]. No associations were found in adults and children regarding age or the P-value of HWE (Table 1).
Forest plots of the association between the ADRB2 rs1042713 polymorphism and risk of asthma in dominant model comparison. GG + GA vs. AA genotype. Each study is shown by an OR estimate with the corresponding 95% CIs. The horizontal lines denote the 95% CIs and the squares represent the point OR estimate of each study. The size of the square is proportional to its inverse-variance weight in the meta-analysis. The diamond represents the pooled meta-analysis effect size estimate. The stratified meta-analysis was performed regarding the ethnicity
Meta-analysis of the ADRB2 rs1042714 polymorphism and asthma risk
There were 52 studies investigating the association between the rs1042714 polymorphism and asthma risk (Additional file 3). In the overall population, significant associations were found in the recessive model comparison (OR = 0.83, 95% CI = 0.70–0.98, I2 = 44%, studies = 52, case = 8242, control = 16,832) (Fig. 3), the homozygote genotype comparison (OR = 0.84, 95% CI = 0.72–0.98, I2 = 25%, studies = 52, cases = 8242, controls = 16,832) (Fig. 4) and the allelic genetic model (OR = 0.91, 95% CI = 0.83–0.99, I2 = 59%, studies = 52, cases = 8242, controls = 16,832) (Additional file 8: Figure S4), but not in the dominant model comparison (Additional file 9: Figure S5 and Table 2). When stratified by age, significant associations were also found in children in the recessive model comparison (OR = 0.59, 95% CI = 0.39–0.88, I2 = 58%, studies = 18, cases = 2498, controls = 2510) (Fig. 3) and the homozygote genotype comparison (OR = 0.63, 95% CI = 0.43–0.92, I2 = 46%, studies = 18, cases = 2498, controls = 2510) (Fig. 4) but not in adults (Table 2). In the analysis stratified by ethnicity, no associations were found in any ethnic group. In the analysis stratified by the P-value of HWE, significant associations were found in the P > 0.05 subgroup in the dominant model comparison (OR = 0.89, 95% CI = 0.80–0.99, I2 = 32%, studies = 35, cases = 5552, controls = 14,306), and the allelic genetic model (OR = 0.91, 95% CI = 0.84–0.99, I2 = 28%, studies = 35, cases = 5552, controls = 14,306). No associations were found in the P < 0.05 subgroup (Table 2).
Forest plots of the association between the ADRB2 rs1042714 polymorphism and risk of asthma in recessive model comparison. GG vs. CC + CG genotype. Each study is shown by an OR estimate with the corresponding 95% CIs. The horizontal lines denote the 95% CIs and the squares represent the point OR estimate of each study. The size of the square is proportional to its inverse-variance weight in the meta-analysis. The diamond represents the pooled meta-analysis effect size estimate. The stratified meta-analysis was performed regarding the age
Forest plots of the association between the ADRB2 rs1042714 polymorphism and risk of asthma in homozygote genotype comparison. GG vs. CC genotype. Each study is shown by an OR estimate with the corresponding 95% CIs. The horizontal lines denote the 95% CIs and the squares represent the point OR estimate of each study. The size of the square is proportional to its inverse-variance weight in the meta-analysis. The diamond represents the pooled meta-analysis effect size estimate. The stratified meta-analysis was performed regarding the age
Meta-analysis of the ADRB2 rs1042711 polymorphism and asthma risk
For the rs1042711 polymorphism, only seven case-control studies provided genotype distribution data (Additional file 4); therefore, no subgroup analysis was conducted. No significant associations were found in the overall population in any of the genetic models. The results are shown in Table 3.
Publication bias
Potential publication bias was investigated using the funnel plot and was further assessed using the Egger’s test. The shape of the funnel plot (Fig. 5) and the Egger’s test (Table 4) did not indicate any evidence of publication bias for the rs1042713, rs1042714 and rs1042711 polymorphisms in any of the genetic model comparisons.
Sensitivity analysis
The sensitivity analysis was conducted by sequentially excluding individual studies to estimate the stability of the results. After sequentially excluding each study, similar statistically results were found (Fig. 6).
Discussion
In this study, the associations between three ADRB2 gene SNPs (rs1042713, rs1042714 and rs1042711) and the risk of asthma were conducted base on the data from 73 studies involving 13,493 asthmatic patients and 22,931 controls. This meta-analysis showed that the rs1042713 polymorphism was not a risk factor for overall asthma susceptibility, which was consistent with most of the previous meta-analyses [15, 16, 25, 27,28,29]; however, the data contradict one of the latest meta-analyses [24]. The difference between these results is seemingly due to the different inclusion criteria. The inclusion criteria in the Xie et al. [24] meta-analysis used P-value of the HWE; however, when stratified by the P-value of the HWE, there was still no association between the Gly16Arg polymorphism and asthma in the HWE (P > 0.05) subgroup in our study and in that of Liang et al. [25]. When reviewing Xie et al.’s literature list, we found that they missed some studies that satisfied their inclusion criteria [31, 54, 77,78,79, 87, 88, 91, 92, 95], which may be a reason for the discrepancies.
Some studies reported that the genotype frequency and allele frequency of the rs1042713 polymorphism vary among different ethnic groups [98, 99]. A search of 1000 Genomes Project or Hapmap data showed an approximately 15% difference in allele frequency for the rs1042713 polymorphism. The Gly16 homozygous genotype frequency is more common in the non-Hispanic White than Chinese and more frequent in non-Hispanic White compared to non-Hispanic Black. Therefore, there is high heterogeneity for the rs1042713 polymorphism in the overall analysis. Even though we excluded some studies that did not meet the HWE, heterogeneity was not decreased. Selective bias in the literature has an important effect on the results of the overall meta-analysis. Large sample sizes can better reflect the truth of the effects of the rs1042713 polymorphism on the asthma risk.
Because the genotype frequency and allele frequency of the rs1042713 polymorphism vary among the different ethnic groups, we divided the ethnic groups according to the studies reported by Ortega et al. [100, 101], such that (1) non-Hispanic Whites of European ancestry were designated as Caucasian, (2) Mexicans and South Americans were designated as Hispanic-Latinos, (3) African Americans and non-Hispanic Blacks from Europe and Africa were designated as non-Hispanic Blacks; and (4) Chinese, Japanese and Korean individuals were considered separate Asian ethnic groups and designated as the East-Asian subgroup. In addition, Indian and Arab descendants were designated as separate ethnic groups because these ethnic subgroups have different genotypes and allele frequencies [98]. The analysis stratified by ethnicity showed a significant risk association in Hispanic-Latinos in the dominant model comparison, consistent with a previous study where a significant association in the South American population was found [25]. In addition, the previous study claimed to divide the cohorts by ethnicity, but for the most part, the cohorts were divided by continent when they combined Indian, Arab, Japanese and Han Chinese individuals as the Asian population [24, 25]. These populations have different genotypic and allelic frequencies for the rs1042713 polymorphism [98]. After the stratified analysis, the heterogeneities in the Arab and Indian population were decreased, and a protective association in the Indian population in the dominant model comparison and a risk association in the Arab population in the dominant model comparison and the homozygote genotype comparison were found. However, there is a need for further studies with larger sample sets.
For the rs1042714 polymorphism in the current meta-analysis, benefitting from the inclusion of more case-control studies, a protective effect was found not only in the pediatric subgroup in the recessive model comparison and the homozygote genotype comparison but also in the overall population in the recessive model comparison, the homozygote genotype comparison and the allelic genetic model. This was consistent with previous reports by Liang et al. [25] and Ammarin et al. [27] that showed a protective effect in the pediatric subgroup in the recessive model comparison and the homozygote genotype comparison, confirming that the Glu27 polymorphism was a protective effect for asthma. A genetic study showed that Glu27 homozygotes had less reactive airways than Gln27 homozygotes, and these results could further suggest a protective role for the Glu27 polymorphism in asthma [102]. In addition, in vitro and ex vivo functional studies indicated that Glu27 allele enhanced resistance to agonist-induced down regulation of the receptor, suggesting a protective role of Glu27 polymorphism in regard to receptor desensitisation [13, 14].
In the analysis stratified by HWE according to the P-value for the rs1042714 polymorphism, significant associations were found in the subgroup with P > 0.05 in the dominant model comparison and the allelic genetic model but not in the P < 0.05 subgroup. These results need to be interpreted with caution. The reason the control group population was not in HWE may be selection for a particular phenotype or that the population was not sufficiently large or random.
For the rs1042711 polymorphism, no significant associations with the risk of asthma were found in any comparison model. More research is needed because only seven case-controls were included in this study. There might not be sufficient statistical evidence to clarify the association between the rs1042711 polymorphism and the risk of asthma.
There could be several potential limitations to this meta-analysis. The first problem relates to the limitations of the literature. All available literature should be included in the meta-analysis, but we only included literature published in English and Chinese, thus neglecting studies published in other languages. Second, even though the existing literature had acceptable quality, detailed information was not provided such as asthma definition varied among different articles and this may be a confounding factor. Using a self- or physician-diagnosis of asthma can be confounded by individuals who do not have asthma such as older subjects with a smoking history who could have COPD. These physician-diagnosed cohorts many times do not have an objective diagnostic basis of asthma based on methacholine BHR or beta agonist responsiveness which could result in confounded and undetected associations. In addition, the participation rates for cases and controls were not reported in the majorities of included studies; thus, our meta-analysis was unable to explore the selection bias. Moreover, with limited information about maternal constitutional and environmental risk factors for asthma (such as smoking history), we could not evaluate the gene-gene and gene-environmental interactions. The different definitions of asthma and the environmental factors in individual studies were obvious sources of clinical heterogeneity and may produce bias. Therefore, moderate-to-high heterogeneities were found in some genetic models for the Gly16Arg polymorphism. Stratification by ethnicity may help to improve homogeneity among studies, but it may also influence statistical power. In addition, some meta-analysis studies claimed to divide the cohorts by ethnicity but it seems like the cohorts were actually being divided by continent for the most part, it will induce the contradictory findings. Third, the Gly16Arg and Gln27Glu polymorphisms tagging for rare variants modulated therapeutic responses and contributed to asthma risk [103]; however, these variants were not specifically genotyped.
Conclusion
In conclusion, the present meta-analysis indicates that the rs1042714 polymorphism is an important genetic protective factor to decrease the risk of developing asthma, especially in children. The rs1042713 polymorphism may be involved in the risk of asthma in the Arab and Hispanic-Latino populations and a protective factor in the Indian population. However, more well-designed and high-quality studies with larger sample sizes should be conducted to support this finding in various ethnic groups.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ADRB2:
-
β2-adrenergic receptor
- AMSTAR:
-
A Measurement Tool to Assess Systematic Reviews
- CI:
-
Confidence intervals
- CNKI:
-
China National Knowledge Infrastructure
- HWE:
-
Hardy-Weinberg equilibrium
- NOS:
-
Newcastle-Ottawa Scale
- OR:
-
Odds ratios
- PRISMA:
-
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- SNP:
-
Single nucleotide polymorphism
References
Busse WW, Lemanske RF Jr. Asthma. N Engl J Med. 2001;344(5):350–62.
Cookson W. The alliance of genes and environment in asthma and allergy. Nature. 1999;402(6760 Suppl):B5–11.
Wills-Karp M, Ewart SL. Time to draw breath: asthma-susceptibility genes are identified. Nat Rev Genet. 2004;5(5):376–87.
Barnes KC. Genomewide association studies in allergy and the influence of ethnicity. Curr Opin Allergy Clin Immunol. 2010;10(5):427–33.
Meyers DA. Genetics of asthma and allergy: what have we learned? J Allergy Clin Immunol. 2010;126(3):439–46 quiz 447-438.
Zhang Y, Moffatt MF, Cookson WO. Genetic and genomic approaches to asthma: new insights for the origins. Curr Opin Pulm Med. 2012;18(1):6–13.
Litonjua AA, Gong L, Duan QL, Shin J, Moore MJ, Weiss ST, Johnson JA, Klein TE, Altman RB. Very important pharmacogene summary ADRB2. Pharmacogenet Genomics. 2010;20(1):64–9.
Dai XH, Zhou JP. Association of IL-13 and beta2 receptor gene polymorphisms with asthma. Shandong Med J. 2008;48(3):119–21.
McGraw DW, Forbes SL, Kramer LA, Liggett SB. Polymorphisms of the 5′ leader cistron of the human beta2-adrenergic receptor regulate receptor expression. J Clin Invest. 1998;102(11):1927–32.
McGraw DW, Liggett SB. Coding block and 5 leader cistron polymorphisms of the beta2-adrenergic receptor. Clin Exp Allergy. 1999;29(Suppl 4):43–5.
Parola AL, Kobilka BK. The peptide product of a 5′ leader cistron in the beta 2 adrenergic receptor mRNA inhibits receptor synthesis. J Biol Chem. 1994;269(6):4497–505.
de Paiva AC, Marson FA, Ribeiro JD, Bertuzzo CS. Asthma: Gln27Glu and Arg16Gly polymorphisms of the beta2-adrenergic receptor gene as risk factors. Allergy Asthma Clin Immunol. 2014;10(1):8.
Green SA, Turki J, Innis M, Liggett SB. Amino-terminal polymorphisms of the human beta 2-adrenergic receptor impart distinct agonist-promoted regulatory properties. Biochemistry. 1994;33(32):9414–9.
Green SA, Turki J, Bejarano P, Hall IP, Liggett SB. Influence of beta 2-adrenergic receptor genotypes on signal transduction in human airway smooth muscle cells. Am J Respir Cell Mol Biol. 1995;13(1):25–33.
Contopoulos-Ioannidis DG, Manoli EN, Ioannidis JP. Meta-analysis of the association of beta2-adrenergic receptor polymorphisms with asthma phenotypes. J Allergy Clin Immunol. 2005;115(5):963–72.
Finkelstein Y, Bournissen FG, Hutson JR, Shannon M. Polymorphism of the ADRB2 gene and response to inhaled beta- agonists in children with asthma: a meta-analysis. J Asthma. 2009;46(9):900–5.
Petrovic-Stanojevic N, Topic A, Nikolic A, Stankovic M, Dopudja-Pantic V, Milenkovic B, Radojkovic D. Polymorphisms of beta2-adrenergic receptor gene in serbian asthmatic adults: effects on response to Beta-agonists. Mol Diagn Ther. 2014;18(6):639–46.
Choudhry S, Ung N, Avila PC, Ziv E, Nazario S, Casal J, Torres A, Gorman JD, Salari K, Rodriguez-Santana JR, et al. Pharmacogenetic differences in response to albuterol between Puerto Ricans and Mexicans with asthma. Am J Respir Crit Care Med. 2005;171(6):563–70.
Israel E, Chinchilli VM, Ford JG, Boushey HA, Cherniack R, Craig TJ, Deykin A, Fagan JK, Fahy JV, Fish J, et al. Use of regularly scheduled albuterol treatment in asthma: genotype-stratified, randomised, placebo-controlled cross-over trial. Lancet. 2004;364(9444):1505–12.
Israel E, Drazen JM, Liggett SB, Boushey HA, Cherniack RM, Chinchilli VM, Cooper DM, Fahy JV, Fish JE, Ford JG, et al. The effect of polymorphisms of the beta (2)-adrenergic receptor on the response to regular use of albuterol in asthma. Am J Respir Crit Care Med. 2000;162(1):75–80.
Liu ZQ, Jiao Y, Liu CJ, Zhang HC, Hu BR. Association between polymorphisms and haplotypes of the beta-2 adrenergic receptor gene and asthma in a Chinese Han population. Int J Clin Exp Med. 2014;7(10):3812–7.
Ramphul K, Lv J, Hua L, Liu QH, Fang DZ, Ji RX, Bao YX. Single nucleotide polymorphisms predisposing to asthma in children of Mauritian Indian and Chinese Han ethnicity. Braz J Med Biol Res. 2014;47(5):394–7.
Martinez-Aguilar NE, Del Rio-Navarro BE, Navarro-Olivos E, Garcia-Ortiz H, Orozco L, Jimenez-Morales S. SPINK5 and ADRB2 haplotypes are risk factors for asthma in Mexican pediatric patients. J Asthma. 2015;52(3):232–9.
Xie H, Cheng Y, Huo Y, Huang G, Su J. Association between beta2-adrenoceptor gene polymorphisms and asthma risk: an updated meta-analysis. PLoS One. 2014;9(7):e101861.
Liang SQ, Chen XL, Deng JM, Wei X, Gong C, Chen ZR, Wang ZB. Beta-2 adrenergic receptor (ADRB2) gene polymorphisms and the risk of asthma: a meta-analysis of case-control studies. PLoS One. 2014;9(8):e104488.
Migita O, Noguchi E, Jian Z, Shibasaki M, Migita T, Ichikawa K, Matsui A, Arinami T. ADRB2 polymorphisms and asthma susceptibility: transmission disequilibrium test and meta-analysis. Int Arch Allergy Immunol. 2004;134(2):150–7.
Thakkinstian A, McEvoy M, Minelli C, Gibson P, Hancox B, Duffy D, Thompson J, Hall I, Kaufman J, Leung TF, et al. Systematic review and meta-analysis of the association between {beta}2-adrenoceptor polymorphisms and asthma: a HuGE review. Am J Epidemiol. 2005;162(3):201–11.
Li X, Zhang Y, Zhang J, Xiao Y, Huang J, Tian C, He C, Deng Y, Yang Y, Fan H. Asthma susceptible genes in Chinese population: a meta-analysis. Respir Res. 2010;11:129.
Chen JL, Huang X, Tan JY, Wan ZD, Wu XW, Li DX. Association between ADRB2 rs1042713 gene polymorphism and susceptibility of asthma in Chinese population: a meta-analysis. Chinese J Immunology. 2015;31(08):1037–9.
Zheng BQ, Wang GL, Yang S, Lu YQ, Liu RJ, Li Y. Study of genetic susceptibility in 198 children with asthma. Zhongguo Dang Dai Er Ke Za Zhi. 2012;14(11):811–4.
Kaneko Y, Masuko H, Sakamoto T, Iijima H, Naito T, Yatagai Y, Yamada H, Konno S, Nishimura M, Noguchi E, et al. Asthma phenotypes in Japanese adults - their associations with the CCL5 and ADRB2 genotypes. Allergol Int. 2013;62(1):113–21.
Karam RA, Sabbah NA, Zidan HE, Rahman HM. Association between genetic polymorphisms of beta2 adrenergic receptors and nocturnal asthma in Egyptian children. J Investig Allergol Clin Immunol. 2013;23(4):262–6.
Tian M, Liang H, Qin QZ, Zhang WX, Zhang SS. ADRB2 polymorphisms in allergic asthma in Han Chinese children. Int Forum Allergy Rhinol. 2016;6(4):367–72.
Kaymak C, Aygun Kocabas N, Aydin N, Oztuna D, Karakaya AE. The relationship between glutathione S-Transferase-P1 and Beta-2 Adrenoreceptor genotypes with asthmatic patients in the Turkish population. Genet Test Mol Biomarkers. 2016;20(9):522–8.
Shea BJ, Reeves BC, Wells G, Thuku M, Hamel C, Moran J, Moher D, Tugwell P, Welch V, Kristjansson E, et al. AMSTAR 2: a critical appraisal tool for systematic reviews that include randomised or non-randomised studies of healthcare interventions, or both. BMJ. 2017;358:j4008.
Potter PC, Van Wyk L, Martin M, Lentes KU, Dowdle EB. Genetic polymorphism of the beta-2 adrenergic receptor in atopic and non-atopic subjects. Clin Exp Allergy. 1993;23(10):874–7.
Reihsaus E, Innis M, MacIntyre N, Liggett SB. Mutations in the gene encoding for the beta 2-adrenergic receptor in normal and asthmatic subjects. Am J Respir Cell Mol Biol. 1993;8(3):334–9.
Martinez FD, Graves PE, Baldini M, Solomon S, Erickson R. Association between genetic polymorphisms of the beta2-adrenoceptor and response to albuterol in children with and without a history of wheezing. J Clin Invest. 1997;100(12):3184–8.
Hopes E, McDougall C, Christie G, Dewar J, Wheatley A, Hall IP, Helms PJ. Association of glutamine 27 polymorphism of beta 2 adrenoceptor with reported childhood asthma: population based study. BMJ. 1998;316(7132):664.
Dewar JC, Wheatley AP, Venn A, Morrison JF, Britton J, Hall IP. Beta2-adrenoceptor polymorphisms are in linkage disequilibrium, but are not associated with asthma in an adult population. Clin Exp Allergy. 1998;28(4):442–8.
Kotani Y, Nishimura Y, Maeda H, Yokoyama M. Beta2-adrenergic receptor polymorphisms affect airway responsiveness to salbutamol in asthmatics. J Asthma. 1999;36(7):583–90.
Holloway JW, Dunbar PR, Riley GA, Sawyer GM, Fitzharris PF, Pearce N, Le Gros GS, Beasley R. Association of beta2-adrenergic receptor polymorphisms with severe asthma. Clin Exp Allergy. 2000;30(8):1097–103.
Gao G, Wang S, Zhang J. Study on beta 2 adrenergic receptor genetic polymorphisms in asthmatics in the people of the Han nationality of northern China. Zhonghua Jie He He Hu Xi Za Zhi. 2000;23(2):93–7.
Wang Z, Chen C, Niu T, Wu D, Yang J, Wang B, Fang Z, Yandava CN, Drazen JM, Weiss ST, et al. Association of asthma with beta (2)-adrenergic receptor gene polymorphism and cigarette smoking. Am J Respir Crit Care Med. 2001;163(6):1404–9.
Shachor J, Chana Z, Varsano S, Erlich T, Goldman E, Dror Y, Yakovy I, Navon R. Genetic polymorphisms of the beta-2 adrenergic receptor in Israelis with severe asthma compared to non-asthmatic Israelis. Isr Med Assoc J. 2003;5(11):821–4.
Lin YC, Lu CC, Shen CY, Lei HY, Guo YL, Su HJ. Roles of genotypes of beta2-adrenergic receptor in the relationship between eosinophil counts and lung function in Taiwanese adolescents. J Asthma. 2003;40(3):265–72.
Binaei S, Christensen M, Murphy C, Zhang Q, Quasney M. Beta2-adrenergic receptor polymorphisms in children with status asthmaticus. Chest. 2003;123(3 Suppl):375S.
Santillan AA, Camargo CA Jr, Ramirez-Rivera A, Delgado-Enciso I, Rojas-Martinez A, Cantu-Diaz F, Barrera-Saldana HA. Association between beta2-adrenoceptor polymorphisms and asthma diagnosis among Mexican adults. J Allergy Clin Immunol. 2003;112(6):1095–100.
Gao JM, Lin YG, Qiu CC, Liu YW, Ma Y, Liu Y. Beta2-adrenergic receptor gene polymorphism in Chinese northern asthmatics. Chin Med Sci J. 2004;19(3):164–9.
Bhatnagar P, Gupta S, Guleria R, Kukreti R. beta2-adrenergic receptor polymorphisms and asthma in the north Indian population. Pharmacogenomics. 2005;6(7):713–9.
Matheson MC, Ellis JA, Raven J, Johns DP, Walters EH, Abramson MJ. Beta2-adrenergic receptor polymorphisms are associated with asthma and COPD in adults. J Hum Genet. 2006;51(11):943–51.
Tsai HJ, Shaikh N, Kho JY, Battle N, Naqvi M, Navarro D, Matallana H, Lilly CM, Eng CS, Kumar G, et al. Beta 2-adrenergic receptor polymorphisms: pharmacogenetic response to bronchodilator among African American asthmatics. Hum Genet. 2006;119(5):547–57.
Munakata M, Harada Y, Ishida T, Saito J, Nagabukuro A, Matsushita H, Koga N, Ohsaki M, Imagawa K, Shiratsuchi T. Molecular-based haplotype analysis of the beta 2-adrenergic receptor gene (ADRB2) in Japanese asthmatic and non-asthmatic subjects. Allergol Int. 2006;55(2):191–8.
Llanes E, Quiralte J, Lopez E, Sastre B, Chacartegui M, del Pozo V, Palomino P, Lahoz C, Cardaba B. Analysis of polymorphisms in olive pollen allergy: IL13, IL4RA, IL5 and ADRB2 genes. Int Arch Allergy Immunol. 2009;148(3):228–38.
Szczepankiewicz A, Breborowicz A, Sobkowiak P, Kramer L, Popiel A. Role of ADRB2 gene polymorphism in asthma and response to beta (2)-agonists in polish children. J Appl Genet. 2009;50(3):275–81.
Ye YM, Kang YM, Kim SH, Lee HY, Kim CW, Park CS, Hong CS, Park HS. Probable role of Beta 2-adrenergic receptor gene haplotype in toluene diisocyanate-induced asthma. Allergy, Asthma Immunol Res. 2010;2(4):260–6.
Qiu YY, Zhang XL, Qin Y, Yin KS, Zhang DP. Beta (2)-adrenergic receptor haplotype/polymorphisms and asthma susceptibility and clinical phenotype in a Chinese Han population. Allergy Asthma Proc. 2010;31(5):91–7.
Al-Rubaish A. beta2-adrenergic receptor gene polymorphisms in normal and asthmatic individuals in the Eastern Province of Saudi Arabia. Ann Saudi Med. 2011;31(6):586–90.
Fu WP, Zhao ZH, Zhong L, Sun C, Fang LZ, Liu L, Zhang JQ, Wang L, Shu JK, Wang XM, et al. Relationship between polymorphisms in the 5′ leader cistron, positions 16 and 27 of the adrenergic beta2 receptor gene and asthma in a Han population from southwest China. Respirology. 2011;16(8):1221–7.
Kohyama K, Abe S, Kodaira K, Yukawa T, Hozawa S, Morioka J, Inamura H, Ota M, Sagara H, Schwartz LB, et al. Arg16Gly beta2-adrenergic receptor gene polymorphism in Japanese patients with aspirin-exacerbated respiratory disease. Int Arch Allergy Immunol. 2011;156(4):405–11.
Thomsen M, Nordestgaard BG, Sethi AA, Tybjaerg-Hansen A, Dahl M. beta2-adrenergic receptor polymorphisms, asthma and COPD: two large population-based studies. Eur Respir J. 2012;39(3):558–66.
Chiang CH, Lin MW, Chung MY, Yang UC. The association between the IL-4, ADRbeta2 and ADAM 33 gene polymorphisms and asthma in the Taiwanese population. J Chin Med Assoc. 2012;75(12):635–43.
Isaza C, Sepulveda-Arias JC, Agudelo BI, Arciniegas W, Henao J, Porras GL, Beltran L. beta (2) -adrenoreceptor polymorphisms in asthmatic and non-asthmatic schoolchildren from Colombia and their relationship to treatment response. Pediatr Pulmonol. 2012;47(9):848–55.
Birbian N, Singh J, Jindal SK, Singla N. Association of beta (2)-adrenergic receptor polymorphisms with asthma in a north Indian population. Lung. 2012;190(5):497–504.
Larocca N, Moreno D, Garmendia JV, Velasquez O, Martin-Rojo J, Talamo C, Garcia A, De Sanctis JB. Beta 2 adrenergic receptor polymorphisms, at codons 16 and 27, and bronchodilator responses in adult Venezuelan asthmatic patients. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2013;157(4):374–8.
Pino-Yanes M, Corrales A, Cumplido J, Poza P, Sanchez-Machin I, Sanchez-Palacios A, Figueroa J, Acosta-Fernandez O, Buset N, Garcia-Robaina JC, et al. Assessing the validity of asthma associations for eight candidate genes and age at diagnosis effects. PLoS One. 2013;8(9):e73157.
Saadi AV, Gupta H, Angural A, Dhanya SK, Mony S, Oberoi D, D'Souza SC, Sahoo RC, Hande MH, Gopinath PM, et al. Single nucleotide polymorphisms of ADRB2 gene and their association with susceptibility for plasmodium falciparum malaria and asthma in an Indian population. Infect Genet Evol. 2013;20:140–7.
Bandaru S, Akka J, Marri VK, Alvala M, Ponnala D, Mundluru HP. Analysis of ADRB2 (Arg16Gly) gene variant with susceptibility, Pharmacogenetic response and disease severity in south Indian asthmatics. Inflammation. 2015;38(6):2146–55.
Leite N, Lazarotto L, Milano GE, Titski AC, Consentino CL, de Mattos F, de Andrade FA, Furtado-Alle L. Beta 2-adrenergic receptor gene association with overweight and asthma in children and adolescents and its relationship with physical fitness. Rev Paul Pediatr. 2015;33(4):381–6.
Ramphul K, Hua L, Bao YX, Li JY, Liu QH, Ji RX, Fang DZ. Identification of IL13 C1923T as a single nucleotide polymorphism for asthma in children from Mauritius. Pediatr Allergy Immunol Pulmonol. 2015;28(2):92–5.
Shah NJ, Vinod Kumar S, Gurusamy U, Annan Sudarsan AK, Shewade DG. Effect of ADRB2 (adrenergic receptor beta2) gene polymorphisms on the occurrence of asthma and on the response to nebulized salbutamol in south Indian patients with bronchial asthma. J Asthma. 2015;52(8):755–62.
Guo X, Zheng H, Mao C, Guan E, Si H. An association and meta-analysis study of 4 SNPs from beta-2 adrenergic receptor (ADRB2) gene with risk of asthma in children. Asian Pac J Allergy Immunol. 2016;34(1):11–20.
Sahi PK, Shastri S, Lodha R, Gupta N, Pandey RM, Kabra SK, Kabra M. ADRB2 polymorphism and salbutamol responsiveness in northern Indian children with mild to moderate exacerbation of asthma. Indian Pediatr. 2016;53(3):211–5.
Weir TD, Mallek N, Sandford AJ, Bai TR, Awadh N, Fitzgerald JM, Cockcroft D, James A, Liggett SB, Pare PD. beta2-adrenergic receptor haplotypes in mild, moderate and fatal/near fatal asthma. Am J Respir Crit Care Med. 1998;158(3):787–91.
Hakonarson H, Bjornsdottir US, Ostermann E, Arnason T, Adalsteinsdottir AE, Halapi E, Shkolny D, Kristjansson K, Gudnadottir SA, Frigge ML, et al. Allelic frequencies and patterns of single-nucleotide polymorphisms in candidate genes for asthma and atopy in Iceland. Am J Respir Crit Care Med. 2001;164(11):2036–44.
Leung TF, Tang NL, Chan IH, Li AM, Ha G, Lam CW, Fok TF. Distribution in allele frequencies of predisposition-to-atopy genotypes in Chinese children. Pediatr Pulmonol. 2002;34(6):419–24.
Telleria JJ, Blanco-Quiros A, Muntion S, Antonio Garrote J, Arranz E, Armentia A, Diez I, Castro J. Tachyphylaxis to beta2-agonists in Spanish asthmatic patients could be modulated by beta2-adrenoceptor gene polymorphisms. Respir Med. 2006;100(6):1072–8.
Xing J, Wang C, Liu JZ, Yan M, Huang KW, et al. Association of receptor gene polymorphisms with asthma in Northern Chinese Han Population. Chinese J Internal Med. 2001;40(3):340–2.
Liao W, Li LW, Zhao CM, Guang LX, Yin XJ, et al. Preliminary study on the relationship between β2-adrenergic receptors genetic polymor- phisms and asthma in children of Han nationality of Chongqing. J Third Military Medical University. 2001;23:968–71.
Dai LM, Wang WZ, Zhang YP, Li W, Zhao ZH, et al. Association of beta2 receptor gene polymorphisms with lung function in asthma patients. Chinese Journal of Tuberculosis and Respiratory Diseases. 2002;25(2):554–5.
Wang W, Wufuer HMTL, Shabiti YLHMJ, Xiang YB, Abula ABLKM. Association between the genetic polymorphisms of β2-adrenergic receptor gene and the asthma susceptibility and clinical phenotypes in Uygur population. J Cardiovascular Pulmonary Dis. 2004;23:147–52.
Feng DX, Ye WX, Zhang XY, Yu H, Diao XY, et al. Study on β2-adrenergic receptor genetic polymorphisms and asthma. J Modern Clin Med Bioengineering. 2004;10:5–7.
Kocabas NA, Kaymak C, Aydin N, Oztuna D, Karakaya AE. Investigation of the beta 2-adrenoceptor (ADRB2) 16 and glutathione S-transferase P1 (GSTP1) gene polymorphisms in Turkish asthma patients. Toxicol Lett. 2007;172:S164–5.
Tuerxun KLBN, Shabiti YLHM, Wang W, Wufuer HMTL. Study on the β2AR polymorphism in asthmatic abnormal black savda patients. Journal of Xinjiang Medical University. 2007;30:945–8.
Cui LY, Liu XH, Gao LX, Fan DS. Study on the association between β2-adrenergic receptor genetic polymorphisms and asthma in the population of Inner Mongolia. Chinese journal of Clinical Medicin. 2007;14:477–81.
Chan IH, Tang NL, Leung TF, Huang W, Lam YY, Li CY, Wong CK, Wong GW, Lam CW. Study of gene-gene interactions for endophenotypic quantitative traits in Chinese asthmatic children. Allergy. 2008;63(8):1031–9.
Zhang XY, Zhao WL, Gui Q, He NH. Relationship between genetic polymorphisms of β2-adrenergic receptor and childhood asthma. Journal of Clinical Pediatrics. 2008;26:399–408.
Xie Y, Yang Z, Chai BC. Relationship of genetic polymorphisms of β2 - adrenergic receptor and asthma in children in Shanghai area. J of Applied Clinical Pediatrics. 2008;23:272–3.
Liu L, Fang LZ, Dai LM. Combination Effect of Gene Polymorphisms in 16 Position of β2-adenergic Receptor and Cigarette Smoking on Asthma in Chinese Han Individuals. Medical Recapitulate. 2009;15(4):1247–78.
Wang JY, Liou YH, Wu YJ, Hsiao YH, Wu LS. An association study of 13 SNPs from seven candidate genes with pediatric asthma and a preliminary study for genetic testing by multiple variants in Taiwanese population. J Clin Immunol. 2009;29(2):205–9.
Lv J, Liu Q, Hua L, Dong X, Bao Y. Association of five single nucleotide polymorphism loci with asthma in children of Chinese Han nationality. J Asthma. 2009;46(6):582–5.
He XQLF, Tan JY, Yang XX. Association of single nucleotide polymorphisms of ADRB2 Arg16Gly with asthma in southern Chinese population. Immunological Journal. 2012;28:687–90.
Ye WXFD, Zhang XY, Yu H, Xu M. Study on the relationship between β2-adrenergic receptor genetic polymorphisms and asthma a in Miao nationality. J Med Res. 2011;40:83–5.
Tatarskyy PFCN, Kucherenko AM, Gulkovskyi RV, Arabskaya LP, et al. Study of possible role of CYP1A1, GSTT1, GSTM1, GSTP1, NAT2 and ADRB2 genes polymorphisms in bronchial asthma development in children. Biopolymers and Cell. 2011;27(1):66–73.
Yang ZZH, Wang W, Yin Y, Zhang L, et al. Effect of β2-adrenergic receptor polymorphisms on childhood asthma and therapeutic efficacy of long acting β2-agonist. J of Clin Pediatrics. 2012;30:739–43.
Almomani BA, Al-Eitan LN, Samrah SM, Al-Quasmi MN, McKnight AJ. Candidate gene analysis of asthma in a population of Arab descent: a case-control study in Jordan. Personalized Med. 2017;14(1):51–61.
Alghobashy AA, Elsharawy SA, Alkholy UM, Abdalmonem N, Abdou MA, Basset MAA, Pasha HF. B2 adrenergic receptor gene polymorphism effect on childhood asthma severity and response to treatment. Pediatr Res. 2018;83(3):597–605.
Maxwell TJ, Ameyaw MM, Pritchard S, Thornton N, Folayan G, Githang'a J, Indalo A, Tariq M, Mobarek A, Evans DA, et al. Beta-2 adrenergic receptor genotypes and haplotypes in different ethnic groups. Int J Mol Med. 2005;16(4):573–80.
Xie HG, Stein CM, Kim RB, Xiao ZS, He N, Zhou HH, Gainer JV, Brown NJ, Haines JL, Wood AJ. Frequency of functionally important beta-2 adrenoceptor polymorphisms varies markedly among African-American, Caucasian and Chinese individuals. Pharmacogenetics. 1999;9(4):511–6.
Ortega VE, Meyers DA. Pharmacogenetics: implications of race and ethnicity on defining genetic profiles for personalized medicine. J Allergy Clin Immunol. 2014;133(1):16–26.
Humes K, Jones NA, Ramirez RR. Overview of race and Hispanic origin: 2010; 2011.
Hall IP, Wheatley A, Wilding P, Liggett SB. Association of Glu 27 beta 2-adrenoceptor polymorphism with lower airway reactivity in asthmatic subjects. Lancet. 1995;345(8959):1213–4.
Ortega VE, Hawkins GA, Moore WC, Hastie AT, Ampleford EJ, Busse WW, Castro M, Chardon D, Erzurum SC, Israel E, et al. Effect of rare variants in ADRB2 on risk of severe exacerbations and symptom control during longacting beta agonist treatment in a multiethnic asthma population: a genetic study. Lancet Respir Med. 2014;2(3):204–13.
Acknowledgements
Not applicable.
Funding
No funding.
Author information
Authors and Affiliations
Contributions
XN conceived and designed the experiments. SZ and WZ searched and assessed the literatures, and extracted the data from literature, respectively. SZ analyzed the data by STATA 11.0 software and prepared the figures SZ and XN drafted and revised the manuscript. All authors have read and approve the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Characteristics of the studies included in the meta-analysis. (XLSX 14 kb)
Additional file 2: Table S2
. Genotype and allele distribution in the meta-analysis for the rs1042713 polymorphism. (XLSX 17 kb)
Additional file 3: Table S3
. Genotype and allele distribution in the meta-analysis for the rs1042714 polymorphism. (XLSX 15 kb)
Additional file 4: Table S4.
Genotype and allele distribution in the meta-analysis for the rs1042711 polymorphism. (XLSX 9 kb)
Additional file 5: Figure S1.
Forest plots of the association between the ADRB2 rs1042713 polymorphism and risk of asthma in recessive model comparison. GG vs. GA + AA genotype. Each study is shown by an OR estimate with the corresponding 95% CIs. The horizontal lines denote the 95% CIs and the squares represent the point OR estimate of each study. The size of the square is proportional to its inverse-variance weight in the meta-analysis. The diamond represents the pooled meta-analysis effect size estimate. The stratified meta-analysis was performed regarding the ethnicity. (TIF 19900 kb)
Additional file 6: Figure S2.
Forest plots of the association between the ADRB2 rs1042713 polymorphism and risk of asthma in homozygote genotype comparison. GG vs. AA genotype. Each study is shown by an OR estimate with the corresponding 95% CIs. The horizontal lines denote the 95% CIs and the squares represent the point OR estimate of each study. The size of the square is proportional to its inverse-variance weight in the meta-analysis. The diamond represents the pooled meta-analysis effect size estimate. The stratified meta-analysis was performed regarding the ethnicity. (TIF 19918 kb)
Additional file 7: Figure S3.
Forest plots of the association between the ADRB2 rs1042713 polymorphism and risk of asthma in the allele comparison. G vs. A allele. Each study is shown by an OR estimate with the corresponding 95% CIs. The horizontal lines denote the 95% CIs and the squares represent the point OR estimate of each study. The size of the square is proportional to its inverse-variance weight in the meta-analysis. The diamond represents the pooled meta-analysis effect size estimate. The stratified meta-analysis was performed regarding the ethnicity. (TIF 19763 kb)
Additional file 8: Figure S4.
Forest plots of the association between the ADRB2 rs1042714 polymorphism and risk of asthma in the allele comparison. G vs. C allele. Each study is shown by an OR estimate with the corresponding 95% CIs. The horizontal lines denote the 95% CIs and the squares represent the point OR estimate of each study. The size of the square is proportional to its inverse-variance weight in the meta-analysis. The diamond represents the pooled meta-analysis effect size estimate. The stratified meta-analysis was performed regarding the ethnicity. (TIF 23295 kb)
Additional file 9: Figure S5.
Forest plots of the association between the ADRB2 rs1042714 polymorphism and risk of asthma in dominant model comparison. GG + CG vs. CC genotype. Each study is shown by an OR estimate with the corresponding 95% CIs. The horizontal lines denote the 95% CIs and the squares represent the point OR estimate of each study. The size of the square is proportional to its inverse-variance weight in the meta-analysis. The diamond represents the pooled meta-analysis effect size estimate. The stratified meta-analysis was performed regarding the age. (TIF 24131 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhao, S., Zhang, W. & Nie, X. Association of β2-adrenergic receptor gene polymorphisms (rs1042713, rs1042714, rs1042711) with asthma risk: a systematic review and updated meta-analysis. BMC Pulm Med 19, 202 (2019). https://doi.org/10.1186/s12890-019-0962-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12890-019-0962-z