Abstract
Background
Mitochondrial diseases are heterogeneous in terms of clinical manifestations and genetic characteristics. The dynamin 1-like gene (DNM1L) encodes dynamin-related protein 1 (DRP1), a member of the GTPases dynamin superfamily responsible for mitochondrial and peroxisomal fission. DNM1L variants can lead to mitochondrial fission dysfunction.
Case presentation
Herein, we report a distinctive clinical phenotype associated with a novel variant of DNM1L and review the relevant literature. A 5-year-old girl presented with paroxysmal hemiplegia, astigmatism, and strabismus. Levocarnitine and coenzyme Q10 supplement showed good efficacy. Based on the patient’s clinical data, trio whole-exome sequencing (trio-WES) and mtDNA sequencing were performed to identify the potential causative genes, and Sanger sequencing was used to validate the specific variation in the proband and her family members. The results showed a novel de novo heterozygous nonsense variant in exon 20 of the DNM1L gene, c.2161C>T, p.Gln721Ter, which is predicted to be a pathogenic variant according to the ACMG guidelines. The proband has a previously undescribed clinical manifestation, namely hemiparesis, which may be an additional feature of the growing phenotypic spectrum of DNM1L-related diseases.
Conclusion
Our findings elucidate a novel variant in DNM1L-related disease and reveal an expanding phenotypic spectrum associated with DNM1L variants. This report highlights the necessity of next generation sequencing for early diagnosis of patients, and that further clinical phenotypic and genotypic analysis may help to improve the understanding of DNM1L-related diseases.
Similar content being viewed by others
Background
Mitochondria are dynamic organelles that undergo constant fusion and fission, both of which are mediated by nuclear DNA (nDNA)-encoded proteins that act on the mitochondrial membrane [1]. A proper dynamic balance between mitochondrial fission and fusion is essential to maintain the morphology of the mitochondrial network and function. Defects in mitochondrial proteins can lead to abnormal fusion and fission, resulting in clinical disease [2]. In mammals, DRP1, a protein encoded by the DNM1L gene, is a core component of the mitochondrial fission process and contains an N-terminal GTPase domain, a middle domain, a non-conserved variable domain (VD), and a C-terminal GTPase effector domain (GED) [3,4,5]. The major functional units are the GTPase domain, which provides the mechanical force for membrane contraction, and the middle domain, which mediates DRP1 oligomerization [6]. In addition, DRP1 is also involved in mediating peroxisomal fission [7].
DRP1 impairment causes two neurological diseases associated with pathogenic variants in DNM1L (MIM*603,850), including encephalopathy due to defective mitochondrial and peroxisomal fission-1 (EMPF1, MIM #614,388) and optic atrophy 5 (MIM#610,708). EMPF1, caused by monoallelic or biallelic pathogenic DNM1L variants, is frequently associated with developmental delay, hypotonia, refractory epilepsy, and even death [8,9,10]. The first reported de novo heterozygous DNM1L variant, c.1184C>A, p.Ala395Asp, caused a disease phenotype manifested by global developmental delay, neonatal encephalopathy, microcephaly, and optic atrophy [10]. The patient died suddenly at 37 days of age with minimal developmental progress. It has been reported that the p.Ala395Asp variant is located in the middle domain of DRP1 and impairs higher order assembly and GTPase activity of DRP1, resulting in abnormal mitochondrial and peroxisomal fission [11]. Recently, an increasing number of heterozygous variants in the DRP1 middle domain have been reported to cause severe encephalopathy with epilepsy and/or failure to thrive [12,13,14]. In contrast, heterozygous variants in the GTPase domain appear to be associated with a milder phenotype limited to isolated optic atrophy [15]. To our knowledge, disease-causing variants in the GED have rarely been reported in humans.
Here, we describe a 5-year-old girl with a novel c.2161C>T, p.Gln721Ter DNM1L variant identified in the GED. She had a different clinical presentation compared to previously reported cases, expanding the clinical presentation of DNM1L-related mitochondrial disease. Furthermore, due to the lack of studies on the phenotype-genotype correlation of DNM1L-related diseases, a review of the literature summarizing the relevance of clinical and genetic features may deepen the understanding of the disease.
Case presentation
Methods
Literature acquisition
The clinical data of the proband were collected from December 2020 to June 2023 in the Children’s Hospital Affiliated to Zhengzhou University, China. Furthermore, we conducted a literature review via PubMed (up to June 2023), using the following search string: “mitochondrial diseases” or “DRP1” or “EMPF1” associated to the keyword “DNM1L” and “children”. Additionally, a manual reference check of the retrieved literature was performed.
Whole-exome sequencing and mtDNA sequencing
This study was approved by the Ethics Committee of Henan Children’s Hospital. After obtaining informed consent, peripheral blood samples were collected from the proband and her family members including parents and sister. Genomic DNA was extracted from 200 µL of peripheral blood from each participant using the Qiagen DNA Blood Midi/Mini kit (Qiagen GmbH, Hilden, Germany) and sheared to approximately 200 bp. The DNA fragments were hybridized and captured by NanoWES according to the manufacturer’s protocol. The libraries were then quantified by qPCR and the size distribution was determined using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). The trio-WES was performed using the Illumina HiSeq2000 sequencer platform (Illumina, San Diego, CA, USA) with 150 bp pair-end sequencing mode. Raw data were obtained after processing with CASAVA v1.82 software (Illumina, San Diego, CA, USA). Sequencing reads were aligned to the human reference genome (hg19/GRCh37) using BWA tool (v0.7.12r1044; http://biobwa.sourceforge.net/) and PCR duplicates were removed by using Picard v1.57 software (http://picard.sourceforge.net/). In general, the test platform detected more than 95% of the target regions with a sensitivity of >99%. Verita Trekker® Variants Detection System (BerryGenomics, Beijing, China) and GATK software (https://software.broadinstitute.org/gatk/) were used for variant calling, followed by ANNOVAR software and Enliven® Variants Annotation Interpretation System (BerryGenomics, Beijing, China) for annotation. In addition, suspicious variants were evaluated according to the variation interpretation guidelines of American College of Medical Genetics and Genomics (ACMG) [16].
Case report and results
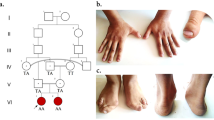
The case is the second child from non-consanguineous Chinese parents, born at term with normal birth parameters and has a healthy older sister. The family pedigree is shown in Fig. 1. She developed normally from the neonatal period until 1.5 years, apart from presenting with astigmatism and strabismus. She was able to call Papa and Mama at 10 months of age, had normal intelligence and was walking at 14 months. At the age of 1.5 years, she was found to have left-sided limb dysfunction preceded by a febrile illness. As the patient had left lower extremity weakness, she had left tip toe walking. The episodic limb dysfunction occurred every 1 to 3 weeks and lasted from 5 min to about 1 day. She was physically immobile during severe episodes, but her symptoms would be relieved after waking up the next day. The girl also had right-sided limb weakness with a right-sided hemiplegic gait and was suspected of having alternating hemiplegia of childhood (AHC). However, sequencing of AHC-related genes showed no significant abnormalities, such as the ATP1A3 gene and the ATP1A2 gene.
Physical examinations of the patient were unremarkable, except for strabismus. Extensive workups were performed for potential etiology including a normal blood ammonia level, lactate level, pyruvate level, serum amino acids, acylcarnitine profiles, liver and kidney function, electromyography (EMG), and electroencephalogram (EEG). Urinary organic acid results were unremarkable. Magnetic resonance imaging (MRI) of the brain at the age of 3 years showed no significant abnormalities. The patient was reported to have hemiparesis preceded by febrile illness or excessive exercise, but long-term treatment with levocarnitine (1 g/d) and coenzyme Q10 (20 mg/d) was started 6 months after the first episode of hemiparesis. After 2 months of treatment with coenzyme Q10 and levocarnitine, she has had no further episodes of hemiplegia and motor function had returned to normal. The patient is now 5 years old, with a weight of 16 kg, a height of 102 cm, a head circumference of 48.5 cm, and is doing well at school.
The trio-WES was used to identify potentially pathogenic genetic variants and Sanger sequencing was applied to confirm the identified variants. The results of the Sanger sequencing were analyzed using Chromas software, while the possible pathogenic effects of variants were predicted as previously described [17]. Furthermore, sequencing of the entire mtDNA genome of the child showed no pathologically significant variants. We identified a heterozygous stop-gain mutation of the DNM1L gene in the proband: c.2161C>T (exon 20, NM_012062.5, chr12:32896294), resulting in a stop-gain mutation NP_036192.2: p.Gln721Ter. The results of Sanger sequencing confirmed its absence in her parents, suggesting that the variant is de novo (Fig. 1). The variant was not found in human disease databases (including the 1000 Genomes Project, EXAC, gnomAD, and dbSNP databases), and was predicted to be disease-causing by online bioinformatics tools (Table 1).
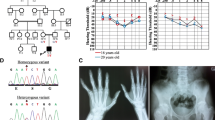
DNM1L c.2161C>T, p.Gln721Ter was a nonsense mutation and was classified as likely pathogenic: PVS1_Moderate + PS2_Moderate + PM2_supporting according to the ACMG guidelines. Other previously reported pathogenic variants were then used for comparison, as described in Fig. 2a. The identified amino acid was highly conserved across different species as shown in Fig. 2c. No other suspected variants were found in the known disease-causing genes that might explain the clinical phenotype of our patients (Supplementary Table 1).
Genetic location of DNM1L gene variants associated with mitochondrial diseases identified to date. A Schematic representation of the location of identified variants in each domain of the DRP1 protein. Changes in amino acid distribution associated with protein domains. Variants reported in this study are shown in red, and “x” indicates the number of cases. B The number of previously reported variants in each domain. C Comparison of the amino acid sequences of the DRP1 protein in different species. The amino acid sequence of the variant site is highly conserved (red font)
Review of the literature
Based on the differences in phenotypes observed in the patients studied, all reported cases of DNM1L variants were reviewed (Supplementary Table 2) [5, 8,9,10, 12,13,14,15, 18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40]. A total of 53 cases were analyzed together with our case. In the described cases, the median age at which patients initially presented with clinical manifestations of specific organs or systems due to DNM1L variants was 12 months (range, neonatal period to 13 years). 40.0% of the cases eventually died (range, 8 days to 20 years), with a minimum age of death of 8 days and an average age of death of 5.7 years.
The phenotypic spectrum of DNM1L variants included neurological, craniofacial, cardiac, and ocular features. Neurological manifestations such as developmental delay/regression, hypotonia, ataxia, peripheral neuropathy, and epilepsy were the main characteristic features of patients with EMPF1. Of the 50 reported cases (except for pedigrees 1, 2, and 14), clinical manifestations included developmental delay/regression (DD and DR) in 74.0% (37/50), epilepsy (GTCS, SRSE, RSE, and FSE) in 68.0% (34/50), and dystonia in 32.0% (16/50). Other symptoms included 9 cases of peripheral neuropathy, 9 cases of microcephaly, 9 cases of encephalopathy, 8 cases of ataxia, 6 cases of pain insensitivity, 6 cases of dysarthria, and 5 cases of strabismus (Table 2). Regarding the auxiliary examinations, 68.0% (34/50) of the cases showed abnormal brain MRI and 44.0% (22/50) of the cases showed abnormal EEG. In addition, 42.0% (21/50) of the cases had elevated lactate.
To date, 33 pathogenic and/or potentially pathogenic variants have been reported in patients. De novo variants were identified in 79.2% (42/53) of the cases, and homozygous variants were identified in only 3.8% (2/53) of cases [5, 9]. As shown in Fig. 2b, more than half of the variants were located in the middle domain of the DRP1 protein, and 67.6% (23/34) of the middle domain variants occurred mainly in exon 11, confirming that exon 11 was the hotspot of variants in the whole DNM1L gene.
Discussion and conclusions
In recent years, mitochondrial genes and nuclear genes related to mitochondrial structure and function have been extensively identified. DNM1L, encoding DRP1, is a member of the dynamin superfamily of GTPases that mediates mitochondrial and peroxisomal fission [3, 7]. Currently, mitochondrial diseases associated with DNM1L are rarely studied. The first case of DNM1L deficiency was reported in 2007 by Waterham et al., who described a case of infanthood-onset of mitochondrial disease with global developmental delay, microcephaly, abnormal brain development, optic atrophy, and persistent hyperlactatemia [10]. Subsequent studies have further broadened the clinical phenotypic spectrum.
The pathogenic variant in DNM1L is associated with a neurological disorder called “EMPF1” and has been described as a fatal encephalopathy. Affected individuals have different phenotypes ranging from severe hypotonia leading to death in the neonatal period to developmental delay/regression (DD/DR), with or without epilepsy [18, 23, 26]. Interestingly, the DNM1L variant in pedigrees 1, 2, and 14 exhibited a milder phenotype, familial isolated optic atrophy, and the results of functional analysis of these variants suggested a dominant negative mechanism [15, 41]. Our patient had an onset at 1.5 years of age and presented with paroxysmal limb motor dysfunction, manifested by unilateral limb weakness, tiptoeing, and a hemiplegic gait on the affected side. This condition has not been seen in previous reports, revealing an extended phenotype of the DNM1L variant. Interestingly, treatment with coenzyme Q10 and Levocarnitine was effective, contrary to reported cases [14, 19, 22, 33]. Furthermore, she was found to have strabismus and visual impairment, consistent with the phenotype of the novel variant of the DNM1L gene identified [15, 19], but without epilepsy throughout the developmental milestones.
Generally, pathogenic variants involving the DRP1 middle domain also appear to be more severe than those affecting the GTPase domain, which mainly manifest as visual abnormalities with or without a tendency to neurological and developmental deficits [5, 8, 15, 24]. Variants in the GTPase domain have been reported in 29 patients, all of whom had a relatively stable course, except for two who developed respiratory failure and died soon after birth [23]. Moreover, we found that the incidence of developmental delay/regression and epilepsy was greater in the middle domain than that in the GTPase domain, and the proportion of epileptic persistence and refractory epilepsy was similarly higher in the middle domain (Table 2). The reason may be that variants in the GTPase domain may disrupt the binding of the GTPase domain, and the presence of multiple interaction sites in the oligomers may hinder their cohesion, resulting in a relatively mild clinical phenotype [5].
The GED plays a role in the activation of the GTPase domain. Previous studies have shown that the GED is involved in the formation and stability of DRP1 homodimeric complexes and that the corresponding GED variant in human DRP1 reduces GTPase activity and decreases intramolecular interactions between the GED and the middle domain [8, 40, 42]. The first patient with the GED variant (p.Tyr691Cys) was found to have neonatal hypotonia that progressed to unprovoked myoclonic seizures, developmental delay, and static encephalopathy accompanied by nystagmus, optic atrophy, and scoliosis. The second case of the GED variant (p.Arg710Gly) showed similar symptoms to those of the p.Tyr691Cys variant. Interestingly, both patients with GED variants exhibited less severe phenotypes such as epilepsy, optic atrophy, and impaired mobility [8, 40]. Here, we describe a patient with a distinct phenotype due to a novel variant in the DNM1L gene located in the GED of the DRP1 protein. As far as we know, this is the third GED variant identified to date. The variant produces a stop-gain mutation (p.Gln721Ter) in the DRP1 protein, resulting in an early termination codon at position 721. Our patient showed no epilepsy or optic atrophy throughout the developmental milestones except for hemiplegia and strabismus, and had the mildest phenotype compared to the patients mentioned above. However, as the interactions, functional and structural roles of the GED of mammalian DRP1 remain unclear and reports of GED variants associated with human disease are lacking, the impact of this domain remains to be further explored. Additionally, based on the difference in age of onset, whether the child eventually presented with epilepsy or encephalopathy needs to be considered in follow-up.
In conclusion, we identified a novel de novo heterozygous DNM1L variant in one individual in a Chinese family. Not only did the case share similar clinical features compared with previous cases, but the newly reported manifestations, such as hemiparesis, broadened the phenotypic spectrum of the DNM1L variant. DNM1L-related diseases have heterogeneous clinical manifestations and genetic features that may affect multiple systems, but the diagnosis of this disease requires genetic testing for confirmation. Furthermore, future functional studies will be essential to confirm that the variant is truly deleterious. Our systematic description of the clinical presentation and progression of patients will help to identify other rare patients with DNM1L variants in a timely manner. Importantly, this report provides a better understanding of the impact of DNM1L variants on phenotypic outcomes, which will inform the clinical diagnosis of DNM1L-related diseases.
Data availability
The original contributions presented in the study are included in the article and further inquiries can be directed to the corresponding author.
Abbreviations
- ACMG:
-
American College of Medical Genetics and Genomics
- DNM1L :
-
Dynamin 1-like
- DRP1:
-
Dynamin-related protein 1
- DD:
-
Developmental delay
- DR:
-
Developmental regression
- EMPF1:
-
Encephalopathy due to defective mitochondrial and peroxisomal fission-1
- FSE:
-
Focal status epilepticus
- GED:
-
GTPase effector domain
- GDD:
-
Global developmental delay
- GTCS:
-
Generalized tonic-clonic seizures
- MRI:
-
Magnetic resonance imaging
- N/A:
-
Not applicable
- ND:
-
No description
- nDNA:
-
Nuclear DNA
- RSE:
-
Refractory status epilepticus
- SRSE:
-
Super refractory status epilepticus
- trio-WES:
-
Trio-whole-exome sequencing
- VD:
-
Variable domain
References
Al Ojaimi M, Salah A, El-Hattab AW. Mitochondrial fission and Fusion: Molecular mechanisms, Biological functions, and related disorders. Membranes 2022, 12(9).
Chen H, Chan DC. Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative Diseases. Hum Mol Genet. 2009;18(R2):R169–176.
Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–87.
Lackner LL. Shaping the dynamic mitochondrial network. BMC Biol. 2014;12:35.
Hogarth KA, Costford SR, Yoon G, Sondheimer N, Maynes JT. DNM1L variant alters baseline mitochondrial function and response to stress in a patient with severe neurological dysfunction. Biochem Genet. 2018;56(1–2):56–77.
Tilokani L, Nagashima S, Paupe V, Prudent J. Mitochondrial dynamics: overview of molecular mechanisms. Essays Biochem. 2018;62(3):341–60.
Schrader M, Bonekamp NA, Islinger M. Fission and proliferation of peroxisomes. Biochim Biophys Acta. 2012;1822(9):1343–57.
Nolden KA, Egner JM, Collier JJ, Russell OM, Alston CL, Harwig MC, Widlansky ME, Sasorith S, Barbosa IA, Douglas AG et al. Novel DNM1L variants impair mitochondrial dynamics through divergent mechanisms. Life Scie Allian. 2022;5(12).
Verrigni D, Di Nottia M, Ardissone A, Baruffini E, Nasca A, Legati A, Bellacchio E, Fagiolari G, Martinelli D, Fusco L, et al. Clinical-genetic features and peculiar muscle histopathology in infantile DNM1L-related mitochondrial epileptic encephalopathy. Hum Mutat. 2019;40(5):601–18.
Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356(17):1736–41.
Chang CR, Manlandro CM, Arnoult D, Stadler J, Posey AE, Hill RB, Blackstone C. A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J Biol Chem. 2010;285(42):32494–503.
Nolan DA, Chen B, Michon AM, Salatka E, Arndt D. A Rasmussen encephalitis, autoimmune encephalitis, and mitochondrial Disease mimicker: expanding the DNM1L-associated intractable Epilepsy and encephalopathy phenotype. Epileptic Disorders: International Epilepsy Journal with Videotape. 2019;21(1):112–6.
Fahrner JA, Liu R, Perry MS, Klein J, Chan DC. A novel de novo dominant negative mutation in DNM1L impairs mitochondrial fission and presents as childhood epileptic encephalopathy. Am J Med Genet Part A. 2016;170(8):2002–11.
Díez H, Cortès-Saladelafont E, Ormazábal A, Marmiese AF, Armstrong J, Matalonga L, Bravo M, Briones P, Emperador S, Montoya J, et al. Severe infantile parkinsonism because of a de novo mutation on DLP1 mitochondrial-peroxisomal protein. Mov Disorders: Official J Mov Disorder Soc. 2017;32(7):1108–10.
Gerber S, Charif M, Chevrollier A, Chaumette T, Angebault C, Kane MS, Paris A, Alban J, Quiles M, Delettre C, et al. Mutations in DNM1L, as in OPA1, result in dominant optic atrophy despite opposite effects on mitochondrial fusion and fission. Brain. 2017;140(10):2586–96.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Medicine: Official J Am Coll Med Genet. 2015;17(5):405–24.
Xu M, Li K, He W. Compound heterozygous mutations in the LTBP2 gene associated with microspherophakia in a Chinese patient: a case report and literature review. BMC Med Genom. 2021;14(1):227.
Whitley BN, Lam C, Cui H, Haude K, Bai R, Escobar L, Hamilton A, Brady L, Tarnopolsky MA, Dengle L, et al. Aberrant Drp1-mediated mitochondrial division presents in humans with variable outcomes. Hum Mol Genet. 2018;27(21):3710–9.
Nasca A, Legati A, Baruffini E, Nolli C, Moroni I, Ardissone A, Goffrini P, Ghezzi D. Biallelic mutations in DNM1L are Associated with a slowly Progressive infantile Encephalopathy. Hum Mutat. 2016;37(9):898–903.
Keller N, Paketci C, Edem P, Thiele H, Yis U, Wirth B, Karakaya M. De novo DNM1L variant presenting with severe muscular atrophy, dystonia and sensory neuropathy. Eur J Med Genet. 2021;64(2):104134.
Liu X, Zhang Z, Li D, Lei M, Li Q, Liu X, Zhang P. DNM1L-Related mitochondrial fission defects presenting as Encephalopathy: a Case Report and Literature Review. Front Pead. 2021;9:626657.
Lhuissier C, Wagner BE, Vincent A, Garraux G, Hougrand O, Van Coster R, Benoit V, Karadurmus D, Lenaers G, Gueguen N, et al. Case report: thirty-year progression of an EMPF1 encephalopathy due to defective mitochondrial and peroxisomal fission caused by a novel de novo heterozygous DNM1L variant. Front Neurol. 2022;13:937885.
Yoon G, Malam Z, Paton T, Marshall CR, Hyatt E, Ivakine Z, Scherer SW, Lee KS, Hawkins C, Cohn RD. Lethal Disorder of mitochondrial fission caused by mutations in DNM1L. J Pediatr. 2016;171:313–316e311.
Longo F, Benedetti S, Zambon AA, Sora MGN, Di Resta C, De Ritis D, Quattrini A, Maltecca F, Ferrari M, Previtali SC. Impaired turnover of hyperfused mitochondria in severe axonal neuropathy due to a novel DRP1 mutation. Hum Mol Genet. 2020;29(2):177–88.
Wei Y, Qian M. Case Report: a Novel de novo mutation in DNM1L presenting with Developmental Delay, Ataxia, and Peripheral Neuropathy. Front Pead. 2021;9:604105.
Chao YH, Robak LA, Xia F, Koenig MK, Adesina A, Bacino CA, Scaglia F, Bellen HJ, Wangler MF. Missense variants in the middle domain of DNM1L in cases of infantile encephalopathy alter peroxisomes and mitochondria when assayed in Drosophila. Hum Mol Genet. 2016;25(9):1846–56.
Sheffer R, Douiev L, Edvardson S, Shaag A, Tamimi K, Soiferman D, Meiner V, Saada A. Postnatal microcephaly and pain insensitivity due to a de novo heterozygous DNM1L mutation causing impaired mitochondrial fission and function. Am J Med Genet Part A. 2016;170(6):1603–7.
Tarailo-Graovac M, Zahir FR, Zivkovic I, Moksa M, Selby K, Sinha S, Nislow C, Stockler-Ipsiroglu SG, Sheffer R, Saada-Reisch A, et al. De novo pathogenic DNM1L variant in a patient diagnosed with atypical hereditary sensory and autonomic neuropathy. Mol Genet Genom Med. 2019;7(10):e00961.
Vanstone JR, Smith AM, McBride S, Naas T, Holcik M, Antoun G, Harper ME, Michaud J, Sell E, Chakraborty P, et al. DNM1L-related mitochondrial fission defect presenting as refractory Epilepsy. Eur J Hum Genetics: EJHG. 2016;24(7):1084–8.
Pan Z, Wu TH, Chen C, Peng P, He YW, Yi WZ, Yin F, Peng J. [DNM1L gene variant caused encephalopathy, lethal, due to defective mitochondrial peroxisomal fission 1: three cases report and literature review]. Zhonghua Er Ke Za Zhi. 2021;59(5):400–6.
Chen X, Li Y, Luo H, Gan J. [Analysis of DNM1L gene variant in a case of fatal encephalopathy caused by mitochondrial peroxidase division deficiency]. Zhonghua yi xue yi chuan xue za zhi = Zhonghua yixue yichuanxue zazhi = Chinese journal of medical genetics 2021, 38(9):887–890.
De Souza Crippa AC, Franklin GL, Takeshita BT, Ghizoni Teive HA. DNM1L mutation presenting as Progressive myoclonic Epilepsy associated with acute febrile infection-related Epilepsy syndrome. Epileptic Disorders: International Epilepsy Journal with Videotape. 2022;24(5):976–8.
Ladds E, Whitney A, Dombi E, Hofer M, Anand G, Harrison V, Fratter C, Carver J, Barbosa IA, Simpson M, et al. De novo DNM1L mutation associated with mitochondrial Epilepsy syndrome with Fever sensitivity. Neurol Genet. 2018;4(4):e258.
Schmid SJ, Wagner M, Goetz C, Makowski C, Freisinger P, Berweck S, Mall V, Burdach S, Juenger H. A De Novo Dominant negative mutation in DNM1L causes sudden Onset Status Epilepticus with subsequent epileptic Encephalopathy. Neuropediatrics. 2019;50(3):197–201.
Mancardi MM, Nesti C, Febbo F, Cordani R, Siri L, Nobili L, Lampugnani E, Giacomini T, Granata T, Marucci G, et al. Focal status and acute encephalopathy in a 13-year-old boy with de novo DNM1L mutation: video-polygraphic pattern and clues for differential diagnosis. Brain Dev. 2021;43(5):644–51.
Ryan CS, Fine AL, Cohen AL, Schiltz BM, Renaud DL, Wirrell EC, Patterson MC, Boczek NJ, Liu R, Babovic-Vuksanovic D, et al. De Novo DNM1L variant in a Teenager With Progressive Paroxysmal Dystonia and Lethal Super-refractory Myoclonic Status Epilepticus. J Child Neurol. 2018;33(10):651–8.
Zaha K, Matsumoto H, Itoh M, Saitsu H, Kato K, Kato M, Ogata S, Murayama K, Kishita Y, Mizuno Y, et al. DNM1L-related encephalopathy in infancy with Leigh syndrome-like phenotype and suppression-burst. Clin Genet. 2016;90(5):472–4.
Vandeleur D, Chen CV, Huang EJ, Connolly AJ, Sanchez H, Moon-Grady AJ. Novel and lethal case of cardiac involvement in DNM1L mitochondrial encephalopathy. Am J Med Genet Part A. 2019;179(12):2486–9.
Minghetti S, Giorda R, Mastrangelo M, Tassi L, Zanotta N, Galbiati S, Bassi MT, Zucca C. Epilepsia partialis continua associated with the p.Arg403Cys variant of the DNM1L gene: an unusual clinical progression with two episodes of super-refractory status epilepticus with a 13-year remission interval. Epileptic Disorders: International Epilepsy Journal with Videotape. 2022;24(1):176–82.
Assia Batzir N, Bhagwat PK, Eble TN, Liu P, Eng CM, Elsea SH, Robak LA, Scaglia F, Goldman AM, Dhar SU et al. De novo missense variant in the GTPase effector domain (GED) of DNM1L leads to static encephalopathy and seizures. Cold Spring Harbor Molecular case Studies 2019, 5(3).
Gerber S, Charif M, Chevrollier A, Chaumette T, Angebault C, Kane S, Paris A, Alban J, Quiles M, Delettre C, et al. Reply: the expanding neurological phenotype of DNM1L-related disorders. Brain. 2018;141(4):e29.
Zhu PP, Patterson A, Stadler J, Seeburg DP, Sheng M, Blackstone C. Intra- and intermolecular domain interactions of the C-terminal GTPase effector domain of the multimeric dynamin-like GTPase Drp1. J Biol Chem. 2004;279(34):35967–74.
Acknowledgements
We thank the patient and family members at all stages of this work.
Funding
This research was funded by the National Natural Science Foundation of China (82000850). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
D.L. and Z.Z. conceived and conducted the study. Z.Z. and X.B. drafted the manuscript. Z.C., J.L., Z.X., X.L., M.X., Q.Z., and Y.Z. contributed to collect clinical data and to the analyzis of data. Y.Y. and D.L. reviewed and edited manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was carried out in accordance with the principles of the Declaration of Helsinki and in a manner consistent with good clinical practice. The study was approved by the Ethics Committee of the Children’s Hospital Affiliated to Zhengzhou University. Signed informed consent was obtained from the patient’s parents.
Consent for publication
Written informed consent for the publication of clinical details was obtained from both parents of the participant.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, Z., Bie, X., Chen, Z. et al. A novel variant of DNM1L expanding the clinical phenotypic spectrum: a case report and literature review. BMC Pediatr 24, 104 (2024). https://doi.org/10.1186/s12887-023-04442-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12887-023-04442-y