Abstract
Background
Incontinentia pigmenti (IP) is an X-liked dominant genodermatosis caused by mutations of the IKBKG/NEMO gene. IP is mostly lethal in males in utero, and only very rare male cases with a somatic mosaic mutation or a 47,XXY karyotype have been reported.
Case presentation
We here report a case of an IKBKG gene deletion in a female infant presenting with a few blisters and erythema in her upper arms at birth. MLPA analysis revealed a rare 94 kb deletion in this patient, encompassing the IKBKG gene and IKBKGP pseudogene. PCR analysis indicated the presence of Alu elements at both ends of the deletion, suggesting non-allelic homologous recombination as an underlying mechanism. Notably, a low-level mosaic deletion was identified in her father’s peripheral blood leukocytes by PCR, suggesting a rare father-to-daughter transmission of IP.
Conclusion
In family studies for an apparently sporadic IP case, parental analysis that includes the father is recommended due to the possibility of male mosaicism.
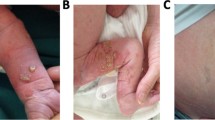
Similar content being viewed by others
Background
Incontinentia pigmenti (IP; MIM #308300) is a rare disorder affecting 1.2 in 100,000 live births [1]. Skin lesions along Blaschko lines are observed in all IP patients and systemic involvement includes visual, dental and neurologic impairment. IP is an X-linked dominant disorder and is typically lethal in utero in males. However, rare affected males are occasionally live-born with severe ectodermal dysplasia and immunodeficiency (MIM#300291). Notably, very rare male cases with typical IP symptoms are similar to their female IP counterparts in that they are live-born harboring somatic mosaicism or with concomitant 47,XXY karyotype, Klinefelter syndrome [2, 3].
IP develops by a loss-of-function variant of the IKBKG/NEMO gene. The protein encoded by this gene, NEMO/IKKγ, is essential for the activation of the nuclear factor-kappa B (NF-κB) transcription factor. The cells lacking in the NEMO/IKKγ protein are sensitive to apoptosis, which contributes to lethality in males and selective skewed X-inactivation of the mutant allele in females [4, 5]. IKBKG is a 23-kb gene consisted of nine coding exons and four alternative non-coding first exons (1A-D), albeit with two promoters. In addition to the genuine IKBKG gene, a processed pseudogene, IKBKGP, encompasses the region between exons 3–10. The IKBKGP pseudogene constitutes a portion of a 35.7 kb segmental duplication that is located oppositely and next to each other. The most frequent pathogenic variant responsible for IP is a recurrent deletion generated by non-allelic homologous recombination (NAHR) due to an aberrant alignment between two 650 bp short interspersed nuclear elements, MER67B. This 11.7 kb deletion removes exons 4–10, thereby producing an early premature codon. The IKBKG region includes many repetitive sequences that are susceptible to rearrangement and other deletions have also been reported [6].
In our present case report, we describe an IP female with a novel Alu-mediated deletion inherited from a father with somatic mosaicism. We also discuss the clinical implications of this case for future genetic testing and counseling.
Case presentation
Clinical phenotype
The newborn female subject of this present study was the first child of a non-consanguineous healthy couple. Her fetal growth had stagnated throughout the late pregnancy period, and she was delivered after a 36-week and 4-day gestation period by cesarean section. She was 2079 g (− 1.3 SD) in weight, 43.5 cm (− 1.4 SD) in height, and had a 32.4 cm (±0 SD) head circumference and 29.0 cm chest circumference. Although her general body condition was good, she showed mild hypoglycemia of 26 mg/dL. A few blisters were observed inside her right upper arm and erythema was evident inside her left upper arm. The antimicrobial agent cefazolin was administrated due to a suspicion of infectious disease. Given her normal hepatic function, herpes virus was considered unlikely. After a few days however, the erythema and blisters had spread to the extremities.
Blood testing for inflammation, infection, and autoimmune disease was negative. A blood cultivation test further indicated no streptococcal, staphylococcal, or fungal infection of the nasal cavity, neck, ear, or affected skin. The use of antimicrobial agents was thus discontinued at 6 days old. The eruptions showed a decrease at 8 days old. Eosinophilia had increased since her birth from a few to more than 20%. At 11 days old, blister-like eruptions and erythema emerged again in a linear distribution on her lower limbs. IP was then strongly suspected based on this clinical progression. Secondary infections of the skin lesions were not present, and brain magnetic resonance imaging was normal. The polus posterior lentis on both eyes showed mild whitening but did not require surgical treatment. The paternal grandmother subsequently remembered that the father of our infant subject had also had minor skin lesions at birth.
Molecular analysis
To further diagnose our current case subject, we performed PCR analysis of the 11.4 kb recurrent deletion in the IKBKG gene that is commonly observed in IP patients [7]. No specific PCR product was obtained in the proband however (data not shown). We next performed sequencing analysis of all coding exons of the IKBKG gene using the Sanger method, but no pathogenic variant was identified (data not shown). We subsequently conducted multiplex ligation-dependent probe amplification (MLPA) (P073-A1; MRC-Holland, Amsterdam, Netherlands) to detect possible rare genomic rearrangements, and identified a large deletion incorporating exons 3–10 of the IKBKG gene, as well as a similar region of the IKBKGP pseudogene, based on a copy number evaluation of the MLPA data (Fig. 1A, B). Neither parent showed a copy number abnormality in this test.
Molecular analysis of the father-to-daughter inheritance of IP in the study case. A Diagram of the quantitative MLPA results. Genomic regions are displayed on the x-axis, whereas the y-axis indicates the ratio to a normal female. The genomic positions of the probes are provided in the instruction manual of the MLPA kit (SALSA P073-A1, MRC-Holland). B Schematic depiction of the location of the deletion relative to the IKBKG gene and IKBKGP pseudogene. The red arrow indicates the deleted region in the study patient. The cyan arrow indicates AluSx1, and the yellow arrow denotes AluY. Black arrowheads indicate segmental duplications. AluSx1, GRCh38/hg38 chrX:154554478–154,554,769; AluY, 154,648,589–154,648,883; IKBKG: 154547620–154,565,033; IKBKGP1: 154639978–154,648,275; deletion: 154554588–154,648,697. C Electropherogram of the HUMARA assay. HpaII undigested (left) and digested (right) PCR products from the patient and her parents are shown. (Above) The red arrowhead indicates the maternal allele, and the cyan arrowhead denotes the paternal allele. (Middle) The father’s X chromosome allele. (Bottom) The mother’s two alleles. D Deletion-specific PCR. P, proband; M, mother of the affected proband; F, father of the affected proband; DW, distilled water; L, 1 kb ladder plus (Invitrogen). The red arrowhead indicates a nested PCR product. E Deletion junction sequence. The green and orange characters indicate the upstream and downstream Alu elements, respectively. The purple character sequences indicate an identical 16 nucleotides. Gray character sequences indicate the region deleted in the variant
The results of a human androgen receptor assay (HUMARA) indicated a skewed X chromosome inactivation in the proband (Fig. 1C) [8]. This inactivated allele was inherited from the father, suggesting either that the proband’s deletion arose on the paternal allele as a de novo event or that the father may also harbor a mosaic deletion that is difficult to detect using MLPA. To examine for the presence of a possible low level mosaic deletion in the father, we established a deletion-specific PCR assay, which is more sensitive than MLPA, by designing the appropriate primers (Fig. 1B, Table 1). We tried some sets of forward and reverse primers located outside the deleted exons, not on the segmental duplication. This PCR approach successfully amplified a 3 kb product in the proband. Notably, a smaller but significant amount of this same PCR amplicon was observed in whole blood leukocyte DNA obtained from her father. Nested PCR was thus performed and confirmed a low-level mosaic deletion in the father (Fig. 1D). Analysis of the paternal blood DNA by quantitative PCR indicated that the mosaic deletion ratio was as low as approximately 3.3%. The Sanger sequence of the PCR product showed that both deletion endpoints were located within the Alu repeat elements. The proximal endpoint belonged to AluSx1 (292 bp), and the distal endpoint to AluY (295 bp), which share an 83.6% sequence similarity. The putative breakpoints were near the center of these elements and an identical 16 nucleotides were observed at the junction (Fig. 1E). These results indicated that the genomic position of the 94 kb deletion was GRCh38/hg38 chrX:154554588–154,648,697.
Discussion and conclusions
We here describe a rare IP case involving a father to daughter inheritance of a novel pathogenic IKBKG deletion. Our analysis indicated that the paternal pathogenic variant was a low-level somatic mosaicism, which was why the child’s father did not suffer lethality at the fetal stage of development. Such a father-to-daughter transmission is very rare with only two such cases reported to date [9]. In both of these prior cases, the female probands manifested typical IP symptoms and the father of each child also had mild IP skin symptoms. DNA testing of the peripheral blood as well as of fibroblasts from each father of those previous patients showed no evidence of an IKBKG mutation, but the sperm DNA carried the proband’s mutation with a 16.7 and 35% germline mosaicism, respectively. This indicated that the mutation arose before differentiation of the primordial germ cells and that the mutant germ cells survived while many mutant cell populations declined in the other somatic tissues under negative selection pressure. In this context, the fact that the father in our current case carried a 3.3% level of mosaicism in his blood may suggest more mutations in his sperm.
In our present case, the father presented with a typical cutaneous rash at birth, but since these skin lesions disappeared with age, he did not realize that he had had IP symptoms until the birth of his daughter. Given that a mosaic mutation can be inherited by offspring, diagnosing an IP male with a mild skin manifestation may be difficult but is important. Even if a proband seems to be a sporadic case without any family history, parental analysis that includes the father is recommended due to the possibility of paternal mosaicism. In terms of genetic counseling in these cases, pediatricians or genetic specialists need to inform the apparently healthy parents the possibility of hidden mosaicism and the recurrence risk of IP in next pregnancy, and also consider how to inform the affected child of future reproductive issues when he or she reaches adulthood.
Besides the typical recurrent deletion of the IKBKG gene, various genomic rearrangements in the IKBKG region, both benign and pathogenic, have also been reported [6]. A total of 7 pathogenic deletions ranging from 4.8–115 kb in size have been described. The breakpoint locations of these cases were different from that of our case. In our present analyses, deletion junctions were analyzed to predict a mechanism from a wide variety of possibilities, including non-homologous end joining, NAHR mediated by Alu, and replication-based processes. Since both endpoints in our present female infant subject were located within 300 bp Alu elements with a 83.6% sequence similarity, Alu-mediated NAHR may have been the underlying mechanism in this case. Among the 7 atypical deletions reported previously, the precise breakpoint was identified only in one case, whose deletion was found to be mediated by Alu-Alu recombination [6]. In contrast, common recurrent exons 3–10 deletions are caused by recombination between the two MER67B elements located in intron 3 and downstream to exon 10 of the IKBKG gene, respectively, and the endpoints are located within 879 bp of MER67B-surrounding regions that have a 100% sequence identity. The 100% identity might explain the difference in the de novo frequencies of these deletions.
Availability of data and materials
The data of mutation identified during the current study are available in the Leiden Open Variation Database repository, Variant ID: #0000868293.
Abbreviations
- IP:
-
Incontinentia pigmenti
- IKBKG :
-
inhibitor of kappa polypeptide gene enhancer in B-cells, kinase gamma; NEMO
nuclear Factor κB, essential modulator
- NF-κB:
-
nuclear factor-kappa B
- IKBKGP :
-
inhibitor of kappa polypeptide gene enhancer in B-cells, kinase gamma pseudogene
- NAHR:
-
non-allelic homologous recombination
- MER67B:
-
medium reiterated 67B
- MLPA:
-
multiplex ligation-dependent probe amplification
- HUMARA:
-
human androgen receptor assay
References
Orphanet. Prevalence and incidence of rare diseases: Bibliographic data. 2021;1(1).
Fusco F, Fimiani G, Tadini G, Michele D, Ursini MV. Clinical diagnosis of incontinentia pigmenti in a cohort of male patients. J Am Acad Dermatol. 2007;56(2):264–7.
Conte M, Pescatore A, Paciolla M, Esposito E, Miano MG, Lioi MB, et al. Insight into IKBKG/NEMO locus: report of new mutations and complex genomic rearrangements leading to incontinentia pigmenti disease. Hum Mutat. 2014;35(2):165–77.
Aradhya S, Bardaro T, Galgóczy P, Yamagata T, Esposito T, Patlan H, et al. Multiple pathogenic and benign genomic rearrangements occur at a 35 kb duplication involving the NEMO and LAGE2 genes. Hum Mol Genet. 2001;10(22):2557–67.
Fusco F, Bardaro T, Fimiani G, Mercadante V, Miano MG, Falco G, et al. Molecular analysis of the genetic defect in a large cohort of IP patients and identification of novel NEMO mutations interfering with NF-κB activation. Hum Mol Genet. 2004;13(16):1763–73.
Fusco F, Paciolla M, Napolitano F, Pescatore A, D’addario I, Bal E, et al. Genomic architecture at the incontinentia pigmenti locus favours de novo pathological alleles through different mechanisms. Hum Mol Genet. 2012;21(6):1260–71.
Bardaro T, Falco G, Sparago A, Mercadante V, Molins EG, Tarantino E, et al. Two cases of misinterpretation of molecular results in incontinentia pigmenti, and a PCR-based method to discriminate NEMO/IKKγ gene deletion. Hum Mutat. 2003;21(1):8–11.
Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51(6):1229–39.
Fusco F, Conte MI, Diociauti A, Bigoni S, Branda MF, Ferlini A, et al. Unusual father-to-daughter transmission of Incontinentia Pigmenti due to mosaicism in IP males. Pediatrics. 2017;140(3):e20162950.
Acknowledgements
We thank the parents of the patient for agreeing to participate in this study.
Funding
This study was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (15H04710 and 24390085 to H.K.) and from the Ministry of Health, Welfare and Labor (16ek0109067h0003 to H.K.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
MK, AS, YI, TK and HK were involved in study design and data interpretation. MK, TK and HK were involved in the data analysis, and were major contributors to the drafting of the manuscript. All authors critically revised the report, commented on drafts of the manuscript, and approved the final manuscript for submission.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Review Board for Human Genome Studies at Fujita Health University. Written informed consent was obtained from the parents of the patient for participation in this study.
Consent for publication
Written informed consent to publish this study was obtained from the parents of the patient.
Competing interests
The authors declare no competing interests in relation to this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kawai, M., Sugimoto, A., Ishihara, Y. et al. Incontinentia pigmenti inherited from a father with a low level atypical IKBKG deletion mosaicism: a case report. BMC Pediatr 22, 378 (2022). https://doi.org/10.1186/s12887-022-03444-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12887-022-03444-6