Abstract
Background
Autism spectrum disorder (ASD) is becoming increasingly prevalent of late. Methylenetetrahydrofolate reductase (MTHFR) has a significant role in folate metabolism. Owing to the inconsistencies and inconclusiveness on the association between MTHFR single nucleotide polymorphism (SNP) and ASD susceptibilities, a meta-analysis was conducted to settle the inconsistencies.
Methods
For this meta-analysis, a total of 15 manuscripts published up to January 26, 2020, were selected from PubMed, Google Scholar, Medline, WangFang, and CNKI databases using search terms “MTHFR” OR “methylenetetrahydrofolate reductase” AND “ASD” OR “Autism Spectrum Disorders” OR “Autism” AND “polymorphism” OR “susceptibility” OR “C677T” OR “A1298C”.
Results
The findings of the meta-analysis indicated that MTHFR C677T polymorphism is remarkably associated with ASD in the five genetic models, viz., allelic, dominant, recessive, heterozygote, and homozygote. However, the MTHFR A1298C polymorphism was not found to be significantly related to ASD in the five genetic models. Subgroup analyses revealed significant associations of ASD with the MTHFR (C677T and A1298C) polymorphism. Sensitivity analysis showed that this meta-analysis was stable and reliable. No publication bias was identified in the associations between MTHFRC677T polymorphisms and ASD in the five genetic models, except for the one with regard to the associations between MTHFRA1298C polymorphisms and ASD in the five genetic models.
Conclusion
This meta-analysis showed that MTHFR C677T polymorphism is a susceptibility factor for ASD, and MTHFR A1298C polymorphism is not associated with ASD susceptibility.
Similar content being viewed by others
Background
Autism spectrum disorder (ASD) is one of the complex neurodevelopmental disorders, which has been increasingly recognized as a public health issue [1]. It affects 9‰ of the entire population of children, and the estimated ratio between male and female (M:F) children is 4:1 [2]. The prevalence rates of ASD in terms of percentages are approximately 1.52‰ in the Middle East [2,3,4,5], 14.7‰ in the USA [6, 7], 1.66‰ in China [8], and 6‰ in Australia [1, 9].
The distinguishing features of ASD include a set of behavioral phenotypes such as social communication deficits, restrictive and repetitive behaviors [10, 11], and worsened quality of life and family functioning for children with ASD and their parents [12]. Brain and nervous system dysfunctions are indicated in ASD [13], which occur as a result of pathophysiological and environmental factors. Folate/homocysteine (Hcy) levels act as a risk factor in ASD [14, 15], indicating the involvement of methylenetetrahydrofolate reductase (MTHFR) in ASD. Therefore, MTHFR has been the focal point of investigation on ASD, as inheritance validates the pathophysiological mechanism of ASD [16,17,18].
MTHFR locus has been mapped to chromosome1 (1p36.3) [19]. Conversion of 5, 10-methylenetetrahydrofolate to 5-methylenetetrahydrofolate is performed by MTHFR, which regulates the intracellular levels of folate and Hcy [15, 20]. Single nucleotide polymorphisms (C677T and A1298C) are associated with the decline in MTHFR activity [21, 22], which is, in turn, correlated with Folate/Hcy levels [23, 24]. Homocysteinemia and low plasma folate are found in individuals with C677T and A1298C alleles [22, 25]. A reduction of approximately 50% ~ 60% in the MTHFR activity is correlated with compound heterozygosity for both C677T and A1298C [19, 22, 26,27,28]. A decline in the enzymatic activity to 35% ~ 70% in homozygotes T is linked to C677T polymorphism in MTHFR [29]. Generally, when compared to C677T mutation, A1298C mutation feebly affects MTHFR activity and Hcy and folate levels [25, 30].
Correlations between single nucleotide polymorphisms (C677T and A1298C) and susceptibility to ASD are still debatable. A correlation between MTHFR C677T polymorphism and a higher susceptibility to ASD has been reported by Boris et al. [22] among Caucasian children [27]. Guo et al. [31] evidenced that MTHFR C677T polymorphism is a risk factor for ASD among Chinese Han children [31]. El-baz et al. [32] recognized a significant correlation between MTHFR C677T polymorphisms and ASD among Egyptian children [32]. Nonetheless, Dos Santos et al. [28] found no correlation between MTHFR C677T polymorphism and ASD [28]. Studies by Khalil et al. [33] and El-baz et al. [32, 34] describe MTHFR A1298C polymorphism to represent a risk factor in correlation with ASD among Egyptian children. On the contrary, Mohammad et al. [35] evidenced that MTHFR A1298C polymorphism variant allele has no link with any independent risk of ASD [35]. In this meta-analysis, updated articles were gathered [26, 32, 36] to authenticate correlations between MTHFR polymorphism (C677T/A1298C) and susceptibility to ASD.
Methods
Search strategy and identification of studies
Scientific literature published before January 26, 2020, in PubMed, Embase, Web of Science, Medline, WanFang datebase, and CNKI database were searched using specific search terms (Supplement file 1). The equivalent Chinese terms were used in the Chinese databases. Moreover, we retrieved related articles from the selected literature references to replenish data that had not been identified in the initial search. All full-text literature were scrutinized to determine whether the papers to be included.
Selection criteria
The following criteria had to be satisfied by the studies to be incorporated in this meta-analysis: (1) Original studies on the correlation between MTHFR polymorphism (C677T/A1298C) and ASD; (2) Cohort or case-control designs; (3) All genotype frequency information is available; (4) Diagnostic criteria of ASD described in the Diagnostic and Statistical Manual of Mental Disorders (4th or 5th edition) [37, 38], and/or Childhood Autism Rating Scale [39]. Certain earlier papers referred to the Manual of Mental Disorders (3rd edition) [40]. The exclusion criteria comprised the following: (1) Researches on the correlation between MTHFR polymorphism (C677T/A1298C) and ASD that are not original; (2) Studies that lack data and complete information; (3) Replicated studies; (4) Review studies.
Data extraction
Two investigators, namely, Yan Li and Shuang Qiu, extracted all the relevant data with the help of a standardized protocol and data collection form. From every qualified study, data such as the name of the first author, year of publication, country, study population (ethnicity), study design, the definition of ASD, sample size of cases and controls, genotyping method, genotype information, and allele frequencies were gathered and documented. Disparities in the study selection were resolved through discussion or consensus with the third investigator (Yawen Liu). The corresponding authors of articles with missing data were emailed for the required data.
Statistical analysis
Odds ratio (OR) and 95% confidence intervals (CI) were deduced to analyse how strongly MTHFR (C677T/A1298C) polymorphism and the risk of ASD were correlated in the five genetic models, viz., allelic, dominant, recessive, heterozygote, and homozygote. Heterogeneity among studies was assessed through Q-test and I2. Random effects model (DerSimonian-Laird methods) [41] was selected to pool data and in case of substantial heterogeneity (Ph < 0.05 and I2 > 50%); else, fixed effect model (Mantel-Haenszel methods) [42] was chosen. Furthermore, subgroup analyses were stratified according to the state with mandatory fortification of folate, population, sample source, and Hardy-Weinberg equilibrium (HWE). The included studies were tested for HWE in the control group utilizing Chi-square tests. Besides, the stability of the results was tested by performing a sensitivity analysis with the sequential omission of each study. To evaluate the potential publication bias in this meta-analysis, Begg’s funnel plot and Egger’s test were conducted. Stata version 12.0 (StataCorp LP, College Station, TX, USA) was used to evaluate all analyses, and p < 0.05 was considered to be statistically significant.
Results
Overall results
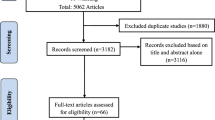
Upon literature search and critical screening, about 15 studies from 125 articles were included in this meta-analysis, as already discussed in the Methods section (Fig. 1). A total of 2609 cases and 7496 controls were enrolled from the 15 articles published on the correlation between MTHFR C677T polymorphism and ASD susceptibility. Of those, only nine articles that included 1961 cases and 1652 controls qualified for the evaluation of the link between MTHFR A1298C and ASD as per the selection criteria. The characteristics of each primary study are summarized and presented in Tables 1 and 2.
Association between MTHFR C677T polymorphism and ASD
Random effect model (Ph < 0.05 or I2 > 50%) was used, and MTHFR C677T polymorphism was found to be remarkably linked to ASD susceptibility in allelic (T vs C: OR = 1.63, 95% CI = 1.30–2.05, p < 0.05), heterozygote (CT vs CC: OR = 1.66, 95% CI = 1.31–2.11, p < 0.05), homozygote (TT vs CC: OR = 2.03, 95% CI = 1.33–3.09, p < 0.05), dominant (TT + CT vs CC: OR = 1.82, 95% CI = 1.39–2.37, p < 0.05), and recessive models (TT vs CT + CC: OR = 1.59, 95% CI = 1.14–2.22, p < 0.05; Table 3, Fig. 2a).
To further clarify the link between MTHFR polymorphisms and the risk of ASD, subgroup analysis was carried out. Firstly, no significant deviation of the correlation among the states with mandatory fortification of folate was recorded. MTHFR C677T polymorphism was not found to be linked to ASD susceptibility: allelic (T vs C: OR = 1.32, 95% CI = 1.00–1.75, p > 0.05), homozygote (TT vs CC: OR = 1.66, 95% CI = 0.94–2.94, p > 0.05), and recessive models (TT vs CT + CC: OR = 1.37, 95% CI = 0.93–2.00, p > 0.05). Nonetheless, it was observed to be associated with ASD susceptibility among the states without mandatory fortification of folate: allelic (T vs C: OR = 2.08, 95% CI = 1.40–3.08, p < 0.05), heterozygote (CT vs CC: OR = 1.95, 95% CI = 1.34–2.82, p < 0.05), homozygote (TT vs CC: OR = 2.78, 95% CI = 1.35–5.73, p < 0.05), dominant (TT + CT vs CC: OR = 2.22, 95% CI = 1.46–3.36, p < 0.05), and recessive models (TT vs CT + CC: OR = 2.23, 95% CI = 1.13–4.38, p < 0.05). Secondly, MTHFR C677T polymorphism was recorded to be correlated with ASD susceptibility in Caucasian population: allelic (T vs C: OR = 1.51, 95% CI = 1.17–1.95, p < 0.05), heterozygote (CT vs CC: OR = 1.62, 95% CI = 1.20–2.18, p < 0.05), homozygote (TT vs CC: OR = 1.92, 95% CI = 1.16–3.16, p < 0.05), and dominant models (TT + CT vs CC: OR = 1.73, 95% CI = 1.25–2.41, p < 0.05). Nonetheless, MTHFR C677T polymorphism was not found to be linked to ASD susceptibility among Asians: homozygote model (TT vs. CC: OR = 2.45, 95% CI = 0.95–6.31, p > 0.05). Thirdly, a hospital-based and population-based sample was adopted for this study. MTHFR C677T polymorphism was found to be linked with ASD susceptibility under five genetic models in hospital- and population-based samples, respectively (all p < 0.05). Fourthly, our results showed that MTHFR C677T polymorphism was consistent/inconsistent with HWE; however, it was significantly associated with ASD susceptibility under five genetic models (all p < 0.05) (Table 3).
Association between MTHFR A1298C polymorphism and ASD
Random effect model (Ph < 0.05 or I2 ≥ 50%) was utilized, and no significant correlation between MTHFR A1298C polymorphism and ASD susceptibility in the five genetic models was identified (allelic, dominant, recessive, heterozygote, and homozygote; all p > 0.05; Table 4, Fig. 2b). As per the subgroup analyses, MTHFR A1298C polymorphism was found to be associated with ASD susceptibility among the states without mandatory fortification of folate: allelic model (C vs. A: OR = 1.84, 95% CI = 1.08–3.14, p < 0.05) and dominant model (CC + AC vs. AA: OR = 2.45, 95% CI = 1.16–5.15, p < 0.05). No significant correlation between MTHFR A1298C polymorphism and ASD susceptibility under the other genetic models in any subgroup was found (all p > 0.05) (Table 4).
Sensitivity analysis and publication bias
The stability of the findings was evaluated through sensitivity analysis conducted by sequentially omitting each study, demonstrating that this meta-analysis is relatively stable and credible (Fig. 3). To evaluate the publication bias, Begg’s funnel plot and Egger’s tests were carried out. No significant publication bias was detected in the correlation between MTHFR C677T polymorphisms and ASD risk in the five genetic models: allelic (PB = 0.029, PE = 0.017), heterozygote (PB = 0.017, PE = 0.008), homozygote (PB = 0.048, PE = 0.053), dominant: (PB = 0.021, PE = 0.010), and recessive models (PB = 0.033, PE = 0.053). However, publication bias was detected among the studies on the correlation between MTHFR A1298C polymorphisms and ASD risk in the following genetic models: allelic (PB = 0.210, PE = 0.010), heterozygote (PB = 0.001, PE = 0.049), homozygote (PB = 0.025, PE = 0.152), dominant (PB = 0.000, PE = 0.049), and recessive models (PB = 0.081, PE = 0.152) (Tables 3 and 4, Fig. 4).
Discussion
Relevant and up to date literature published prior to January 26, 2020 were selected for examining the correlation between MTHFR polymorphism (C677T and A1298C) and ASD risk in this meta-analysis. The findings of this study exhibit that MTHFR C677T polymorphism is a susceptibility factor of ASD, but MTHFR A1298C polymorphism is not linked with ASD susceptibility.
Several meta-analytic studies on the correlation between C677T polymorphism of MTHFR and ASD risk have been conducted. Frustaci et al. [24] studied six articles [22, 27, 28, 35, 43, 44], which consisted of 877 cases and 939 controls, mainly Caucasians, and found a remarkable correlation between C677T polymorphism of MTHFR and ASD risk [24]. Pu et al. [25] investigated eight articles [9, 18, 22, 27, 28, 31, 35, 43] involving 1672 cases and 6760 controls, also mainly Caucasians, evidenced a significant risk on the T allele mutation of MTHFR C677T in ASD [25]. Rai et al. [26] investigated 1978 cases and 7257 controls (Caucasians: 1355 cases and 6460 controls; Asians: 623 cases and 797 controls) in 13 studies [18, 22, 27, 28, 31, 33, 35, 43, 44, 46, 48, 50] and found that C677T polymorphism of MTHFR is a risk factor for ASD susceptibility as well [26]. Similarly, the current meta-analysis enrolled 2609 cases and 7496 controls (Caucasian: 1786 cases and 6499 controls, Asian: 823 cases and 997 controls) from 15 selected literature [9, 18, 22, 26,27,28, 31, 32, 33, 35, 43, 47, 48, 50], further confirmed the association between C677T polymorphism of MTHFR and ASD susceptibility.
A previous meta-analysis, conducted on the correlation between A1298C polymorphism of MTHFR and ASD risk [25] (included five literatures; 1470 cases and 1060 controls; Caucasians: 1332 cases and 922 controls, Asians: 138 cases and 138 controls, respectively) [18, 22, 35, 43, 44] reported that A1298C polymorphism of MTHFR is remarkably linked to reduced ASD risk but only in the recessive model [25].
In the present meta-analysis, eight of the selected articles [18, 22, 32, 35, 36, 43, 44, 47, 50] had enrolled 1961 cases and 1652 controls (Caucasians: 1387 cases and 991 controls, Asians: 574 cases and 661 controls), and it was recognized that A1298C polymorphism of MTHFR was not correlated with ASD susceptibility. However, Khalil et al. (42 cases and 48 controls) [49] and El-Baz et al. (31 cases and 39 controls) [32] revealed that MTHFR A1298C polymorphism represented a risk factor in association with ASD. This disagreement may be caused by small samples in the study.
There are several limitations for this study. First, the subgroup analyses of environmental risk factors, sex, and gene-environment interactions were not performed owing to insufficient information. Second, this meta-analysis was mainly focused on Caucasians and Asians, thus limiting the generalization of the findings to other ethnicities. Third, in agreement with the findings of Frustaci et al. [24], Pu et al. [25] and Rai et al. [26], heterogeneity exists in this exploration. Fourth, publication bias was found in the association between MTHFR A1298C polymorphisms and ASD risk.
Conclusion
To conclude, this meta-analysis confirms that C677T polymorphism of MTHFR is remarkably linked with ASD risk. Nevertheless, the findings agree that the A1298C polymorphism of MTHFR is not significantly correlated with ASD. Exploring gene-gene and gene-environment interactions could throw more light on the genetic link between MTHFR variants and ASD risk.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ASD:
-
Autism spectrum disorder
- MTHFR:
-
Methylenetetrahydrofolate reductase
- SNPs:
-
Single nucleotide polymorphisms
- HWE:
-
Hardy-Weinberg equilibrium
- OR:
-
Odds ratio
- CI:
-
Confidence interval
- AIC:
-
Akaike’s information criterion
- LD:
-
Linkage disequilibrium
References
Do B, Lynch P, Macris EM, Smyth B, Stavrinakis S, Quinn S, et al. Systematic review and meta-analysis of the association of autism Spectrum disorder in visually or hearing impaired children. Ophthalmic Physiol Opt. 2017;37:212–24.
Szatmari P, Liu X-Q, Goldberg J, Zwaigenbaum L, Paterson AD, Woodbury-Smith M, et al. Sex differences in repetitive stereotyped behaviors in autism: implications for genetic liability. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:5–12.
Eapen V, Mabrouk AA, Zoubeidi T, Yunis F. Prevalence of pervasive developmental disorders in preschool children in the UAE. J Trop Pediatr. 2007;53:202–5.
Al-Farsi YM, Al-Sharbati MM, Al-Farsi OA, Al-Shafaee MS, Brooks DR, Waly MI. Brief report: prevalence of autistic spectrum disorders in the Sultanate of Oman. J Autism Dev Disord. 2011;41:821–5.
Richards C, Jones C, Groves L, Moss J, Oliver C. Prevalence of autism spectrum disorder phenomenology in genetic disorders: a systematic review and meta-analysis. Lancet Psychiatry. 2015;2:909–16.
Irimia A, Torgerson CM, Jacokes ZJ, Van Horn JD. The connectomes of males and females with autism spectrum disorder have significantly different white matter connectivity densities. Sci Rep. 2017;7:46401.
Autism, Investigators DDMNSYP. Prevalence of autism spectrum disorders : Autism and developmental disabilities monitoring network, six sites, United States, 2000. MMWR Surveill Summ. 2007;56:12–28.
Wan Y, Hu Q, Li T, Jiang L, Du Y, Feng L, et al. Prevalence of autism spectrum disorders among children in China: a systematic review. Shanghai Arch Psychiatry. 2013;25:70–80.
Bourke J, de Klerk N, Smith T, Leonard H. Population-based prevalence of intellectual disability and autism spectrum disorders in Western Australia: a comparison with previous estimates. Medicine (Baltimore). 2016;95:e3737.
Diaz-Beltran L, Esteban FJ, Varma M, Ortuzk A, David M, Wall DP. Cross-disorder comparative analysis of comorbid conditions reveals novel autism candidate genes. BMC Genomics. 2017;18:315.
Ch'ng C, Kwok W, Rogic S, Pavlidis P. Meta-analysis of gene expression in autism spectrum disorder. Autism Res. 2015;8:593–608.
Anderson KA, Shattuck PT, Cooper BP, Roux AM, Wagner M. Prevalence and correlates of postsecondary residential status among young adults with an autism spectrum disorder. Autism. 2014;18:562–70.
Dillenburger K, Jordan JA, McKerr L, Keenan M. The millennium child with autism: early childhood trajectories for health, education and economic wellbeing. Dev Neurorehabil. 2015;18:37–46.
Main PA, Angley MT, Thomas P, O'Doherty CE, Fenech M. Folate and methionine metabolism in autism: a systematic review. Am J Clin Nutr. 2010;91:1598–620.
Melnyk S, Fuchs GJ, Schulz E, Lopez M, Kahler SG, Fussell JJ, et al. Metabolic imbalance associated with methylation dysregulation and oxidative damage in children with autism. J Autism Dev Disord. 2012;42:367–77.
Schaevitz LR, Berger-Sweeney JE. Gene-environment interactions and epigenetic pathways in autism: the importance of one-carbon metabolism. ILAR J. 2012;53:322–40.
Eapen V. Genetic basis of autism: is there a way forward? Curr Opin Psychiatry. 2011;24:226–36.
Lyall K, Schmidt RJ, Hertz-Picciotto I. Maternal lifestyle and environmental risk factors for autism spectrum disorders. Int J Epidemiol. 2014;43:443–64.
Goyette P, Sumner JS, Milos R, Duncan AM, Rosenblatt DS, Matthews RG, et al. Human methylenetetrahydrofolate reductase: isolation of cDNA, mapping and mutation identification. Nat Genet. 1994;7:195–200.
James SJ, Melnyk S, Jernigan S, Pavliv O, Trusty T, Lehman S, et al. A functional polymorphism in the reduced folate carrier gene and DNA hypomethylation in mothers of children with autism. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1209–20.
James SJ, Cutler P, Melnyk S, Jernigan S, Janak L, Gaylor DW, et al. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am J Clin Nutr. 2004;80:1611–7.
Boris M, Goldblatt A, Galanko J, James SJ. Association of MTHFR gene variants with autism. J Am Phys Surg. 2004;9(4):106–8.
Jacob RA, Gretz DM, Taylor PC, James SJ, Pogribny IP, Miller BJ, et al. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr. 1998;128:1204.
Frustaci A, Neri M, Cesario A, Adams JB, Domenici E, Dalla Bernardina B, et al. Oxidative stress-related biomarkers in autism: systematic review and meta-analyses. Free Radic Biol Med. 2012;52:2128–41.
Pu D, Shen Y, Wu J. Association between MTHFR gene polymorphisms and the risk of autism spectrum disorders: a meta-analysis. Autism Res. 2013;6:384–92.
Rai V. Association of methylenetetrahydrofolate reductase (MTHFR) gene C677T polymorphism with autism: evidence of genetic susceptibility. Metab Brain Dis. 2016;31(4):727–735. https://doi.org/10.1007/s11011-016-9815-0.
Pasca SP, Dronca E, Kaucsar T, Craciun EC, Endreffy E, Ferencz BK, et al. One carbon metabolism disturbances and the C677T MTHFR gene polymorphism in children with autism spectrum disorders. J Cell Mol Med. 2009;13:4229–38.
dos Santos PA, Longo D, Brandalize AP, Schüler-Faccini L. MTHFR C677T is not a risk factor for autism spectrum disorders in South Brazil. Psychiatr Genet. 2010;20:187–9.
Pan X, Lu Y, Long Y, Yao D. Genetic polymorphisms of methylenetetrahydrofolate reductase (MTHFR) gene Ala222Val and susceptibility to ovary cancer: a systematic review and meta-analysis. Tumour Biol. 2014;35:2133–9.
van der Put NM, Gabreels F, Stevens EM, Smeitink JA, Trijbels FJ, Eskes TK, et al. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet. 1998;62:1044–51.
Guo T, Chen H, Liu B, Ji W, Yang C. Methylenetetrahydrofolate reductase polymorphisms C677T and risk of autism in the Chinese Han population. Genet Test Mol Biomarkers. 2012;16:968–73.
El-Baz F, El-Aal MA, Kamal TM, Sadek AA, Othman AA. Study of the C677T and 1298AC polymorphic genotypes of MTHFR gene in autism spectrum disorder. Electron Physician. 2017;9:5287–93.
Khalil NMR. Evaluation of MTHFR genetic polymorphism as a risk factor in Egyptian autistic children and mothers. Afr J Psychiatry. 2014;18.
Ashwood P, Krakowiak P, Hertz-Picciotto I, Hansen R, Pessah I, Van de Water J. Elevated plasma cytokines in autism spectrum disorders provide evidence of immune dysfunction and are associated with impaired behavioral outcome. Brain Behav Immun. 2011;25:40–5.
Mohammad NS, Jain JM, Chintakindi KP, Singh RP, Naik U, Akella RR. Aberrations in folate metabolic pathway and altered susceptibility to autism. Psychiatr Genet. 2009;19(4):171–6. https://doi.org/10.1097/YPG.0b013e32832cebd2.
Zhao D, Sun C, Yang X, Zhang Z, Li N, Hou L, et al. Association of methylenetetrahydrofolate reductase in gene C677T and A1298C polymorphism among children with autism Chin. J Sch Health. 2013;34:52–5.
Pitcher TM, Piek JP, Barrett NC. Timing and force control in boys with attention deficit hyperactivity disorder: subtype differences and the effect of comorbid developmental coordination disorder. Hum Mov Sci. 2002;21:919–45.
Kosterman R, Hill KG, Lee JO, Meacham MC, Abbott RD, Catalano RF, et al. Young adult social development as a mediator of alcohol use disorder symptoms from age 21 to 30. Psychol Addict Behav. 2014;28:348–58.
Rimal H, Pokharel A. Prevalence of attention deficit hyperactivity disorder among school children and associated co-morbidities - a hospital based descriptive study. Kathmandu Univ Med J. 2016;14:226–30.
Strawbridge WJ, Deleger S, Roberts RE, Kaplan GA. Physical activity reduces the risk of subsequent depression for older adults. Am J Epidemiol. 2002;156:328–34.
Kelley GA, Kelley KS. Statistical models for meta-analysis: a brief tutorial. World J Methodol. 2012;2:27–32.
Kahan BC. Accounting for centre-effects in multicentre trials with a binary outcome - when, why, and how? BMC Med Res Methodol. 2014;14:20.
James SJ, Melnyk S, Jernigan S, Cleves MA, Halsted CH, Wong DH, et al. Metabolic endophenotype and related genotypes are associated with oxidative stress in children with autism. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:947–56.
Divyakolu STY, Thomas W, Thumoju S, Sreekanth SR, Vasavi M, OmSai VR, Nagaratna V, Hasan Q, Ahuja YR. Evaluation of C677T polymorphism of the Methylenetetrahydrofolate Reductase (MTHFR) gene in various neurological disorders. J Neurol Disord. 2013;2.
Park J, Ro M, Pyun JA, et al. MTHFR 1298A>C is a risk factor for autism spectrum disorder in the Korean population. Psychiatry Res. 2014;215(1):258–9. https://doi.org/10.1016/j.psychres.2013.11.006.
Sener EF, Oztop DB, Ozkul Y. MTHFR gene C677T polymorphism in autism spectrum disorders. Genet Res Int. 2014;2014:698574.
Shawky RM, El-baz F, Kamal TM, Elhossiny RM, Ahmed MA, El Nady GH. Study of genotype–phenotype correlation of methylene tetrahydrofolate reductase (MTHFR) gene polymorphisms in a sample of Egyptian autistic children. Egypt J Med Hum Genet. 2014;15:335–41.
Liu X, Solehdin F, Cohen IL, et al. Population- and family-based studies associate the MTHFR gene with idiopathic autism in simplex families. J Autism Dev Disord. 2011;41(7):938–944. https://doi.org/10.1007/s10803-010-1120-x.
Meguid NA, Dardir AA, Khass M, Hossieny LE, Ezzat A, El Awady MK. MTHFR genetic polymorphism as a risk factor in Egyptian mothers with down syndrome children. Dis Markers. 2008;24:19–26. https://doi.org/10.1155/2008/214027.
Schmidt RJ, Hansen RL, Hartiala J, et al. Prenatal vitamins, one-carbon metabolism gene variants, and risk for autism. Epidemiology. 2011;22(4):476-485. https://doi.org/10.1097/EDE.0b013e31821d0e30.
Acknowledgments
The authors also thank all the participants in the study.
Funding
This work was supported by the Ministry of Science and Technology of the People’s Republic of China grant 2015DFA31580, ScienAce and Technology Department of Jilin Province grant 20150101130JC, Jilin Provincial Key Laboratory of Neuronal Plasticity grant 20140622001JC and 20160622020JC, and China Postdoctoral Science Foundation grant 2013 M530989. The funding bodies had no role in the design of the study, the collection, analysis, or interpretation of the data, or writing the manuscript.
Author information
Authors and Affiliations
Contributions
Conception and design: YL, YC, and YWL; provision of study material: YL, YC, SQ, YG, JS, and ZL; collection and assembly of data: YL, SQ; data analysis and interpretation: YL, SQ, ZL, YC, and YWL; manuscript writing: YL; revision of the language/article: all authors; final approval of the manuscript: all authors.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1
: Supplement file 1. Search strategy: For this meta-analysis, a total of 15 manuscripts published up to January 26, 2020, were selected from PubMed, Google Scholar, Medline, WangFang, and CNKI databases using search terms “MTHFR” OR “methylenetetrahydrofolate reductase” AND “ASD” OR “Autism Spectrum Disorders” OR “Autism” AND “polymorphism” OR “susceptibility” OR “C677T” OR “A1298C”.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, Y., Qiu, S., Shi, J. et al. Association between MTHFR C677T/A1298C and susceptibility to autism spectrum disorders: a meta-analysis. BMC Pediatr 20, 449 (2020). https://doi.org/10.1186/s12887-020-02330-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12887-020-02330-3