Abstract
Background
Rotavirus antigenemia and RNAemia (the presence of rotavirus RNA in serum) have been commonly identified among paediatric patients with acute gastroenteritis. In this study we examined the association between rotavirus antigenemia and clinical features, and sought to determine the genotypes of rotaviruses detected in paired stool and serum samples.
Methods
Paired stool and serum samples were obtained from children hospitalised for acute gastroenteritis in Belém, Brazil, between June 2012 and June 2015. The 20-point Vesikari scoring system was used to assess the disease severity upon a retrospective medical record review. Stool and serum samples were primarily screened for the presence of rotavirus antigen using a commercial ELISA assay. The rotavirus isolates from stool and serum samples were genotyped by using the classical reverse-transcriptase polymerase chain reaction (RT-PCR) and/or through nucleotide sequencing of VP4 and VP7 genes. Viral load was estimated using real-time RT-PCR.
Results
In total rotavirus antigen was detected in 109 (24.2%) stool samples from 451 children, whereas antigenemia occurred in 38.5% (42/109) of these patients. We demonstrated that patients positive for rotavirus RNA in paired stool and serum samples were more likely to have a higher frequency of vomiting episodes in a 24-h period (p = 0.0035). Our findings also suggested that children not vaccinated against rotavirus are more likely to develop antigenemia, as compared to those given at least one vaccine dose (p = 0.0151). G12P [8] and G2P [4] genotypes were predominant throughout the study period, accounting for 52.3% (57/109) and 27.5% (30/109) of the typed isolates, respectively. Ten stool-serum pairs could be typed for VP4 and VP7 genes. Seven of these pairs showed concordant results with G2P [4] genotype being detected in stool and serum samples, whereas discrepancies between genotypes (G2P [4]/G2P[NT] and G12P [8]/G2P[NT]) were seen in three pairs.
Conclusions
Rotavirus antigenemia and RNAemia occur in a significant number of children hospitalised for acute gastroenteritis in Belém, Brazil, and may contribute to a greater disease severity, particularly translated into a greater number of vomiting episodes. This study documented a high concordance of genotypes detected in a subgroup of paired stool and serum samples.
Similar content being viewed by others
Background
Worldwide rotavirus still remains a major cause of deaths and hospitalisations due to gastroenteritis among children younger than five years, even though almost 100 countries have incorporated to date rotavirus vaccination into their national immunisation programs (NIPs) [1, 2]. According to recent estimates from the Global Burden of Disease Study, 2015 [3], around 260,000 deaths are attributable to rotavirus annually, most of which (~ 90%) occurring in the low-income countries of Asia and Africa.
Typically the clinical course of rotaviral enteritis can show a wide spectrum of symptoms ranging from mild, watery diarrhoea to severe gastroenteritis with vomiting, fever, abdominal pain and dehydration which may last 3 to 8 days [4, 5]. Although rotaviruses primarily infect mature enterocytes of the upper small intestine, there is increasing evidence pointing to their potential for spreading beyond the gastrointestinal tract, leading to the appearance of unusual extra-intestinal manifestations, namely febrile seizures and central nervous diseases such as encephalitis. [6,7,8].
It has been described that rotavirus RNA (which may imply viraemia) and antigen(s) (namely VP6) are detected in 64–93% and 33–90% in serum of children with rotavirus gastroenteritis, respectively [9,10,11]. Systemic spreading of rotavirus has been evidenced through the detection of antigen and/or RNA in multiple organs and tissues, possibly translating into the occurrence of a variety of medically relevant extra-intestinal diseases [12]. Although yet a controversial issue, some studies support the notion that viraemia and/or antigenemia may result in greater clinical severity, particularly with regards to the intensity/frequency of fever, convulsion and vomiting episodes [10, 13, 14]
As based on a binary classification system that focuses on the outer layer VP7 and VP4 proteins there are currently 28 G (of glycoprotein) and 39 P (protease-sensitive) genotypes, respectively. Of these, only 12 G and 15 P types are commonly associated with infections in humans, particularly those strains bearing G1P [8], G2P [4], G3P [8], G4P [8], G9P [8] and G12P [8] specificities, which altogether account for greater than 80% of circulating rotaviruses [4, 15, 16]. The issue of whether or not rotavirus genotypes may be a determinant of extra-intestinal spread remains to be fully elucidated. While some authors have reported that children infected with G1 type are more prone to develop antigenemia, as compared with other types [14, 17], there have been studies in Bangladesh, India and the United States showing that no significant difference was seen between infecting genotypes among children with or without antigenemia/ viraemia [6, 9, 12, 18].
In the current post-rotavirus vaccination scenario, where two live, attenuated, oral rotavirus vaccines [Rotarix (GlaxoSmithKline Biologicals, Rixensart, Belgium); and RotaTeq (Merck & Co. Inc. Kenilworth, NJ)] are licensed for use in over 100 countries [19], studies aiming at demonstrating the real world impact of vaccination in terms of morbidity and mortality are strongly recommended. To date, post-licensure evaluations of vaccination impact on rotavirus gastroenteritis-related morbidity and mortality have in general been conducted based on the detection of antigen in stools. It has however been demonstrated that concomitant attempts of detecting rotavirus antigenemia in post-licensure epidemiological studies are likely to enhance surveillance of rotavirus cases, besides providing a more accurate assessment of vaccine effectiveness [20].
Aim and objectives
This study aimed primarily at determining if children hospitalised for acute gastroenteritis with rotavirus RNA and/or rotavirus antigen in stools and serum samples were more prone to develop severe gastroenteritis symptoms, as compared to those with RNA and/or rotavirus antigen detected in stools only. We also sought to assess whether or not rotavirus genotypes recovered from stool samples are homologous to those detected in sera.
Methods
Patients and specimens collection
This was a prospective, hospital-based study involving children with acute gastroenteritis attending public/private hospitals in Belém, Northern Brazil, between June 2012 and June 2015: from June 2012 to March 2013 at Clínica Pediátrica do Pará (which has closed as from March 2013 onwards); and from April 2013 to June 2015 at Clínica Pio XII). The recruitment of these children took place in these large paediatric hospitals, which then accounted for over 50% of all hospital admissions for gastroenteritis in the Belém area, as based on our previous local surveillance studies [21].
The study was approved by the Independent Ethics Committee of the Instituto Evandro Chagas, Health Surveillance Secretariat, Brazilian Ministry of Health, and was conducted in accordance with the principles of the Declaration of Helsinki, as well as in compliance with the Good Clinical Practice guidelines.
Children eligible to participate in the study were those aged at least 12 weeks and born after March 6, 2006, when rotavirus vaccination was introduced nationally in Brazil [22]. For surveillance purposes a case of acute gastroenteritis was defined as the passage of three or more looser-than-normal or watery stools within the 24 h before presentation, requiring at least one overnight stay and intravenous rehydration therapy. We did not consider eligible for recruitment both nosocomial gastroenteritis cases and children with gastroenteritis lasting ≥14 days at presentation. Information on demographics, medical/vaccination history, feeding practices and specific symptoms were obtained upon receipt of signed informed consent forms from parents/guardians. The 20-point scoring system, as proposed by Ruuska and Vesikary[23], was used to grade the severity of clinical symptoms (diarrhoea, fever and vomiting) upon a retrospective medical record review; gastroenteritis episodes reaching clinical scores of 1–10, ≥11 and ≥ 15 were classified as mild/moderate, severe and very severe, respectively.
Both serum and stool samples for rotavirus detection were collected from each eligible patient within the first 48 h of hospitalisation, stored thereafter in coolers and promptly transported - under controlled temperature within the range of 2-8 °C - to the Virology Section at Instituto Evandro Chagas, where these specimens were kept at -20 °C until processing. We excluded from analyses those patients (n = 105) from whom either blood or stool samples only were obtained. All stool samples were routinely screened for the presence of VP6 rotavirus antigen using a previously validated [24] sandwich-type commercial ELISA assay (RIDASCREEN R Rotavirus; R-Biopharma, Darmsdadt, Germany), according to manufacturer’s instructions. The same method was used to detect rotavirus antigen in undiluted sera, even though evidence of antigenemia was determined if the sample yielded an optical density, at a wavelength of 450 nm, equal to the mean OD of negative controls plus ≥2 standard deviations, in accordance with procedures described elsewhere [6, 17, 25].
Following viral genomic RNA extraction from ELISA-positive stool and serum specimens, amplification of the gene segments encoding for the VP4 and VP7 antigens was performed through a classical two-step reverse-transcription polymerase chain reaction (RT-PCR), using first round consensus primers 4Con3-4Con2 and Beg9-End9, respectively, as described elsewhere [26, 27]. Stool samples were subjected to a second, nested-RT-PCR step including well-established oligonucleotide primers targeted at G (G1, G2, G3, G4, G9 and G12) and P (P [4], P [6], P [8], and P [9]) rotavirus genotypes [26, 28, 29]. In addition, a second, nested-PCR step was performed with sera using VP7F and VP7R primers, according to the methods described earlier [30]. The resulting bands could be visualised following gel electrophoresis and staining with SYBR Safe™ DNA gel stain. In order to measure the rotavirus load in stool and serum samples we performed a reverse-transcriptase quantitative PCR (qRT-PCR), using the primers and TaqMan probe targeting a highly conserved region of the non-structural protein 3 (NSP3) of rotavirus, essentially as described before [31]. The reaction was performed on an ABI 7500 genetic analyser (Applied Biosystems, Foster City, CA).
To determine the electropherotypes the extracted dsRNAs from stool samples were also further electrophoresed in a 5% polyacrylamide gel, followed by silver staining as previously described [32].
Nucleotide sequencing was carried out with rotavirus-positive serum samples that could not be genotyped previously using conventional RT-PCR. Eleven pairs of stool and serum specimens were subjected to nucleotide sequencing for VP7 and VP4 genes with a Big Dye Terminator cycle sequencing kit v 3.1 (Applied Biosystems, Foster City, CA). Electrophoresis was performed in the ABI Prism 3130xl automatic sequencer (Applied Biosystems) and the sequences obtained were aligned and edited using the BioEdit Sequence Alignment Editor program (v. 7.0.5.2). Neighbour-joining method was used to perform the phylogenetic analysis, in which distance was calculated from aligned sequences. The nucleotide sequences reported in this study have been deposited in the GenBank sequence database and assigned the following accession numbers: MH456983 to MH456986.
Statistical analyses were carried out using the BioEstat v5.0 software package [33]. Comparisons of individual laboratory results, clinical symptoms and severity scores between patients with and without rotavirus antigenemia were performed using χ2 tests or G test when appropriate for categorical data. The Student’s t-test or the Mann-Whitney U test was used for comparison of two independent sets of continuous, quantitative data. The Spearman’s rank correlation coefficients (rs) were used to measure the strength of the relationship between either antigenemia or RNAemia concentrations and clinical and laboratory parameters. The Spearman’s rank correlation was also used to assess the relationship between viral load in stool samples and levels of antigen in sera. We considered P values of less than 0.05 as being statistically significant.
Results
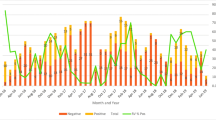
During June 2012 through June 2015, we identified 3740 children who were age-eligible to potentially participate in the study. Of these, a total of 556 paediatric inpatients with acute gastroenteritis were enrolled to participate in this study. Paired serum and stool samples could be obtained from 451 subjects. Paired samples from the former subgroup were tested by ELISA and qRT-PCR (quantitative RT-PCR) as demonstrated in Fig. 1. By ELISA, 109 (24.2%) stool samples out of the 451 subjects were rotavirus-positive; and among these rotavirus-positive children 38.5% (42/109) had detectable rotavirus antigen in their sera (antigenemia). Conversely, antigenemia was detected in four patients with rotavirus-negative stool samples. The analysis of stool samples by qRT-PCR yielded rotavirus RNA in 105 of patients and also in serum among 41 of these children, representing 39% of RNAemia in this subgroup. Stool samples from 242 patients could not be tested (NT) by qRT-PCR In addition, RNAemia was detected in 8 patients among those with stool samples either negative or not tested by PCR.
In order to explore potential indicators of antigenemia, we compared the clinical characteristics and the vaccination status between rotavirus-related acute gastroenteritis patients with rotavirus-positive (n = 42) and rotavirus-negative (n = 67) sera, by ELISA (Table 1). As based on the Vesikari scoring system, a trend for greater clinical severity was seen among children with rotavirus-positive serum, as compared to those with rotavirus-negative serum (p = 0.1557). With regards to specific clinical parameters, the only statistically significant difference was observed for the number of vomiting episodes in a 24-h period: patients positive for rotavirus RNA in paired stool and sera were more likely to have ≥3 vomiting episodes/24 h (p = 0.0035). Our findings also suggest that children not vaccinated against rotavirus were more likely to have antigenemia than those given at least one vaccine dose (P = 0.0151). Extra-intestinal involvement of rotavirus infection was not seen in this study.
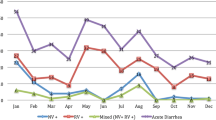
Genotyping was done on the 109 rotavirus-positive stool samples to determine G and P types, as shown in Fig. 2. Altogether G12P [8] and G2P [4] rotavirus strains were largely predominant throughout the study period, accounting for 52.3% (57/109) and 27.5% (30/109) of the isolates, respectively. The circulation of G2P [4] strains was observed from June 2012 through July 2013, with peak incidence rates in July (83.3% of genotyped strains) and August 2012 (84.6%), decreasing sharply thereafter until July 2013, in line with the overall rotavirus positivity rates. Rotavirus strains with G12P [8] genotype specificities emerged in July 2013, with rates increasing significantly in the following months and reaching incidence rates as high as 75% in September 2013 and 88.9% in July 2014. A variety of other rotavirus genotypes co-circulated at very low frequencies, concurrently with the predominant G2P [4] and G12P [8] strains, particularly during July 2013 through June 2015. G1P [8] rotavirus strains accounted for an overall 2.7% prevalence rate but were found to circulate in August 2012 and January 2014. The remainder of the circulating strains belonged to G12P [4], G2P [8], G3P [8], and G9P [8] genotypes and accounted for < 4% of all typed isolates. Partially typed or fully untypeable rotavirus strains were found during July 2013–June 2015 only, altogether accounting for around 10% of all isolates.
Table 2 shows a comparison between G- and- P- type-specificities in paired stool-serum samples from ten individual patients admitted for rotavirus-related gastroenteritis. There were fully concordant results in seven of the eight stool-serum pairs in which rotavirus G2P [4] genotype was identified. Among these, a potential discrepancy was seen in one pair with identification of G2P [4] and G2P[NT] genotypes in stool and serum samples, respectively. Rotavirus strains with discordant G-type and P-type specificities were detected in two pairs: G12P [8] and G2P[NT] genotypes in stools and sera, respectively. Among the G2 concordant pairs all electropherotypes displayed identical short patterns, while a long profile was associated with the two discordant G12P [8] and G2P[NT] pairs. In order to confirm the latter discrepancy in the stool-serum pairs, as determined by RT-PCR, a sequence analysis of the VP7 gene was performed on these two samples (Fig. 3).
Phylogenetic dendogram of partial VP7 gene of G2P [4] and G12P [8] strains from stool and serum samples (highlighted in bold) during June 2012–June 2015 in Belém, Brazil. Bootstrap values (2000 replicates) are shown at the branch nodes; values < 70% are not shown. Strains detected in serum samples are indicated with empty triangles while strains detected in stool samples are indicated with black circles
Phylogenetic analysis of the partial VP7 gene of G2 sequences from serum samples of subjects PID 445 and PID 446 showed high sequence identities (97.8–100%) with strains of human origin from Brazil, Russia, Bangladesh, Italy and Thailand. In addition, G12 genotypes detected concurrently in stool samples from PID 445 and PID 446 patients were shown to be closely related (95.6–98.5%) to human strains from Bangladesh, Thailand and Belgium. Serum samples from ten diarrhoeic children with rotavirus-positive stools yielded clear strong amplicons of 881 base pairs (bp) and 876 bp, of the genes encoding VP7 and VP4 proteins, respectively, as depicted in Additional file 1: Figure S1.
As demonstrated in Fig. 4, there was a moderate negative correlation between the stool viral load as determined by qRT- PCR cycle Ct values, and the rotavirus antigen levels expressed as optical densities (OD) in sera tested by ELISA [n = 96 paired samples; Pearson’s correlation coefficient (rs) = − 0.3240; 95% CI − 0.1262 to − 0.4971; and p value = 0.0013].
Discussion
In spite of the current scenario where two licensed vaccines are increasingly being made available for introduction in many countries around the world, there remains to be fully elucidated the mechanisms of pathogenesis in rotavirus infection, particularly that underlying virus spreading beyond the gastrointestinal tract into the blood stream [6]. The current study’s main focus lies on the potential impact of rotavirus antigenemia on clinical manifestations in a cohort of children hospitalised for acute gastroenteritis in Belém, Brazil. Since current available evidence points to the fact that antigenemia and RNAemia are common events among children infected with rotavirus, we chose to use ELISA (targeted mainly at the VP6 protein) and qRT-PCR/RT-PCR in order to determine the presence and characterise rotavirus strains in both faecal and serum samples.
Noticeably in our cohort, rotavirus antigen was detected in 24% of the stool specimens tested by ELISA, an overall positivity rate found to be higher than that of 17% reported in a study also conducted during the early years post-rotavirus vaccine introduction in Brazil [34]. The distinct field/laboratory methodologies used in these studies, however, may have accounted for such difference in prevalence rates. A finding that deserves further consideration is the fact that one fifth of the children in our study cohort developed severe rotavirus-related gastroenteritis, despite a rotavirus vaccination coverage rate of around 90% in our setting. Since previous studies in our region have suggested that children who undergo a full vaccination schema are less likely to develop rotavirus infection requiring hospitalisation [35], an incomplete immunisation schedule could possibly have accounted for such unexpected high rate of severe rotavirus-related gastroenteritis. Nonetheless, we cannot rule out in this context the potential challenge represented by the globally emerging G12P [8] genotype, which was found to be the dominant circulating rotavirus strain during a two-year surveillance period (June 2013–July 2015) in our study.
Our results showed that antigenemia was present in around 40% of children hospitalised for acute RV-related gastroenteritis in Belém, a rate similar to those reported in previous studies conducted elsewhere which were in the range of 33–75% [10, 13, 17, 18, 36]. Our data showed that 40% of patients with a rotavirus-positive stool sample were found to have rotavirus RNA in serum, a rate lower than those rates from studies conducted elsewhere which also included either blood samples or plasma and – unlike our study – were conducted before universal introduction of rotavirus vaccine [14]. Interestingly, our study suggests that ELISA and RT-PCR had similar sensitivities for the detection of antigenemia (positivity rate, 9.3%) and RNAemia (11.4%), respectively. These results appear to be in line with those from studies in India where comparable – even though higher – rates of antigenemia (71.2%) and RNAemia (81.9%) were detected among children with rotavirus-positive stool samples [37]. Unlike previous studies [36] which have recently reported a higher prevalence rate of RNAemia (70.8%), as compared to antigenemia (33.3%), in peripheral blood mononuclear cells (PBMC) and plasma, respectively, of children with acute diarrhoea in USA. Moreover, these authors utilised a different RT-PCR system where primers targeted specifically the rotavirus NSP4 gene.
As also reported elsewhere [17], we have demonstrated that rotavirus RNA levels detected in serum samples (11.4%) were significantly lower than RNA levels found in stool samples (50.7%), supporting the general notion that rotaviruses replicate primarily in the mature, differentiated enterocyte of the small intestine [38].
In our study we performed a correlation analysis of paired stool and serum samples obtained from 96 paediatric patients which provided evidence that the stool rotavirus copy numbers correlate significantly with the ELISA OD values in sera (Fig. 4). These findings are in line with those from Ramani et al. [8, 18] who also stated a positive correlation between the levels of rotavirus antigen in sera with the stool viral load. These authors have hypothesised that a higher stool viral load (and severe disease) would account for a major damage of the intestinal epithelium, resulting in the spread of rotavirus beyond the intestines into the circulatory system.
In the present study we were able to assess whether or not VP7 and VP4 genotype specificities were concordant in comparing paired stool-serum samples from ten paediatric patients hospitalised for rotavirus-related gastroenteritis during the first year of surveillance. Rotavirus strains bearing G2 and P [4] genotype specificities – which were found to be dominant during June 2012–July 2013 – could be identified in seven of the ten pairs, suggesting that rotaviruses infecting the gut were the same strains which disseminated beyond the intestine. These concordant results appear to be consistent with findings from a few other studies conducted elsewhere, even though discrepancy of genotypes in stool and serum samples (G12P [8] and G2P[NT] pairs) of individual patients was also see in our two of our study patients. These latter findings raise the hypothesis that identification of distinct genotypes in two specimens from a single patient may reflect either consecutive infections at different time points or the occurrence of intrinsic difference in growth characteristics and/or tissue tropism, leading to speculate that during co-infections a preferential extra-intestinal spread of some strains may occur [25, 39, 40]. In this context, for example, Hemming et al. [14] have suggested that both antigenemia and RNAemia are particularly common in severe rotavirus gastroenteritis caused by G1P [8] genotype. Nonetheless, the issue of whether or not the potential for rotavirus antigenemia/viraemia is limited to specific genotype(s) remains largely controversial [41]. In our study, the discrepancy among genotypes present in different specimens from the same individual was clearly confirmed through sequence analysis of the VP7 gene of the two samples, as also reported elsewhere [25]. Additionally in support to the discordant genotypes was the identification of short and long electrophoretic profiles for the G2 and G12 strains present in the same specimen, respectively.
In the present study we also sought to determine whether children with antigenemia presented with more severe rotavirus disease than children without antigenemia. It was observed that patients with rotavirus antigen in serum were more likely to develop frequent vomiting, as compared to patients without antigenemia. In contrast, we could not find any significant difference between the groups in regards to the severity of other clinical parameters including fever, duration of vomiting, diarrhoea and white blood cell count. Unlike other studies conducted elsewhere [8, 18], our findings did not show any significant difference in the overall disease severity, as graded by the Vesikari clinical scoring system, between children with antigenemia and children without antigenemia. Of note, our observation that rotavirus antigenemia may specifically correlate with increased severity of vomiting is consistent with findings from recent studies in Finland and India [14, 37]. Since the rotavirus-induced emesis involves secretion of serotonin from enterochromaffin cells with stimulation of vagal afferent nerves connected with the brain stem, it has been speculated that antigenemia/RNAemia might reflect an infection deeper into the gut, possibly leading to a more powerful activation of the brain areas essential for vomiting [14, 42, 43].
Of potential importance in the current post-rotavirus vaccine introduction scenario, our findings suggest that children not vaccinated against rotavirus appear to be more likely to develop antigenemia, as compared to those given at least one vaccine dose. However, further studies are needed to confirm this finding since such a difference may have occurred by chance owing to the small sample size in our study.
Although a significant proportion of children in our study were found to develop antigenemia/RNAemia, we could not detect any clinical manifestations which might be suggestive of extra-intestinal involvement of rotavirus infection. This supports the notion that while viraemia constitutes a common event, it does not appear that systemic infection is frequently associated with clinically significant non-gastrointestinal disease. Furthermore, it has also been postulated that subclinical extra-intestinal infections often occur in children and may occasionally progress to an overt clinical condition [6, 12, 44, 45].
The findings of this study are subject to some limitations. First, our antigenemia results should be interpreted with caution, since all currently available ELISA commercial kits are designed to detect rotavirus antigen in stool samples and had not been validated for use with either human or plasma samples. Nonetheless, our data suggest that ELISA and RT-PCR display similar sensitivities for the detection of antigenemia and RNAemia, respectively. Of note in this regard, earlier studies have suggested that ELISA may potentially offer a new practical diagnostic tool for the detection of rotavirus from serum/plasma samples in situations where a stool sample is not readily available (e.g. severe dehydration) [13]. Second, we were unable to demonstrate the presence of infectious virus particles in the serum samples using cell cultures for rotavirus isolation, which might reflect a true viremia. It should be pointed out however that RNA detection in blood has been reported as a strong indicator of the presence of viraemia among children with rotavirus gastroenteritis [6]. In addition, the current study did not include the search for differences at both nucleotide and amino acid levels of the paired strains under comparison, an aspect which is worth considering further.
In summary, our results confirm and extend previous well documented findings that rotavirus antigenemia/ RNAemia occurs routinely among children with acute rotavirus gastroenteritis and may contribute to a greater disease severity by increasing the number of vomiting episodes. Of particular interest in the current context of growing rotavirus vaccine introduction, it is has been pointed out that detection of antigenemia/RNAemia may represent an additional, useful approach to improve the sensitivity of post-licensure epidemiological studies of vaccine effectiveness [20]. In addition, the detection of rotavirus antigenemia/RNAemia seems also warranted as it remains to be determined whether any currently used rotavirus vaccine strains may eventually spread beyond the intestine.
Conclusions
To our knowledge this is the first report in Brazil showing that rotavirus antigenemia and RNAemia are detectable in a significant proportion of infants and young children admitted to hospital due to acute gastroenteritis. According to the Vesikari scoring system a trend for greater clinical severity was seen among children with rotavirus-positive serum, as compared to those with rotavirus-negative serum. A statistically significant difference between these groups was particularly seen with regards to the number of vomiting episodes. It has also been demonstrated a positive correlation between the levels of rotavirus antigen in sera, as determined by ELISA, with the stool viral load, as assessed by qRT-PCR. Both G2P [4] and G12P [8] rotavirus genotypes accounted for most of the isolates throughout the study period; in addition, we have documented a high concordance of genotypes detected in a subgroup of paired stool and serum samples. Further studies on rotavirus antigenemia/RNAemia are warranted to better assess the effectiveness of currently used rotavirus vaccines, as well as to determine whether licensed vaccines may eventually spread beyond the intestines.
Availability of data and materials
Raw data of the study are available from the corresponding author on reasonable request. For this, a permission from MCAJ would be needed. The nucleotide sequences of VP7 genome segments of two representative G2 strains and two representative G12 strains collected in this study in Belém, Brazil, are available in the GenBank, through accession numbers MH456983 to MH456986.
References
Lanata CF, Fischer-Walker CL, Olascoaga AC, Torres CX, Aryee MJ, Black RE, et al. Global causes of diarrheal disease mortality in children <5 years of age: a systematic review. PLoS One. 2013;8:e72788.
Liu J, Platts-Mills JA, Juma J, Kabir F, Nkeze J, Okoi C, et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet. 2016;388:1291–301.
GBD Diarrhoeal Disease Collaborators. Estmates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis. 2017;17:909–48.
Estes MK, Greenberg HB. Rotaviruses. In: Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B, et al. Fields Virology. 6th edition. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins, Philadelphia; 2013. p. 1347–1401.
Staat MA, Azimi PH, Berke T, Roberts N, Bernstein DI, Ward RL, et al. Clinical presentations of rotavirus infection among hospitalized children. Pediatr Infect Dis J. 2002;21:221–7.
Blutt SE, Matson DO, Crawford SE, Staat MA, Azimi P, Bennett BL, et al. Rotavirus antigenemia in children is associated with viremia. PLoS Med. 2007;4:e121.
Ramig RF. Pathogenesis of intestinal and systemic rotavirus infection. J Virol. 2004;8:10213–20.
Sugata K, Taniguchi K, Yui A, Miyake F, Suga S, Asano Y, et al. Analysis of rotavirus antigenemia and extraintestinal manifestations in children with rotavirus gastroenteritis. Pediatrics. 2008;122:392–7.
Ahmed K, Bozday IG, Mitui MT, Ahmed S, Kabir L, Buket D, et al. Circulating rotaviral RNA in children with rotavirus antigenemia. J Negat Results Biomed. 2013;12:1–8.
Blutt SE, Kirkwood CD, Parreño V, Warfield KL, Ciarlet M, Estes MK, et al. Rotavirus antigenaemia and viraemia: a common event? Lancet. 2003;362:1445–9.
Jalivand S, Marashi SM, Tafakhori A, Shoja Z. Extraintestinal involvement of rotavirus infection in children. Arch Iran Med. 2015;18:604–5.
Alfajaro MM, Cho K-O. Evidences and consequences of extra-intestinal spread of rotaviruses in humans and animals. Virus Dis. 2014;25:186–94.
Fischer TK, Ashley D, Kerin T, Reynolds-Hedman E, Gentsch J, Widdowson M-A, et al. Rotavirus antigenemia in patients with acute gastroenteritis. JID. 2005;192:913–9.
Hemming M, Huhti L, Räsänen S, Salminen M, Vesikari T. Rotavirus antigenemia in children is associated with more severe clinical manifestations of acute gastroenteritis. Pediatr Infect Dis J. 2014;33:366–71.
Bányai K, László B, Duque J, Steele AD, Nelson EA, Gentsch JR, et al. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine. 2012;30(Suppl 1):A122–30.
Trojnar E, Sachsenröder J, Twardziok S, Reetz J, Otto PH, Johne R. Identification of an avian group a rotavirus containing a novel VP4 gene with a close relationship to those of mammalian rotaviruses. J Gen Virol. 2013;94:136–42.
Ray P, Fenaux M, Sharma S, Malik J, Subodh S, Bhatnagar S, et al. Quantitative evaluation of rotaviral antigenemia in children with acute rotaviral diarrhea. JID. 2006;194:588–93.
Ramani S, Paul A, Saravanabavan A, Menon VK, Arumugan R, Sowmyanarayanan TV, et al. Rotavirus antigenemia in Indian children with rotavirus gastroenteritis and asymptomatic infections. CID. 2010;51:1284–9.
WHO. Rotavirus vaccines. WHO position paper – 2013. Wkly Epidemiol Rec. 2013;88:49–64.
Patel M, Rench MA, Boom JA, Tate JE, Sahni LC, Hull JA, et al. Detection of rotavirus antigenemia in routinely obtained serum specimens to augment surveillance and vaccine effectiveness evaluations. Pediatr Infect Dis J. 2010;29:836–9.
Linhares AC, Macias-Parra M, Sáez-Llorens X, Vergara R, Jimenez E, Velázquez FR, et al. Rotavirus gastroenteritis in Latin America: a hospital-based study in children under 3 years of age. Trials Vaccinol. 2012;1:36–41.
Linhares AC, Justino MCA. Rotavirus vaccination in Brazil: effectiveness and health impact seven years post-introduction. Expert Rev Vaccines. 2014;13:43–57.
Ruuska T, Vesikari T. Rotavirus disease in Finnish children: use of numerical scores for clinical severity of diarrhoeal episodes. Scand J Infect Dis. 1990;22:259–67.
Eing BR, May G, Baumeister HG, Kühn JE. Evaluation of two enzyme immunoasssays for detection of human rotaviruses in fecal specimens. J Clin Microbiol. 2001;39:4532–4.
Chitambar SD, Tatte VS, Dhongde R, Kalra V. High frequency of rotavirus viremia in children with acute gastroenteritis: discordance of strains detected in stool and sera. J Med Virol. 2008;80:2169–76.
Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, et al. Identification of group a rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–73.
Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, et al. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276–82.
Banerjee I, Ramani S, Primrose B, Iturriza-Gomara M, Gray JJ, Brown DW, et al. Modification of rotavirus multiplex RT-PCR for the detection of G12 strains based on characterization of emerging G12 rotavirus strains from South India. J Med Virol. 2007;79:1413–21.
Gouvea V, Santos N. Timenetsky M do C. VP4 of bovine and porcine group a rotaviruses by PCR. J Clin Microbiol. 1994;32:1333–7.
Iturriza-Gómara M, Isherwood B, Desselberger U, Gray J. Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999. J Virol. 2001;75:3696–705.
Zeng SQ, Halkosalo A, Salminen M, Szakal ED, Puustinen L, Vesikari T. One-step quantitative RT-PCR for the detection of rotavirus in acute gastroenteritis. J Virol Methods. 2008;153:238–40.
Pereira HG, Azeredo RS, Leite JP, Barth OM, Sutmoller F, de Farias V, et al. Comparison of polyacrylamide gel electrophoresis (PAGE), immuno-electron microscopy (IEM) and enzyme immunoassay (EIA) for the rapid diagnosis of rotavirus infection in children. Mem Inst Oswaldo Cruz. 1983;78:245–50.
Ayres M, Ayres Júnior M, Ayres DL, Santos AS. Bioestat 5.0 – Aplicações estatísticas nas áreas das ciências biomédicas. Belém: ONG Mamirauá; 2007.
Guerra SF, Linhares AC, Mascarenhas JD, Oliveira A, Justino MC, Soares LS, et al. Rotavirus strain surveillance for three years following the introduction of rotavirus vaccine into Belém. Brazil J Med Virol. 2015;87:1303–10.
Justino MCA, Campos EMNA, Mascarenhas JDP, Soares LS, Soares SGS, Soares TS, et al. Detecção de antígenos de rotavírus no soro de crianças hospitalizadas por gastrenterite aguda em Belém, Estado do Pará, Brasil. Rev Pan-Amaz Saúde. 2016;7:153–8.
Moon S, Wang Y, Dennehy P, Simonsen KA, Zhang J, Jiang B. Anteigenemia, RNAemia, and innate immunity in children with acute rotavirus diarrhea. FEMS Immunol Med Microbiol. 2012;64:382–91.
Das S, Sahoo GC, Das P, Singh UK, Jaiswal AK, Singh P, et al. Evaliuating the impact of breastfeeding on rotavirus antigenemia and disease severity in Indian chidren. PLoS One. 2016;11:e0146243.
Crawford SE, Patel DG, Cheng E, Berkova Z, Hyser JM, Ciarlet M, et al. Rotavirus viremia and extraintestinal viral infection in the neonatal rat model. J Virol. 2006;80:4820–32.
Chiappini E, Arista S, Moriondo M, De Grazia S, Giammanco GM, Azzari C, et al. VP7 and VP4 sequence analyses of rotavirus strains from Italian children with viraemia and acute diarrhoea. JPGN. 2010;50:114–6.
Iturriza-Gómara M, Autcherlonie IA, Zaw W, Molyneaux P, Desselberger U, Gray J. Rotavirus gastroenteritis and central nervous system (CNS) infection: characterization of the VP7 and VP4 genes of rotavirus strains isolated from paired fecal and cerebrospinal fluid samples from a child with CNS disease. J Clin Microbiol. 2002;40:4797–9.
Blutt SE, Conner ME. Rotavirus: the gut and beyond! Curr Opin Gastroenterol. 2007;23:39–43.
Hagbom M, Istrate C, Engblom D, Karlsson T, Rodriguez-Diaz J, Buesa J, et al. Rotavirus stimulates release of serotonin (5-TG) from human enterochromaffin cells and activates brain structures involved in nausea and vomiting. PLoS Pathog. 2011;7:e1002115.
Hansen MB, Witte AB. The role of serotonin in intestinal luminal sensing and secretion. Acta Physiol (Oxf). 2008;193:311–23.
Fuchigami T, Goto K, Hasegawa M, Saito K, Kida T, Hashimoto K, et al. A 4-year-old girl with clinically mild encephalopathy with a reversible splenial lesion associated with rotavirus infection. J Infect Chemother. 2013;19:149–53.
Cioc AM, Nuovo GJ. Histologic and in situ viral findings in the myocardium in cases of sudden, unexpected death. Mod Pathol. 2002;15:914–22.
Acknowledgements
We are indebted to the nursing staff and paediatricians at Clínica Pediátrica and Clínica Pio XII, Belém, Pará, Brazil. We also wish to thank all the participating children and parents/guardians.
Funding
This study received financial support from the Evandro Chagas Institute (IEC), Health Surveillance Secretariat, which supported the study team to perform sample collection, analysis, interpretation of the data obtained and writing the manuscript. The National Council for Scientific and Technological Development (CNPq) provided financial support to purchase laboratory kits for use in the analysis. TSM, FPF and CBGV received grants from the IEC-CNPq’s Junior Scientific Initiation Program for high school students. RJSB received a grant from the Coordination for the Improvement of Higher Education Personnel (CAPES) within the IEC’s Virology Post-Graduation Program.
Author information
Authors and Affiliations
Contributions
MCAJ and ACL designed the study. MCAJ, EAC, MJCP, TSM, FPF, OMB, CBGV and RJSB collected the data. JDPM, LSS and SFSG performed the laboratory procedures. IPF, MCAJ and ACL made the interpretation of statistical analysis. MCAJ and ACL wrote the first draft of the paper with input from all the authors who each approved the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Independent Ethics Committee of the Instituto Evandro Chagas, Health Surveillance Secretariat, Brazilian Ministry of Health (protocol 0039/2011), and was conducted in accordance with the principles of the Declaration of Helsinki, as well as in compliance with the Good Clinical Practice guidelines. Children were recruited for the study after informed written consent was obtained from the parent or guardian.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Figure S1. Agarose gel electrophoresis of RT-PCR VP7 (A) and VP4 (B) gene products obtained from the sera of ten rotavirus-positive children with gastroenteritis in Belém, Brazil. (PPTX 121 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Justino, M.A., Campos, E.A., Mascarenhas, J.P. et al. Rotavirus antigenemia as a common event among children hospitalised for severe, acute gastroenteritis in Belém, northern Brazil. BMC Pediatr 19, 193 (2019). https://doi.org/10.1186/s12887-019-1535-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12887-019-1535-2