Abstract
Background
Thyroid eye disease (TED) is the most common autoimmune disease and usually occurs in patients with hyperthyroidism. In this disease, eye-related tissue, such as eye muscles, eyelids, tear glands, etc., become inflated, which causes the eyes and eyelids to become red, swollen, and uncomfortable. The pathophysiology of this disease is still poorly known.
Aim
This study aims to discover potential biomarkers and regulatory pathways of TED which will not only help to diagnose the disease and understand orbital involvement in thyroid dysfunction but also provide an insight for better therapeutics.
Methods
We applied a data-driven approach by combining gene biomarkers both from published literature and computationally predicted from microarray gene expression data. Further, the DAVID tool is used for Gene Ontology-based enrichment analysis.
Results
We obtained a total of 22 gene biomarkers, including 18 semi-automatically curated from the literature and 4 predicted using data-driven approaches, involved in the pathogenesis of TED that can be used as potential information for therapeutic targets. Further, we constructed a regulatory pathway of TED biomarkers comprises of 310 connected components, and 1134 interactions using four prominent interaction databases.
Conclusion
This constructed pathway can be further utilized for disease dynamics and simulation studies.
Similar content being viewed by others
Introduction
Thyroid eye disease (TED), also known as Graves’ eye disease (GED), or Graves’ ophthalmopathy (GO), is an autoimmune condition in which immune cells attack the thyroid gland due to secretion of excess amount of thyroid-stimulating hormone (TSH). These excess hormones increase metabolism (hypermetabolism) which is characterized by fast heartbeat, palpitation, profuse sweating, high blood pressure, heat intolerance, weight loss, etc. [1, 2] The hyperthyroidism leads to the eye protruded from eye orbit due to inflammatory disorder and also leads to permanent facial disfigurement. It causes swelling of muscle, fat, tissues, i.e. periorbital tissues leading to proptosis [3]. In fact, autoimmune attack generally targets the eye muscles because these tissues contain proteins that seem similar to the immune system as those of the thyroid gland [4, 5]. According to the ReportLinker report, the global treatment market of TED is expected to pump up from USD 211.49 Million in 2019 to USD 344.19 Million by the end of 2025 [6]. In another report published by MarketWatch News, the TED market projected a compound annual growth rate (CACG) of 8.53% during the forecast period of 2020–2026 [7].
In TED, tissues around the eye are attacked, leading to inflammation and swelling, which causes redness and pain, puffiness around the eyes, erythema, conjunctivitis, proptosis, and upper eyelid retraction [8]. Environmental factors like smoking and radioactive iodine are more predisposed to TED [9]. There are many other factors but smoking has a major influence [10]. Smoking induces thyroid functional changes, like a decrease or increase in thyroid hormones and also the risk of thyroid cancer [11]. Like thyroid disease, women are more vulnerable to TED than males with the female to male ratio of 4:1 [12]. Immune cells attack the periorbital tissues that lead to the expansion of eye muscles or fat [2, 12]. Hyperthyroidism leads to an overactive thyroid gland, i.e., more secretion of TSH. The TSH helps to maintain body metabolism, and it’s over secretion develops several consequences like high blood pressure, fatigue, weight loss, and irritabilitye [13]. Further, autoimmune cells attack the enlarged thyroid gland as well as eye muscles and periorbital tissue. These tissues contain proteins that appear similar to the thyroid gland, like thyroid-stimulating hormone receptor (TSH-R), target as immune assault [14]. The progressive eye swelling may trigger acute pressure inside the eye socket, pressure-pain which worsens movements of the eye, decreased vision when swollen tissues push on the optic nerve [2].
The TED is diagnosed by a blood test, thyroid computed tomography (CT) scan, magnetic resonance imaging (MRI) scan, radioactive iodine uptake test, low thyroid TSH test, positive thyroid-stimulating immunoglobulin (TSI) test, increase free thyroxine (T4), and elevated anti-peroxidase [15, 16]. Since TED occurs due to the immune system’s attack on the healthy tissues, therefore, treatment of the thyroid gland does not improve eye disease [17]. Some of the prevalent methods of treatment of Graves’ disease are anti-thyroid drugs, such as Thionmaids, Methimazole, Teprotumumab (under clinical trial and study by FDA) [18] or beta-blockers (such as Propranolol, Atenolol, Metoprolol), use of radioactive iodine, or surgery [19]. These treatments are based on age and the degree of illness of the patient [20]. The use of anti-thyroid drugs, which reduces the release of thyroids hormones, is the least invasive method to treat Graves’ disease. Radioactive iodine therapy is the most common method to treat Graves’ disease in the United States. The definite treatment of Graves’ disease is thyroidectomy, a surgery to remove the thyroid gland [21, 22]. As far as TED is concerned, mild cases may be treated with sunglasses, artificial tears, or ointments. However, more serious cases may be treated with corticosteroids which reduce swelling of tissues around the eyes. Orbital therapy and orbital decomposition surgery are also used to treat TED [23, 24].
The molecular mechanism underpinning TED is gradually becoming clearer due to advancements in both experimental and computational techniques. The availability of large-scale biological data (i.e., multi-omics) offers a paradigm shift from sub-optimal treatment to optimal targeted therapy [25]. Biomarkers are of pivotal importance which serves as a useful noninvasive tool in the clinical armamentarium for disease studies including its diagnosis, prevention, drug target identification, designing drug for a particular receptor, and biological processes to a therapeutic intervention [26]. It can be genes, mRNAs, and metabolites. In the case of TED, a set of biomarkers were identified in immunogenetics, hormones, antibodies, cytokines, urine, orbital fat, and tear. Peroxisome proliferation activation receptor gamma (PPAR-γ) activation leads to an eye’s protrusion. Triiodothyronine (T3) and Thyroxine (T4) dysfunction secretion and antibodies (TRAbs) levels are associated with the severity of TED [9, 27]. IL-1β, IL-10, IL-8, C-C chemokine ligand 20 (CCL20), IL-17 are the pro-inflammatory cytokines. Polymorphism of IL-10 is associated with the incidence of TED [28]. Soluble vascular cell adhesion molecules-1 (sVCAM-1) and intercellular adhesion molecules-1 (ICAM-1) are elevated in the blood of TED patients [29]. CLAT-4 immunoglobulin functions as an immune checkpoint and downregulates immune responses [30]. Further, CD152, an expression product of CTLA-4 genes, downregulates T-cell activation [31]. IL-1β and IL-6 level in orbital fat associated with the smoking status of TED patients [32]. IFN-γ is differentially upregulated in TED, and platelet-derived growth factor (PDGF) responsible for the physiological event in TED [32]. PDGF and IL-1β induce adipogenesis through the enzyme cyclooxygenase-2 (COX-2). Levels of TSHR-Ab are a useful tool for the measurement of TED [33]. PAI-1 and SERPINE1 regulate the proteolytic activity [34]. Wong and colleagues [35] reported the association of IL-1A and other IL genes with Graves’s Ophthalmopathy (GO). Tear proteins, like S100A4 and PIP, also serve as biomarkers to predict ocular and systemic disease progression [36]. PTPN22 (protein tyrosine phosphatase) shows negative regulation of T-cell activation [37]. NF-kB (nuclear factor kappa-B) is a transcriptional factor activated by various cytokines. Other genetic factors like TLR-9, CD86, CD103, glucocorticoid receptors, CTLA-4, TNF-alpha, HLA-DRB-1 are also associated with TED [38]. A review on biomarkers associated with TED can be found in Turck et al. [32] and Longo et al. [34]
Even though few studies have been carried out to identify gene biomarkers in TED, but its study at the genome-wide scale is lacking. Further, the possible regulatory pathway of biomarkers in TED has not been studied earlier. Hence, in the present study, we applied a data-driven approach to detect gene biomarkers in TED, which combines biomarkers both from reported literature as well as we computationally predicted using microarray gene expression profiles. Further, a regulatory pathway of biomarkers is constructed followed by various Gene Ontology-based enrichment analyses. This regulatory pathway can be further utilized for disease dynamics, molecular docking, and simulation studies.
Methods
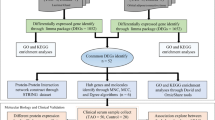
The thyroid eye disease (TED) is a complex disease having an overlap with other Grave’s disease and there are several molecular players involved. Here, we constructed a systematic network of gene regulatory pathways of TED. The network is based in semi-automated curate literature-based information reported to be gene biomarkers of TED and predicted information using high-throughput gene expression data. The constructed network has been topological, gene ontology (GO) and tissue-specifically analyzed to translate it into meaningful disease-specific and tissue-specific markers and can be utilized as targets in the diagnosis and therapy of TED. The methodological pipeline adopted in this study is depicted in Fig. 1.
Extraction of experimentally determined biomarkers
To extract TED genes from published literature, we used the Entrez System of the National Center for Biotechnology Information (NCBI) and performed a query with the MeSH (Medical Subject Headings) terms “((thyroid eye disease) AND biomarker)”. The Entrez (https://www.ncbi.nlm.nih.gov/) is an online cross-database search system that helps the users to perform a global query in NCBI’s various genomics, genes, proteins, genetics, literature, and other health sciences databases. The search results provide a summary of hits in various NCBI databases. We recorded the list of genes from the NCBI-Gene database and a list of published literature in NCBI-PubMed for manual screening of biomarker genes in TED. The manually screened biomarker genes were cross-checked by two independent reviewers.
Prediction of gene biomarkers using gene expression profiles
To predict gene biomarkers of TED, we took microarray gene expression data from the NCBI-GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE58331). In this dataset, RNA was extracted and analyzed with Affymetrix from anterior orbit and lacrimal gland tissues of the collected biopsies. The data contains gene expression of various inflammatory diseases including TED, but we considered only TED gene expression data from anterior orbit tissue for this study. The considered datasets contain 27 TED samples and 22 control samples, both from anterior tissues of men as well as women.
Construction of gene regulatory pathway
Molecular interaction data of the identified gene biomarkers were collected from four major databases including STRING [39], GeneMania [40], TRRUST [41], and KEGG Pathway [42]. These interaction data were merged to construct a consolidated gene regulatory pathway for further downstream analysis.
GO enrichment analysis
Gene Ontology (GO) enrichment is performed to study various enrichment analyses of a given gene set, i.e., a list of gene biomarkers, that finds which GO terms are either over-represented or under-represented using stored annotations in the database. We utilized DAVID 6.8 tool [43] for studying GO enrichment analysis, functional category analysis, disease enrichment, and disease class enrichment analysis, and KEGG pathway enrichment analysis. The DAVID 6.8 server can be accessed from https://david.ncifcrf.gov/.
Results
After performing the query with the MeSH term “((thyroid eye disease) AND biomarker)” in NCBI’s Entrez System, we obtained a list of 24 genes in NCBI’s Gene database along with a list of 285 publications in PubMed. We manually screened these 24 genes to verify their role as biomarkers in TED. Further, we also manually screened all the obtained publications to obtain more reported biomarkers in TED. Finally, after the manual screening of each gene and reported literature, we shortlisted a list of 18 experimentally verified genes reported to be involved in the pathogenesis of TED, as shown in Table 1 along with PubMed ID (PMID) of supporting literature, and its involvement in various pathways. To predict gene biomarkers using microarray gene expression data of TED (GEO Accession No. GSE58331), we executed the GEO2R tool on both TED samples and normal samples. In other words, we performed a case-control analysis to find out differentially expressed genes (DEGs) that can be utilized as disease biomarkers. We applied the fold-change statistics to compute DEGs, along with several other statistics including p-value, and false discovery rate (FDR). To filter significant genes that may be the potential gene biomarkers of the disease under study, we applied a threshold of p-value <=0.05 and have at least two-fold change (i.e., − 1.0 > = logFC > = + 1.0) in their gene expression, as generally applied and suggested by researchers [44,45,46,47]. Further, duplicate genes and genes with missing names and statistical values were removed from the list. In this way, we obtained a list of 63 genes for down-stream analysis. To further narrow down and perform significant analysis of predicted gene biomarkers, we shortlisted only those genes which are transcription factors (TFs), and associated with autoimmune or thyroid disease, and/or involved in autoimmune or thyroid disease KEGG pathway, and/or GO enriched with the term “thyroid” or “autoimmune” as a biological process, shown in Table 2. For this purpose, we utilized the TRRUST v2 database [41]. Hence, we obtained a total of 22 gene biomarkers, including 18 semi-automatically curated from the literature and 4 predicted, involved in the pathogenesis of TED that can be used as potential information for therapeutic targets.
To construct a consolidated gene regulatory path of TED biomarkers, we retrieved and merged the interaction of identified 22 biomarkers from four major databases. All the duplicate interactions were eliminated. The network in Fig. 2 shows the identified regulatory pathway of TED biomarkers consists of 310 connected components, 1134 interactions, the average number of neighbors of 7.3, two self-loops, and a clustering coefficient of 0.234. This network is scale-free and follows power-law. To further narrow down the list of the most significant genes for TED, we performed GO enrichment analysis, functional category analysis, disease enrichment, and disease class enrichment analysis, and KEGG pathway enrichment analysis using the DAVID tool. The terms and keywords used for this enrichment analysis and their subsequent results are presented in Table 3. To focus our analysis on TED, we considered GO enrichment terms as “inflammatory response” and “immune response”, and analysis results show that most of the identified gene biomarkers are enriched with either of these two terms or both (Table 3). Similarly, in disease enrichment class analysis, most of the disease terms are enriched with “vision” or “immune” or both (Table 3). In the KEGG pathways analysis, most of the genes are either enriched with the term “autoimmune thyroid disease” or “cytokine-cytokine receptor interaction” (Table 3). Hence, these deeper analysis results provide that the identified gene biomarkers are involved in the pathogenesis of TED.
Discussion
The disease biomarkers is an important noninvasive tool in the clinical armamentarium for the study of diseases. This study focuses on the discovery of potential biomarkers and regulatory pathways in TED that can be utilized for disease diagnosis and helps us know insight for better therapeutics. Our analysis of combined gene biomarkers, both curated from published literature and computationally predicted using microarray gene expression data, discern its involvement in TED and other thyroid-associated ocular diseases. Our data-driven analysis identified a list of 9 Cytokines (CXCL8, CXCL10, IL10, IL17A, IL22, IL4, IL6, IL13, and IFNG) which are reported to increase the volume of orbital tissue [32]. Further, enrichment analysis identified 11 genes involved in the inflammatory response, 11 genes involved in immune response, and 10 genes are enriched in cytokine-cytokine receptor interaction (refer to Table 3). Cytokine IL10 is pro-inflammatory and its polymorphism has been reported to be associated with the incidence of TED [28]. IFNG is also reported to be differentially upregulated in TED [32]. Levels of TSHR-Ab are a useful tool for the measurement of TED [33]. Our major analysis results, in-line with the reported literature, are summarized in the following section.
TSHR as an autoantigen in TED
TSHR is a glycoprotein hormone receptor and a member of the G protein-coupled receptor family. It has a ligand-binding extracellular domain, intracellular domain, and a transmembrane domain. The thyroid-stimulating immunoglobulins (TSI), also called thyroid-stimulating antibodies (TSAb), and TSH bind to TSHR which leads to activation of the receptor and unregulated production of thyroid hormones [48]. Besides thyroid epithelium, TSHR is found in several connective tissues and adipose depots. The mRNA expression level of TSHR is higher in orbital fibroblasts from TED patients. In TED, TSIs can activate TSHR that signals the production of IL-6 [49]. The KEGG pathway enrichment analysis discerns that TSHR is enriched in various pathways including autoimmune thyroid disease (hsa05320), Thyroid hormone synthesis (hsa04918), and cAMP signaling pathway (hsa04024). `.
TSHR-IGF1R cross-talk
In several studies, it is reported that TSHR is the main target of stimulatory autoantibodies in the pathogenesis of TED, and stimulatory IGF1R autoantibodies cross-talk with TSH [50,51,52]. In fact, signaling initiated from either of these two receptors can be controlled by inhibiting the activity of IGFR1 [53]. These two make a physically and functionally interactive complex within orbital fibroblasts, and inhibition of IGF1R reduces TSH-dependent signalling [25]. Smith et al. [54] call TSHR-IGF1R cross talk as “partners of crime”, while Wiersinga [55] calls it “an unfortunate marriage between TSHR and IGFR1”. Teprotumumab is an IGF1R inhibitor that interrupts the key molecular mechanism of TED pathogenesis and is reported to have significant potential to reduce disease manifestations [25, 56, 57].
Cytokines and chemokines in TED
Cytokines and chemokines may induce the expression level of immunomodulatory proteins in orbital fibroblasts and may contribute to disease progression [58]. Cytokines are small proteins that are important in certain diseases, especially immune response, inflammation, and host response to infection. They are involved in various cell signaling including autocrine, paracrine, and endocrine signaling, known as immune-modulating agents. Interleukins (IL), chemokines, interferons (TFNs), and tumor necrosis factors (TNFs) are known as cytokines. Chemokines are small cytokines that produce various types of cells as immune cells that include four subfamilies: CXC, CC, XC3C, and XC. In TED, orbital tissue remodeling is carried out due to cytokine-dependent fibroblast activation. The literature reports that cytokines (IL-4, IL-6, IL-10, IL-13, IL-17A, IL-22, TNFA, IFNG) and chemokines (CXCL8, CXCL10) were found in extraocular muscles and fat of TED patient [59], and differential modulation of CXCL8 versus CXCL10 by cytokines [59]. In our study, some of the identified cytokines (IL10, IL17A, IL22, IL4, IL6, IL13, and IFNG) and chemokines (CXCL8, CXCL10) gene biomarkers of TED are aligned with these findings.
Conclusion
Thyroid eye disease (TED) is an autoimmune disease and hyperthyroidism where the tissue around the eye is attacked, leading to inflammation and swelling, which causes redness and pain, puffiness around the eyes, erythema, conjunctivitis, proptosis, and upper eyelid retraction. Among the several factors, smoking has a major influence on TED. Like hyperthyroidism, women are more vulnerable to TED than males with a female to male ratio of 4:1. Due to advancements in high-throughput and computational techniques, the molecular mechanism underpinning TED is gradually becoming clearer. The availability of large-scale biological data (i.e., multi-omics) offers the better discovery of biomarkers which serves as a useful noninvasive tool in the clinical armamentarium for disease studies including its diagnosis, prevention, drug target identification, designing drug for a particular receptor, and biological processes to a therapeutic intervention.
In this study, we applied a data-driven approach to detect gene biomarkers in TED, which combines biomarkers from both reported in the literature as well as we computationally predicted. Further, a regulatory pathway of biomarkers is constructed followed by various Gene Ontology-based enrichment analyses. This regulatory pathway provides an insight into the regulation mechanism in TED. Our study reports 22 gene biomarkers involved in the pathogenesis of TED that can be used as potential information for therapeutic targets. Further, we constructed a regulatory pathway of TED biomarkers consists and performed GO enrichment analysis, functional category analysis, disease enrichment, and disease class enrichment analysis, and KEGG pathway enrichment analysis using the DAVID tool. In this future work, you may perform a deeper analysis of biomarkers and constructed networks, perform molecular docking, and simulation studies against identified potential biomarkers.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the Entrez (https://www.ncbi.nlm.nih.gov/) and NCBI-GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE58331).
References
Perros P, Neoh C, Dickinson J. Thyroid eye disease. Bmj. 2009;338(mar06 1):b560. https://doi.org/10.1136/bmj.b560.
KEC. Kellogg Eye Center, Michigan Medicine, University of Michigan. URL: https://www.umkelloggeye.org/conditions-treatments/thyroid-eye-disease. Access on January 10, 2020.
Boboridis KG, Uddin J, Mikropoulos DG, Bunce C, Mangouritsas G, Voudouragkaki IC, Konstas AG. Critical appraisal on orbital decompression for thyroid eye disease: a systematic review and literature search. Adv Ther. 2015;32(7):595–611. https://doi.org/10.1007/s12325-015-0228-y.
De Groot LJ, Larsen PR, Hennemann G. Graves’ disease and the manifestations of thyrotoxicosis. Thyroid and Its Diseases; 1996. p. 371–416.
Cohen IR, Miller A. Autoimmune disease models. Academic Press; 2014.
ReportLinker. Thyroid Eye Disease Treatment Market Research Report by Drug, by Route of Administration, by Diagnosis, by Treatment, by Distribution - Global Forecast to 2025. Report Linker, Published: April 13, 2020.
MarketWatch. Thyroid Eye Disease Market projecting a CAGR of 8.53% during the forecast period, 2020-2026. MarketWatch news, Published: March 31, 2020.
Douglas RS, Gupta S. The pathophysiology of thyroid eye disease: implications for immunotherapy. Curr Opin Ophthalmol. 2011;22(5):385–90. https://doi.org/10.1097/ICU.0b013e3283499446.
McAlinden C. An overview of thyroid eye disease. Eye and Vision. 2014;1(1):9. https://doi.org/10.1186/s40662-014-0009-8.
Vestergaard P. Smoking and thyroid disorders-a meta-analysis. Eur J Endocrinol. 2002;146(2):153–61. https://doi.org/10.1530/eje.0.1460153.
Thornton J, Kelly SP, Harrison RA, Edwards R. Cigarette smoking and thyroid eye disease: a systematic review. Eye. 2007;21(9):1135–45. https://doi.org/10.1038/sj.eye.6702603.
Gharib S, Moazezi Z, Bayani MA. Prevalence and severity of ocular involvement in graves’ disease according to sex and age: a clinical study from Babol, Iran. Caspian J Intern Med. 2018;9(2):178–83. https://doi.org/10.22088/cjim.9.2.178.
Lee HJ, Li CW, Hammerstad SS, Stefan M, Tomer Y. Immunogenetics of autoimmune thyroid diseases: a comprehensive review. J Autoimmun. 2015;64:82–90. https://doi.org/10.1016/j.jaut.2015.07.009 13. Sabra MM, Di Cristofano A. 89th Annual Meeting of the American Thyroid Association October 30–November 3, 2019 Sheraton grand Chicago, Chicago.
Khong JJ, McNab AA, Ebeling PR, Craig JE, Selva D. Pathogenesis of thyroid eye disease: review and update on molecular mechanisms. Br J Ophthalmol. 2016;100(1):142–50. https://doi.org/10.1136/bjophthalmol-2015-307399.
Raef H, Ladinsky M, Arem R. Concomitant euthyroid Graves' ophthalmopathy and isolated ocular myasthenia gravis. Postgrad Med J. 1990 Oct 1;66(780):849–52. https://doi.org/10.1136/pgmj.66.780.849.
Bothun ED, Scheurer RA, Harrison AR, Lee MS. Update on thyroid eye disease and management. Clin Ophthalmol. 2009;3:543–51. https://doi.org/10.2147/opth.s5228.
Lehmann GM, Garcia-Bates TM, Smith TJ, Feldon SE, Phipps RP. Regulation of lymphocyte function by PPARγ: relevance to thyroid eye disease-related inflammation. PPAR Res. 2008;2008:1–12. https://doi.org/10.1155/2008/895901.
Douglas RS. Teprotumumab, an insulin-like growth factor-1 receptor antagonist antibody, in the treatment of active thyroid eye disease: a focus on proptosis. Eye (Lond). 2019;33(2):183–98. https://doi.org/10.1038/s41433-018-0321-y.
Abraham P, Acharya S. Current and emerging treatment options for graves’ hyperthyroidism. Ther Clin Risk Manag. 2010;6:29.
Gillespie EF, Smith TJ, Douglas RS. Thyroid eye disease: towards an evidence base for treatment in the 21st century. Curr Neurol Neurosci Rep. 2012;12(3):318–24. https://doi.org/10.1007/s11910-012-0256-9.
Verity DH, Rose GE. Acute thyroid eye disease (TED): principles of medical and surgical management. Eye (Lond). 2013;27(3):308–19. https://doi.org/10.1038/eye.2012.284.
NIH-NIDDKD. National Institute of Health – National Institute of Diabetes and Digestive and Kidney Diseases. URL: https://www.niddk.nih.gov/health-information/endocrine-diseases/graves-disease, accessed on January 28, 2020.
Barrio-Barrio J, Sabater AL, Bonet-Farriol E, Velázquez-Villoria Á, Galofré JC. Graves’ ophthalmopathy: VISA versus EUGOGO classification, assessment, and management. J Ophthalmol. 2015 Jul;2015:1–16. https://doi.org/10.1155/2015/249125.
Ismailova DS, Belovalova IM, Grusha YO, Sviridenko NY. Orbital decompression in the system of treatment for complicated thyroid eye disease: case report and literature review. Int Med Case Rep J. 2018;11:243–9. https://doi.org/10.2147/IMCRJ.S164372.
Patel A, Yang H, Douglas RS. A new era in the treatment of thyroid eye disease. Am J Ophthalmol. 2019;208:281–8. https://doi.org/10.1016/j.ajo.2019.07.021.
Mayeux R. Biomarkers: potential uses and limitations. NeuroRx. 2004;1(2):182–8. https://doi.org/10.1602/neurorx.1.2.182.
Nicolì F, Lanzolla G, Mantuano M, et al. Correlation between serum anti-TSH receptor autoantibodies (TRAbs) and the clinical feature of Graves' orbitopathy. J Endocrinol Invest. 2021;44(3):581–5. https://doi.org/10.1007/s40618-020-01353-y.
Tu Y, Fan G, Zeng T, Cai X, Kong W. Association between polymorphism within interleukin related genes and graves' disease: a meta-analysis of 22 case-control studies. Oncotarget. 2017;8(58):98993–9002. https://doi.org/10.18632/oncotarget.20114.
Krassas GE, Pontikides N, Doukidis D, Heufelder G, Heufelder AE. Serum levels of tumor necrosis factor-α, soluble intercellular adhesion molecule-1, soluble vascular cell adhesion molecule-1, and soluble interleukin-1 receptor antagonist in patients with thyroid eye disease undergoing treatment with somatostatin analogues. Thyroid. 2001;11(12):1115–22. https://doi.org/10.1089/10507250152740957.
Xu W, Hiếu T, Malarkannan S, Wang L. The structure, expression, and multifaceted role of immune-checkpoint protein VISTA as a critical regulator of anti-tumor immunity, autoimmunity, and inflammation. Cell Mol Immunol. 2018;15(5):438–46. https://doi.org/10.1038/cmi.2017.148.
Xiaoheng C, Yizhou M, Bei H, Huilong L, Xin W, Rui H, Lu L, Zhiguo D. General and specific genetic polymorphism of cytokines-related gene in AITD. Mediat Inflamm. 2017;2017:1–8. https://doi.org/10.1155/2017/3916395.
Turck N, Eperon S, De Los Angeles Gracia M, Obéric A, Hamédani M. Thyroid-associated orbitopathy and biomarkers: where we are and what we can hope for the future. Dis Marker. 2018;2018.
Diana T, Kahaly GJ. Thyroid Stimulating Hormone Receptor Antibodies in Thyroid Eye Disease-Methodology and Clinical Applications. Ophthalmic Plast Reconstr Surg. 2018;34(4S Suppl 1):S13–9.
Longo CM, Higgins PJ. Molecular biomarkers of Graves' ophthalmopathy. Exp Mol Pathol. 2019;106:1–6. https://doi.org/10.1016/j.yexmp.2018.11.004.
Wong KH, Rong SS, Chong KK, Young AL, Pang CP, Chen LJ. Genetic associations of interleukin-related genes with graves’ ophthalmopathy: a systematic review and meta-analysis. Sci Rep. 2015;5(1):16672. https://doi.org/10.1038/srep16672.
Hagan S, Martin E, Enríquez-de-Salamanca A. Tear fluid biomarkers in ocular and systemic disease: potential use for predictive, preventive and personalised medicine. EPMA J. 2016;7(1):15. https://doi.org/10.1186/s13167-016-0065-3.
Clarke F, Purvis HA, Sanchez-Blanco C, Gutiérrez-Martinez E, Cornish GH, Zamoyska R, Guermonprez P, Cope AP. The protein tyrosine phosphatase PTPN22 negatively regulates presentation of immune complex derived antigens. Sci Rep. 2018;8(1):1–1.
Khalilzadeh O, Noshad S, Rashidi A, Amirzargar A. Graves' ophthalmopathy: a review of immunogenetics. Current Genom. 2011;12(8):564–75. https://doi.org/10.2174/138920211798120844.
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acid Res. 2016;gkw937.
Zuberi K, Franz M, Rodriguez H, Montojo J, Lopes CT, Bader GD, Morris Q. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 2013;41(W1):W115–22. https://doi.org/10.1093/nar/gkt533.
Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, Lee S, Kang B, Jeong D, Kim Y, Jeon HN, Jung H, Nam S, Chung M, Kim JH, Lee S. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46(D1):D380–6. https://doi.org/10.1093/nar/gkx1013.
Ogata H, Goto S, Fujibuchi W, Kanehisa M. Computation with the KEGG pathway database. Biosystems. 1998;47(1–2):119–28. https://doi.org/10.1016/S0303-2647(98)00017-3.
Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44.
Raza K, Parveen R. Reconstruction of gene regulatory network of Colon Cancer using information theoretic approach. In: Proc. of 4th International Conference (CONFLUENCE-2013): The Next Generation Information Technology Summit 2013; 2013. p. 461–6. https://doi.org/10.1049/cp.2013.2357.
Raza K. Reconstruction, topological and gene ontology enrichment analysis of cancerous gene regulatory network modules. Curr Bioinforma. 2016;11(2):243–58. https://doi.org/10.2174/1574893611666160115212806.
Raza K. Analysis of microarray data using artificial intelligence based techniques. Handbook of Research on Computational Intelligence Applications in Bioinformatics. USA: IGI Global; 2016. p. 216–39. https://doi.org/10.4018/978-1-5225-0427-6.ch011.
Jabeen A, Ahmad N, Raza K. Differential expression analysis of ZIKV infected human RNA sequence reveals potential genetic biomarkers. Lecture Notes in Bioinformatics, Springer, vol. 11465; 2019. p. 1–12. https://doi.org/10.1007/978-3-030-17938-0_26.
Wang Y, Smith TJ. Current concepts in the molecular pathogenesis of thyroid-associated ophthalmopathy. Invest Ophthalmol Vis Sci. 2014;55(3):1735–48. https://doi.org/10.1167/iovs.14-14002.
Chen B, Tsui S, Smith TJ. IL-1 beta induces IL-6 expression in human orbital fibroblasts: identification of an anatomic-site specific phenotypic attribute relevant to thyroid-associated ophthalmopathy. J Immunol. 2005;175:1310–9.
Weightman DR, Perros P, Sherif IH, Kendall-Taylor P. Autoantibodies to IGF-1 binding sites in thyroid associated ophthalmopathy. Autoimmunity. 1993;16(4):251–7.
Krieger CC, Place RF, Bevilacqua C, Marcus-Samuels B, Abel BS, Skarulis MC, Kahaly GJ, Neumann S, Gershengorn MC. TSH/IGF-1 receptor cross talk in Graves' ophthalmopathy pathogenesis. J Clin Endocrinol Metabol. 2016;101(6):2340–7. https://doi.org/10.1210/jc.2016-1315.
Krieger CC, Boutin A, Perry J, et al. β-Arrestin 1 dependent TSH/IGF1 receptor crosstalk in graves’ orbital fibroblasts. Thyroid. 2018;28(suppl 1):A-6.
Mohyi M, Smith TJ. 40 YEARS OF IGF1: IGF1 receptor and thyroid-associated ophthalmopathy. J Mol Endocrinol. 2018;61(1):T29–43. https://doi.org/10.1530/JME-17-0276.
Smith TJ, Hegedüs L, Douglas RS. Role of insulin-like growth factor-1 (IGF-1) pathway in the pathogenesis of Graves' orbitopathy. Best Pract Res Clin Endocrinol Metab. 2012;26(3):291–302. https://doi.org/10.1016/j.beem.2011.10.002.
Wiersinga WM. Autoimmunity in Graves' ophthalmopathy: the result of an unfortunate marriage between TSH receptors and IGF-1 receptors? J Clin Endocrinol Metabol. 2011;96(8):2386–94. https://doi.org/10.1210/jc.2011-0307.
Chen H, Mester T, Raychaudhuri N, Kauh CY, Gupta S, Smith TJ, Douglas RS. Teprotumumab, an IGF-1R blocking monoclonal antibody inhibits TSH and IGF-1 action in fibrocytes. J Clin Endocrinol Metab. 2014;99(9):E1635–40. https://doi.org/10.1210/jc.2014-1580.
Douglas RS, Sile S, Thompson EH. Teprotumumab treatment effect on proptosis in patients with active thyroid eye disease: results from a phase 3, randomized, double-masked, placebo-controlled, parallel-group, multicenter study. Proceedings of the American Association of clinical endocrinologists. Endocr Pract. 2019;25.
Prabhakar BS, Bahn RS, Smith TJ. Current perspective on the pathogenesis of graves’ disease and ophthalmopathy. Endocr Rev. 2003 Dec 1;24(6):802–35. https://doi.org/10.1210/er.2002-0020.
Hiromatsu Y, Yang D, Bednarczuk T, Miyake I, Nonaka K, Inoue Y. Cytokine profiles in eye muscle tissue and orbital fat tissue from patients with thyroid-associated ophthalmopathy. J Clin Endocrinol Metabol. 2000 Mar 1;85(3):1194–9. https://doi.org/10.1210/jcem.85.3.6433.
Acknowledgements
N.A
Funding
N.A.
Author information
Authors and Affiliations
Contributions
HZ and ZW conceived the idea, WX and YW collected the data, HZ, WX, YW and ZW performed the experiment, and wrote paper. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This research work does not involve direct human participation. Hence, ethical approval and consent to participate is not application.
Consent for publication
Not applicable.
Competing interests
Authors declare that there is any competing interest in the publication of this manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zou, H., Xu, W., Wang, Y. et al. A data-driven approach for the discovery of biomarkers associated with thyroid eye disease. BMC Ophthalmol 21, 166 (2021). https://doi.org/10.1186/s12886-021-01903-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12886-021-01903-9