Abstract
Background
Osteosarcoma (OS) is one of the most common aggressive bone malignancy tumors in adolescents. With the application of new chemotherapy regimens, finding new and effective anti-OS drugs to coordinate program implementation is urgent for the patients of OS. Oridonin had been proved to mediate anti-tumor effect on OS cells, but its mechanism has not been fully elucidated.
Methods
The effects of oridonin on the viability, clonal formation and migration of 143B and U2OS cells were detected by CCK-8, colony formation assays and wound-healing test. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was used to explore the mechanism of oridonin on OS. Western blot (WB), real-time quantitative PCR (qRT-PCR) were used to detect the expression levels of apoptosis and ferroptosis-relative proteins and genes. Annexin V-FITC apoptosis detection kit and flow cytometry examination were used to detect the level of apoptosis. Iron assay kit was used to evaluate the relative Fe2+ content. The levels of mitochondrial membrane potential and lipid peroxidation production was determined by mitochondrial membrane potential detection kit and ROS assay kit.
Results
Oridonin could effectively inhibit the survival, clonal formation and metastasis of OS cells. The KEGG results indicated that oridonin is associated with the malignant phenotypic signaling pathways of proliferation, migration, and drug resistance in OS. Oridonin was capable of inhibiting expressions of BAX, cl-caspase3, SLC7A11, GPX4 and FTH1 proteins and mRNA, while promoting the expressions of Bcl-2 and ACSL4 in 143B and U2OS cells. Additionally, we found that oridonin could promote the accumulation of reactive oxygen species (ROS) and Fe2+ in OS cells, as well as reduce mitochondrial membrane potential, and these effects could be significantly reversed by the ferroptosis inhibitor ferrostatin-1 (Fer-1).
Conclusion
Oridonin can trigger apoptosis and ferroptosis collaboratively in OS cells, making it a promising and effective agent for OS therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteosarcoma (OS), a common bone malignancy in adolescents, has a high mortality and disability rates [1]. Surgical resection was a primary method for treatment of OS in the past, and the use of neoadjuvant chemotherapy has raised the survival rate to 60–70% [2]. However, in recent years, chemotherapy causes great physical and psychological suffering to patients [3]. Meanwhile, the emergence of multi-drug resistance poses a serious challenge to the chemotherapy regimen [4, 5], targeting drug resistance-related mechanisms may be one of the keys to OS therapy [6]. Therefore, solving the problem of chemotherapeutic drug resistance is crucial for the treatment of OS.
With the advantages of multi-target and multi-pathway treatment increasing [7], the traditional Chinese medicine can slow the growth of cancers, reduce drug resistance and lessen the negative effects from radiotherapy and chemotherapy significantly [8], which had become one of the key directions of anti-tumor drug research. Rabdosia rubescens, a traditional Chinese herbal medicine with the functions of clearing heat, detoxifying, activating blood circulation and anti-cancer, which had been used in the treatment of chronic bronchitis, chronic hepatitis, esophageal cancer, liver cancer, breast cancer, and other diseases [9,10,11]. Oridonin, the primary anticancer component in Rabdosia rubescens, was demonstrated to inhibit the proliferation and metastasis of OS while increasing the sensitivity of OS to doxorubicin, making it a promising drug for clinical treatment of OS [12,13,14,15]. However, the specific mechanism of inhibiting OS resistance is still unclear.
Studies have shown that programmed cell death (PCD) is closely related to chemotherapy resistance of cancers [16]. Apoptosis is the main kind of PCD, and tumor development, metastasis and drug resistance of OS cells were all closely related to apoptosis as reported [17, 18]. Inhibition of apoptosis, one of the classic markers of cancer, is related to drug resistance and may be an important therapeutic target to overcome the drug resistance [19]. Ferroptosis, a new form of PCD [20], characterized by hazardous lipid peroxidation and mitochondrial dysfunction [21], had been proved closely related to inhibition of cancer cells proliferation, migration, and invasion [22] Studies have shown that ferroptosis can affect the efficacy of cancer treatment and reverse the resistance to chemotherapy, targeted therapy, and immunotherapy by regulating GPX4 signaling pathways, iron metabolism, and lipid metabolism [16, 23, 24]. Activating ferroptosis to fight against cancer was a treatment with high safety and selectivity [25]. Meanwhile, there was correlation between apoptosis and ferroptosis [26, 27]. Multiple studies had shown that simultaneous targeting of apoptosis and ferroptosis can effectively bypassing chemoresistance, which was an effective strategy to inhibit OS cells [28,29,30,31]. Therefore, we hypothesize that oridonin can cause apoptosis and ferroptosis simultaneously, thereby enhancing its inhibitory effects on osteosarcoma proliferation, metastasis, and drug resistance.
In this study, we would explore the inhibitory effect of oridonin on the proliferation and migration of OS cells, as well as its promoting effect on apoptosis and iron death of OS cells, so as to provide a new idea for the treatment of OS.
Materials and methods
Cell culture
Human OS cell lines including 143B, U2OS were preserved by molecular biology laboratory, Henan Luoyang Orthopedic Hospital (Henan Provincial Orthopedic Hospital), (Henan, China). 143B and U2OS cells were cultured in McCoy’s 5A medium (#C3020–0500, VivaCell) which supplemented with 10% fetal bovine serum (#10270–106, Gibco) and 1% penicillin/streptomycin (#C3420–0100, VivaCell), and cultured at 37 °C in a humidified atmosphere with 5% CO2.
Reagents and antibodies
Oridonin (HPLC ≥98%) was purchased from Shanghai yuanye Bio-Technology Co., Ltd. (#B20310), then the drug was dissolved in DMSO and stored in − 20 °C for usage. Ferrostatin-1 (Fer-1) was purchased from MedChemExpress (#HY-100579, MCE), and stored in − 20 °C for usage. All primary antibody used in the experiments included Actin (1:10000, #60008–1-Ig, Proteintech), Bcl-2 (1:1000, #61193, Proteintech), BAX (1:5000, #50599–2-Ig, Proteintech), cl-caspase3 (1:10000, #19677–1-AP, Proteintech), Tublin (1:1000, #66031–1-Ig, Proteintech), GAPDH (1:2000, #CL594–60004, Proteintech), GPX4 (1:5000, #67763–1-IG, Proteintech), SLC7A11 (1:1000, #26864–1-AP, Proteintech), FTH1 (1:5000, #11682–1-AP, Proteintech), and ACSL4 (1:10000, #81196–1-RR, Proteintech), they were stored in − 20 °C for usage. Ferrostatin-1 (Fer-1) (#HY-100579, MCE), an effective ferroptosis inhibitor [32], was dissolved in dimethylsulfoxide (DMSO) to create stock solutions and stored at − 20 °C.
Cell viability assay
143B and U2OS cells were exposed to oridonin (0, 5, 10, 20 μM) for 24 h and 48 h in 96-well plates with a density of 7000 cells per well. Cell proliferation was detected by CCK-8 (#C0038, Beyotime) assays according to the manufactures instructions. The optical density (OD) value was detected by a microplate reader (#Epoch, BioTek). The IC50 value was calculated by non-linear regression fitted via GraphPad Prism (Version 9) software.
Plate colony formation assays
143B and U2OS cells were exposed to oridonin (0, 5, 10 μM) for 10 days in 6-well plates with a density of 400 cells per well. When 143B and U2OS cells formed sufficiently large colonies, they were fixed with 4% paraformaldehyde (#BL539A, Biosharp) for 20 min at RT and stained with 0.5% crystal violet solution (#C8470, Solarbio) incubate for 2 min at RT. The colony numbers were counted under the microscope (#IX73, OLYMPUS).
Wound-healing test
143B and U2OS cells were exposed to oridonin (0, 5, 10 μM) for 10 days in 6-well plates at a density of 5 × 104. Each well was scraped a scratch with a 20 μL pipette tip vertically when the cells reached 100% confluence, then the plates were washed with PBS three times and changed to fresh serum-free medium. Microscope (#IX73, OLYMPUS) was used to image the cells in the same vision at hours 0, 12 and 24 h to observe and photograph the changes of scratch width. The migration rate was calculated by a formula: (scratch distance (0 h) – scratch distance (12 h / 24 h)) scratch distance (0 h).
Kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis
Potential targets of oridonin through DRUGBANK (https://go.drugbank.com/) database were searched, and 487 targets were obtained after deleting duplicates. 1263 correlated targets of OS were collected and filtered from Genecards (https://www.genecards.org/) database, OMIM (https://www.omim.org/) database, TTD (https://db.idrblab.net/ttd/) database, DisGENET (https://www.disgenet.org/) database. Intersection 115 targets of the two were imported into the DAVID (https://david.ncifcrf.gov/) database for KEGG enrichment analysis, resulting in 162 enrichment findings. Among these, 20 relevant signaling pathways were selected and displayed through the Microbiology Information Online platform (http://www.bioinformatics.com.cn/).
Flow cytometry examination
143B and U2OS cells were cultured in 6-well plates for 24 h, after which they were divided into control and oridonin groups (10 μM and 20 μM), and then continued to be cultured for another 24 h before collecting the cells. The positive control group cells were incubated on ice with an Apoptosis Positive Control Solution (500 μL) for 30 minutes, followed by one wash with PBS. All groups were resuspended in 1 × Binding Buffer. The negative control group was divided into four tubes, three of which were respectively added with Annexin V-FITC (5 μL), propidium iodide (PI) (10 μL), or both, and incubated at RT in the dark for 5 minutes. The positive control and oridonin groups were simultaneously added with Annexin V-FITC (5 μL) and propidium iodide (PI) (10 μL), then incubated at room temperature in the dark for 5 minutes. Subsequently, cells from each group were subjected to apoptosis analysis using a FACScan flow cytometer (#FACSAria II, BD).
Western blot analysis (WB)
143B and U2OS cells were exposed to oridonin (0, 5, 10 μM) for 24 h in 6-well plates cells. The cultures were discarded medium and washed with 1 × PBS (#G4202, Servicebio) three times. The 143B and U2OS cells were collected and mixed with cell lysates 80 uL (#P0013B, Beyotime) incubated for 1 h on ice, then sonicated and centrifuged (30 min, 14,000 g, 4 °C). BCA method was performed for the whole protein quantification of the cells by BCA kit (#PC0020, Solarbio). Via sodium dodecyl sulfate polyacrylamide gel electrophoresis and transmembrane, the 143B and U2OS cells’ protein was transferred onto polyvinylidene difluoride membranes. The membranes were incubated with the indicated antibodies prior to detection using enhanced chemiluminescent reagent. Then the membranes were blocked with 5% non-fat milk for 1 h at RT and incubated with the primary antibody overnight at 4 °C. After incubating the secondary antibody (anti-rabbit/mouse IgG HRP) for 1 h and wash it 10 min for three times at RT, the membranes were visualized by WB exposure machine (Amersham Imager 680/Tanon 4800 Multi) after treatment of ECL chemiluminescence kit (#P0018FS, Beyotime).
Real-time quantitative PCR (qRT-PCR)
Total RNA was extracted by RNAzol reagent (#15596018, ThermoFisher) and reversed to cDNA (Takara). The qRT-PCR examination was performed by the Fluorescent quantitative PCR instrument (#CFX96 deep well, Biorad) using a SYBR Select Master Mix kit (#4472908, Life Technology) according to the instructions. The relative expressions of the target genes were normalized against GAPDH and analyzed using the 2−△△Ct method [33]. Supplementary Table 1 displays the detailed sequences of primers.
Assessment of Fe2+ content
143B and U2OS cells were exposed to oridonin (0, 5, 10 μM) for 24 h in 15 dish plates, after which the Iron assay Kit (#BC5415, Solarbio) was used to evaluate the relative Fe2+ content directed by the manufacturer. The value of OD in 593 nm was measured on microplate reader (#Epoch, BioTek). The standard curve was drawn against the standard samples, then the relative Fe2+ level was calculated.
Measurement of ROS
After 143B and U2OS cells adhered, exposed them to the medium containing oridonin (0, 5 μM and 10 μM) Serum-free McCoy’s 5A for 24 h in 6-well plates Cells. ROS assay kit (#R6033, UElandy) was used to test the level of ROS which directed by the manufacturer. Before treating all cells with 1 mL the ROS fluorescent probe-2, 7-dichlorofuorescin diacetate (DCFH-DA, 10 μM) for 30 min at 37 °C avoid light, the positive control group was first treated with 1 mL ROS positive inducer-ROSUP (100 μM). Then 143B and U2OS cells were washed twice with serum-free cell culture medium, then viewed them under fluorescence microscope (#BX53, OLYMPUS).
Measurement of mitochondrial membrane potential
Mitochondrial membrane potential detection kit (JC-1) (#J6004S, UElandy) was used to determine the mitochondrial membrane potential. Hoechest33342 (#H4047, UElandy) was used to tag the DNA of living cells. When 143B and U2OS cells adhered, exposed them to oridonin (0, 5 μM and 10 μM) for 24 h in 6-well plate. After removing the media, 143B and U2OS cells were washed with PBS once. 500 μL serum-free cell culture medium, 500 μL JC-1 solution and 500 μL Hoechest33342 were added to each well, the positive control group was additionally treated with 500 μL CCCP (50 μM), then mixing them and incubated for 20 min. The 143B and U2OS cells were washed twice by JC-1 dye buffer after incubation, then added 2 ml cell culture medium to it. Cells were observed under fluorescence microscope (#BX53, OLYMPUS).
Statistical analysis
Image J software was used to analysis the results of WB. GraphPad Prism (Version 9) was operated to evaluate all data, which were shown as the mean ± SD. Two-sample t test was used for independent samples and one-way analysis of variance (ANOVA) was used for multiple samples. The differences were considered statistically significant with a significant level (p) less than 0.05.
Results
Oridonin inhibited the proliferation and migration of OS cells
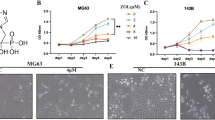
To discover the anti-cancer effects of oridonin on cell viability of OS cells, 143B and U2OS cells were treated at indicated time points (24 or 48 h) with various concentration of oridonin (0, 5 μM, 10 μM and 20 μM). Our results indicated that oridonin exhibited significant cytotoxic effects to OS cells, the cell viability of 143B and U2OS cells was inhibited in a dose-dependent manner compared to the untreated group (Fig. 1A, B). The half maximal inhibitory concentrations (IC50) of oridonin on these cells at 24 h were 9.716 μM and 10.68 μM respectively. We used 143B and U2OS cells with of oridonin (5 and 10 μM) treatment in the following experiments. The result of plate colony formation assays (Fig. 1C, D) and wound healing test (Fig. 1E, F) demonstrated that the ability of OS cells proliferation and migration were controlled by oridonin. In conclusion, these results suggested that oridonin may dose-dependently attenuated the proliferation and metastasis of OS cells.
Oridonin inhibits OS cells viability. A-B 143B and U2OS cells lines were treated with oridonin (0, 5, 10 and 20 μM) for 24 and 48 h. Cell viability was measured by CCK-8 assays. (n = 5, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, vs. normal). C-D 143B and U2OS cells treated with oridonin (0, 5, 10 μM). Colony formation was evaluated by colony formation assays. (n = 3, **** p < 0.0001, vs. normal). E-F 143B and U2OS cells lines were treated with oridonin. (0, 5, 10 μM) for 0 h, 12 h and 48 h. Cells migration was evaluated by wound-healing tests (4X). (n = 3, *** p < 0.001, **** p < 0.0001, vs. normal)
Oridonin induced apoptosis in OS cells
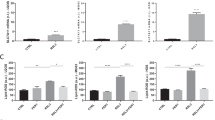
Based on the results of the KEGG enrichment analysis (Fig. 2A), we discovered that oridonin not only affects the signal pathways related to proliferation and metastasis of OS cells, but also participates in the tumor immunity and drug resistance of OS cells, such as EGFR tyrosine kinase inhibitor resistance, platinum drug resistance, T cell receptor signaling pathway and anti-programmed cell death 1 (PD-L1) expression and programmed death-ligand 1 (PD-1) checkpoint pathway in cancer. To further explore the possible mechanism of oridonin’s involvement in tumor suppression and drug resistance, we first examined the effect of oridonin on apoptosis of 143B and U2OS cells. 143B and U2OS cells were subjected to oridonin (0, 5 μM and 10 μM) for 24 h before monitoring the markers associated with apoptosis. The results of WB and qRT-PCR demonstrated that pro-apoptotic Bax and cleaved caspase-3 were increased, while anti-apoptotic Bcl-2 expression was clearly inhibited (Fig. 2B, C). The results of apoptosis investigations using flow cytometry revealed that when 143B and U2OS cells were exposed to oridonin (0, 10 μM and 20 μM) for 24 h, the percentage of apoptotic cells rose in a concentration-dependent way (Fig. 2D-G). In general, our data demonstrated that oridonin increased apoptosis in OS cells.
Oridonin induced apoptosis in OS cells. A The enrichment analysis of KEGG pathways resulted in 162 pathways (p value < 0.05), select 20 of them to draw the signal path bar chart. B-C WB and qRT-PCR was used to analysis the expression levels of Bcl-2, Bax and cleaved caspase3 in 143B and U2OS cells exposed to oridonin (0–5-10 μM) for 24 h, Tublin and GAPDH as reference (n = 3, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, vs. normal). D-G The 143B and U2OS cells were treated with oridonin (0–10-20 μM) for 24 h, detected apoptotic cells by Annexin V-FITC and PI double staining and quantified (n = 3, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, vs. normal)
Oridonin triggered the ferroptosis in OS cells
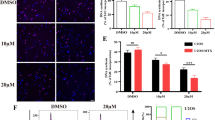
Studies have shown that ferroptosis is related to tumor inhibition, T cell immunity and PD-L1/ PD-1 treatment resistance, etc. [34, 35]. Therefore, the study further explored the effect of oridonin on ferroptosis of OS cells. The SLC7A11, GPX4, FTH1 and ACSL4 were all strongly associated with the occurrence of ferroptosis, and we therefore examined their expression in OS cells treated with oridonin (0, 5 μM and 10 μM) for 24 h. The results of WB and qRT-PCR revealed that oridonin could dramatically downregulate the expression of SLC7A11, GPX4, and FTH1 expression, while upregulate the expression of ACSL4 (Fig. 3A-C). Ferrous ion colorimetry was used to determine the intracellular iron. Compared with the untreated group, Fe2+ level was increased in oridonin-treated groups (5 μM and 10 μM) with dose-dependent performance (Fig. 3D). In addition, we observed that the green fluorescence obviously increased in a dose-dependent approach, illustrating that oridonin increased the ROS accumulation (Fig. 3E, F) and reduced the mitochondrial membrane potential (Fig. 3G, H) in these two OS cells. These results supported the fact that oridonin might induced the ferroptosis in OS cells.
Oridonin could triggered the ferroptosis in OS cells. A-C The expressions of ACSL4, SLC7A11, FTH1 and GPX4 in oridonin treated U2OS and 143B cells analyzed by WB and qRT-PCR, Actin and GAPDH as reference. (n = 3, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, vs. normal). D The accumulation of Fe2+ was detected in the oridonin treated cells by a microplate reader. (*p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, vs. normal). E-F ROS were detected by DCFH-DA probe in the oridonin treated cells, the accumulation of ROS was illustrated by green fluorescence (40X). Cells treated with ROSUP were used as positive control (*p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, vs. normal). G-H The mitochondrial membrane potential detected by JC-1 probe, dye Hoechest33342 was used for nuclear localization (100X). Cells treated with CCCP were used as positive control (*p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, vs. normal)
Oridonin promoted OS cells death may in a ferroptosis-dependent manner
To demonstrate the role of ferroptosis in the anti-OS activity of oridonin, 143B and U2OS cells were pretreated with Fer-1 (1 μM) to suppress the ferroptosis generated by oridonin in OS cells. The upregulation of FTH1 and GPX4 relevant to oridonin were rescued by Fer-1 (Fig. 4A-C). Fer-1 could relieve the oridonin-mediated cytotoxicity, increased the IC50 value of oridonin act on 143B and U2OS cells to 12.15 μM and 14.02 μM, respectively (Fig. 4D-G). Meanwhile, Fer-1 inhibited oridonin-induced intracellular ROS accumulation (Fig. 4H, I) and reversed the mitochondrial membrane potential suppression (Fig. 4J, K). Collectively, the results presented above suggested that oridonin-triggered ferroptosis might be one of the primary mechanisms by which oridonin suppressesd the growth of OS cells.
Fer-1 rescued the anti-tumor effects of oridonin in OS cells. A-C The expressions of ferroptosis markers FTH1 and GPX4 in oridonin-treated 143B and U2OS cells were increased after added Fer-1. Actin as reference. (n = 3, * p < 0.05, ** p < 0.01, *** p < 0.001, ****p < 0.0001, vs. normal). D-G The 143B and U2OS cells were co-treated with oridonin and Fer-1, and the cell viability was examined by CCK-8 assays. (n = 3, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, vs. normal). H-K Compared the intracellular contents of ROS (20X) (H-I) in the oridonin and co-treated OS cells, meanwhile, observed the change of mitochondrial membrane potential (40X) (J-K). Cells treated with ROSUP and CCCP were used as positive control (n = 3, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, vs. normal)
Discussion
OS is one of the most common aggressive bone malignancy tumors in the adolescence, with a significant mortality and disability rate. Despite advances in conventional and targeted therapies, the survival rate of OS has not improved significantly in last ten years [36]. Each year, approximately 2–3 million people died from OS on a global scale [2], the high metastatic potential and therapy resistance of OS cited as the primary causes [37]. Therefore, in this study, we focused on finding a drug which can simultaneously inhibit the proliferation, metastasis and drug resistance of OS.
Previous studies have demonstrated that the traditional Chinese medicine treatment has the advantage of being multi-target and multi-pathway, which has distinct advantages in drug resistance and tumor metastasis inhibition [38]. Oridonin is derived from the traditional Chinese herb Rabdosia rubescens and had been shown to have a wide variety of anticancer effects and was regarded a promising antitumor agent [39,40,41]. Network pharmacology had a certain usefulness in tumor therapy mining [42, 43]. In the study, network pharmacology was applied to explore the potential pathways of oridonin and the results have showed that oridonin was associated with multiple phenotypes and pathways associated with proliferation and metastasis of OS. Then we further searched the relevant literature and found that the previous studies have fully investigated the mechanism of oridonin in inhibiting the proliferation and metastasis of OS. Yang et al. found that oridonin inhibited 143B cells proliferation via upregulating the Dkk-1 expression and/or enhancing the function of GSK3β to downregulate Wnt/β-catenin signal transduction [15]. Yang et al. found that oridonin inhibited TGF-β-induced phosphorylation of Smad 2/3 and prevented Smad dimer translocation into the nucleus to inhibit the metastasis of OS cells [44]. In addition, Song et al. and Xiao-Hui et al. also found oridonin can induce apoptosis through Akt and MAPKs signaling pathways and inhibit the progression of OS [45, 46]. We next further confirmed the inhibition of oridonin on OS cells by cytotoxicity, plate colony formation assays, wound-healing tests and detection of the apoptosis-related indexes in the study.
Except that, the results of KEGG enrichment analysis displayed that oridonin was associated with EGFR tyrosine kinase inhibitor resistance, platinum drug resistance, T cell receptor signaling pathway and (anti-programmed cell death 1) PD-L1 expression and (programmed death-ligand 1) PD-1 checkpoint pathway in cancer and other cancer chemotherapy-related pathways in OS cells, which suggested that oridonin might be involved in the drug resistance of OS. Studies had found that the effect of oridonin on drug resistance in OS cells might related to the synergistic pro-apoptotic effect of oridonin and these chemotherapy drugs [12, 47, 48]. Liliya et al. have illustrated that oridonin could promote apoptosis, enhance the chemotherapy effect of doxorubicin in OS and reduce the clinical dosage of doxorubicin significantly. Given that the etiology of OS is complex, where multiple cell death mechanisms involved in OS’s pathological process [49]. It appears likely that the effective OS inhibitors should target multiple cell death processes simultaneously, which could also help address the problem of drug resistance [50]. In addition to apoptosis, there might exist other mechanisms for oridonin to reverse drug resistance in OS cells, which need to be further explored.
In addition, as a new type of PCD, ferroptosis had manifested as an imbalance of iron homeostasis, accumulation of ROS, mitochondrial malfunction, and so on, which plays a key role in tumor inhibition and reversal of chemotherapy, targeted therapy resistance and immunotherapy resistance [16, 23]. For example, inhibition of STAT3 could regulate GPX4, SLC7A11 and FTHI to induce ferroptosis, which can restore the sensitivity of gastric cancer cells to chemotherapy drugs [50]. Tuo et al. found that ferroptosis could overcome the resistance to EGFR-tyrosine kinase inhibitors [51]. Meanwhile, overcoming the immune checkpoint inhibitors resistance remained a great challenge in the treatment of cancers. Studies have shown that ferroptosis was involved in T cell immunity and sensitized resistant tumors to anti-PD-1 therapy [34, 35]. However, it is still unclear about whether ferroptosis was involved in the anti-OS activity of oridonin. In this study, the results showed that oridonin could reduce the expression of SLC7A11, GPX4, and FTH1, while increasing the expression of ACSL4. Furthermore, compared with the control group, oridonin could promote the production of ROS, increase Fe2+ level and reduce mitochondrial membrane. In addition, ferroptosis inhibitor Fer-1 could reverse the drop in ferroptosis-related indices and significantly alleviate the toxic effect on OS cells caused by oridonin. To sum up, the study was first to demonstrate that oridonin inhibits OS in a ferroptosis-depend way.
Ferroptosis and apoptosis are two types of programmed cell death, involved in ferroptosis and apoptosis were extremely essential to the growth of different tumors, which also conferred resistance to anticancer drugs [52,53,54]. Targeting ferroptosis and apoptosis pathways were potential therapeutics for cancer, many medicines had been reported to play anti-cancer activities via the combination routes [55]. According to the study, oridonin exhibits an anti-OS activity might via a combination of ferroptosis and apoptosis, which might be an effective approach to inhibit the development and alleviate drug resistance in OS cells.
However, there are still some limitations in this study. Firstly, drug-resistant OS cell lines have not been established in the study. Secondly, the effects of oridonin on apoptosis and ferroptosis of drug-resistant OS cell lines have not been explored. This will be the focus of our next research direction.
Conclusion
In conclusion, the study confirmed that oridonin could inhibit the apoptosis and ferroptosis pathway of OS cells to restrict the growth and metastasis of OS which provided a novel strategy for coordinating the action of several cell death modes in the treatment of OS. Meanwhile, it provided a solid fundation for promoting the application of oridonin in clinical treatment as an antitumor drug and adjuvant chemotherapy drug.
Availability of data and materials
No datasets were generated or analysed during the current study.
Abbreviations
- OS:
-
Osteosarcoma
- PCD:
-
Programmed cell death
- Fer-1:
-
Ferrostatin-1
- BCL-2:
-
B-cell lymphoma-2
- BAX:
-
BCL2-Associated X
- cl-caspase3:
-
Cleaved Caspase-3
- SLC7A11:
-
Solute Carrier Family 7 Member 11
- GPX4:
-
Glutathione peroxidase 4
- FTH1:
-
Ferritin heavy chain 1
- ROS:
-
Reactive Oxygen Species
- PD-L1:
-
Anti-programmed cell death 1
- PD-1:
-
Programmed death-ligand 1
References
Andersen GB, Knudsen A, Hager H, Hansen LL, Tost J. miRNA profiling identifies deregulated miRNAs associated with osteosarcoma development and time to metastasis in two large cohorts. Mol Oncol. 2018;12(1):114–31. https://doi.org/10.1002/1878-0261.12154.
Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320-5. https://doi.org/10.1093/annonc/mdq276.
Gill J, Gorlick R. Advancing therapy for osteosarcoma. Nat Rev Clin Oncol. 2021;18(10):609–24. https://doi.org/10.1038/s41571-021-00519-8.
Meltzer PS, Helman LJ. New horizons in the treatment of osteosarcoma. N Engl J Med. 2021;385(22):2066–76. https://doi.org/10.1056/NEJMra2103423.
Bielack SS, Blattmann C, Borkhardt A, Csóka M, Hassenpflug W, Kabíčková E, et al. Osteosarcoma and causes of death: a report of 1520 deceased patients from the cooperative osteosarcoma study group (COSS). Eur J Cancer. 2022;176:50–7. https://doi.org/10.1016/j.ejca.2022.09.007.
Li S, Liu F, Zheng K, Wang W, Qiu E, Pei Y, et al. CircDOCK1 promotes the tumorigenesis and cisplatin resistance of osteogenic sarcoma via the miR-339-3p/IGF1R axis. Mol Cancer. 2021;20(1):161. https://doi.org/10.1186/s12943-021-01453-0.
Zhang Y, Lou Y, Wang J, Yu C, Shen W. Research status and molecular mechanism of the traditional Chinese medicine and antitumor therapy combined strategy based on tumor microenvironment. Front Immunol. 2020;11:609705. https://doi.org/10.3389/fimmu.2020.609705.
Zhang X, Qiu H, Li C, Cai P, Qi F. The positive role of traditional Chinese medicine as an adjunctive therapy for cancer. Biosci Trends. 2021;15(5):283–98. https://doi.org/10.5582/bst.2021.01318.
Ma YC, Ke Y, Zi X, Zhao W, Shi XJ, Liu HM. Jaridonin, a novel ent-kaurene diterpenoid from Isodon rubescens, inducing apoptosis via production of reactive oxygen species in esophageal cancer cells. Curr Cancer Drug Targets. 2013;13(6):611–24. https://doi.org/10.2174/15680096113139990030.
Li F, Fan J, Wu Z, Liu RY, Guo L, Dong Z, et al. Reversal effects of Rabdosia rubescens extract on multidrug resistance of MCF-7/Adr cells in vitro. Pharm Biol. 2013;51(9):1196–203. https://doi.org/10.3109/13880209.2013.784342.
Sartippour MR, Seeram NP, Heber D, Hardy M, Norris A, Lu Q, et al. Rabdosia rubescens inhibits breast cancer growth and angiogenesis. Int J Oncol. 2005;26(1):121–7.
Kazantseva L, Becerra J, Santos-Ruiz L. Oridonin enhances antitumor effects of doxorubicin in human osteosarcoma cells. Pharmacol Rep. 2022;74(1):248–56. https://doi.org/10.1007/s43440-021-00324-1.
Kadota S, Basnet P, Ishii E, Tamura T, Namba T. Antibacterial activity of trichorabdal a from Rabdosia trichocarpa against helicobacter pylori. Zentralbl Bakteriol. 1997;286(1):63–7. https://doi.org/10.1016/s0934-8840(97)80076-x.
Du Y, Zhang J, Yan S, Tao Z, Wang C, Huang M, et al. Oridonin inhibits the proliferation, migration and invasion of human osteosarcoma cells via suppression of matrix metalloproteinase expression and STAT3 signalling pathway. J BUON. 2019;24(3):1175–80.
Liu Y, Liu YZ, Zhang RX, Wang X, Meng ZJ, Huang J, et al. Oridonin inhibits the proliferation of human osteosarcoma cells by suppressing Wnt/β-catenin signaling. Int J Oncol. 2014;45(2):795–803. https://doi.org/10.3892/ijo.2014.2456.
Zhu X, Li S. Ferroptosis, necroptosis, and Pyroptosis in gastrointestinal cancers: the chief culprits of tumor progression and drug resistance. Adv Sci (Weinh). 2023;10(26):e2300824. https://doi.org/10.1002/advs.202300824.
Schmitt CA. Senescence, apoptosis and therapy--cutting the lifelines of cancer. Nat Rev Cancer 2003;3(4):286–295. https://doi.org/10.1038/nrc1044.
Nössing C, Ryan KM. 50 years on and still very much alive: 'Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics'. Br J Cancer. 2023;128(3):426–31. https://doi.org/10.1038/s41416-022-02020-0.
Cabanos HF, Hata AN. Emerging insights into targeted therapy-tolerant Persister cells in Cancer. Cancers (Basel). 2021;13(11):2666. https://doi.org/10.3390/cancers13112666.
Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22(4):266–82. https://doi.org/10.1038/s41580-020-00324-8.
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–72. https://doi.org/10.1016/j.cell.2012.03.042.
Liu X, Du S, Wang S, Ye K. Ferroptosis in osteosarcoma: a promising future. Front Oncol. 2022;12:1031779. https://doi.org/10.3389/fonc.2022.1031779.
Zhang C, Liu X, Jin S, Chen Y, Guo R. Ferroptosis in cancer therapy: a novel approach to reversing drug resistance. Mol Cancer. 2022;21(1):47. https://doi.org/10.1186/s12943-022-01530-y.
Friedmann Angeli JP, Krysko DV, Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat Rev Cancer. 2019;19(7):405–14. https://doi.org/10.1038/s41568-019-0149-1.
Su Y, Zhao B, Zhou L, Zhang Z, Shen Y, Lv H, et al. Ferroptosis, a novel pharmacological mechanism of anti-cancer drugs. Cancer Lett. 2020;483:127–36. https://doi.org/10.1016/j.canlet.2020.02.015.
Chen Q, Cao Y, Li H, Liu H, Liu Y, Bi L, et al. Sodium nitroprusside alleviates nanoplastics-induced developmental toxicity by suppressing apoptosis, ferroptosis and inflammation. J Environ Manag. 2023;345:118702. https://doi.org/10.1016/j.jenvman.2023.118702.
Yu H, Li JM, Deng K, Zhou W, Li KH, Wang CX, et al. GPX4 inhibition synergistically boosts mitochondria targeting nanoartemisinin-induced apoptosis/ferroptosis combination cancer therapy. Biomater Sci. 2023;11(17):5831–45. https://doi.org/10.1039/d3bm00601h.
Fu J, Li T, Yang Y, Jiang L, Wang W, Fu L, et al. Activatable nanomedicine for overcoming hypoxia-induced resistance to chemotherapy and inhibiting tumor growth by inducing collaborative apoptosis and ferroptosis in solid tumors. Biomaterials. 2021;268:120537. https://doi.org/10.1016/j.biomaterials.2020.120537.
Wang Y, Zhang L, Zhao G, Zhang Y, Zhan F, Chen Z, et al. Homologous targeting nanoparticles for enhanced PDT against osteosarcoma HOS cells and the related molecular mechanisms. J Nanobiotechnology. 2022;20(1):83. https://doi.org/10.1186/s12951-021-01201-y.
He C, Jiang Y, Guo Y, Wu Z. Amplified Ferroptosis and apoptosis facilitated by differentiation therapy efficiently suppress the progression of osteosarcoma. Small. 2023:e2302575. https://doi.org/10.1002/smll.202302575.
He T, Lin X, Yang C, Chen Z, Wang L, Li Q, et al. Theaflavin-3,3′-Digallate plays a ROS-mediated dual role in Ferroptosis and apoptosis via the MAPK pathway in human osteosarcoma cell lines and xenografts. Oxidative Med Cell Longev. 2022;2022:8966368. https://doi.org/10.1155/2022/8966368.
Miotto G, Rossetto M, Di Paolo ML, Orian L, Venerando R, Roveri A, et al. Insight into the mechanism of ferroptosis inhibition by ferrostatin-1. Redox Biol. 2020;28:101328. https://doi.org/10.1016/j.redox.2019.101328.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–8. https://doi.org/10.1006/meth.2001.1262.
Wang W, Green M, Choi JE, Gijón M, Kennedy PD, Johnson JK, et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569(7755):270–4. https://doi.org/10.1038/s41586-019-1170-y.
Jiang Z, Lim SO, Yan M, Hsu JL, Yao J, Wei Y, et al. TYRO3 induces anti-PD-1/PD-L1 therapy resistance by limiting innate immunity and tumoral ferroptosis. J Clin Invest. 2021;131(8):e139434. https://doi.org/10.1172/JCI139434.
Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33(27):3029–35. https://doi.org/10.1200/JCO.2014.59.4895.
Pakos EE, Nearchou AD, Grimer RJ, Koumoullis HD, Abudu A, Bramer JA, et al. Prognostic factors and outcomes for osteosarcoma: an international collaboration. Eur J Cancer. 2009;45(13):2367–75. https://doi.org/10.1016/j.ejca.2009.03.005.
Tang JL, Liu BY, Ma KW. Traditional Chinese medicine. Lancet. 2008;372(9654):1938–40. https://doi.org/10.1016/S0140-6736(08)61354-9.
Cai M, Yao Y, Yin D, Zhu R, Fu T, Kong J, et al. Enhanced lysosomal escape of cell penetrating peptide-functionalized metal-organic frameworks for co-delivery of survivin siRNA and oridonin. J Colloid Interface Sci. 2023;646:370–80. https://doi.org/10.1016/j.jcis.2023.04.126.
Hwang TL, Chang CH. Oridonin enhances cytotoxic activity of natural killer cells against lung cancer. Int Immunopharmacol. 2023;122:110669. https://doi.org/10.1016/j.intimp.2023.110669.
Zhou F, Gao H, Shang L, Li J, Zhang M, Wang S, et al. Oridonin promotes endoplasmic reticulum stress via TP53-repressed TCF4 transactivation in colorectal cancer. J Exp Clin Cancer Res. 2023;42(1):150. https://doi.org/10.1186/s13046-023-02702-4.
Zhang X, Feng H, Li Z, Guo J, Li M. Aspirin is involved in the cell cycle arrest, apoptosis, cell migration, and invasion of Oral squamous cell carcinoma. Int J Mol Sci. 2018;19(7):2029. https://doi.org/10.3390/ijms19072029.
Poornima P, Kumar JD, Zhao Q, Blunder M, Efferth T. Network pharmacology of cancer: from understanding of complex interactomes to the design of multi-target specific therapeutics from nature. Pharmacol Res. 2016;111:290–302. https://doi.org/10.1016/j.phrs.2016.06.018.
Sun Y, Jiang X, Lu Y, Zhu J, Yu L, Ma B, et al. Oridonin prevents epithelial-mesenchymal transition and TGF-β1-induced epithelial-mesenchymal transition by inhibiting TGF-β1/Smad2/3 in osteosarcoma. Chem Biol Interact. 2018;296:57–64. https://doi.org/10.1016/j.cbi.2018.09.013.
Wang XH, Zhang SF, Bao JT, Liu FY. Oridonin synergizes with Nutlin-3 in osteosarcoma cells by modulating the levels of multiple Bcl-2 family proteins. Tumour Biol. 2017;39(6):1010428317701638. https://doi.org/10.1177/1010428317701638.
Jin S, Shen JN, Wang J, Huang G, Zhou JG. Oridonin induced apoptosis through Akt and MAPKs signaling pathways in human osteosarcoma cells. Cancer Biol Ther. 2007;6(2):261–8. https://doi.org/10.4161/cbt.6.2.3621.
Xu J, Li Y, Kang M, Chang C, Wei H, Zhang C, et al. Multiple forms of cell death: a focus on the PI3K/AKT pathway. J Cell Physiol. 2023;238(9):2026–38. https://doi.org/10.1002/jcp.31087.
Zhang J, Wang N, Zhou Y, Wang K, Sun Y, Yan H, et al. Oridonin induces ferroptosis by inhibiting gamma-glutamyl cycle in TE1 cells. Phytother Res. 2021;35(1):494–503. https://doi.org/10.1002/ptr.6829.
Wang W, Zhang N. Oridonin inhibits Hela cell proliferation via downregulation of glutathione metabolism: a new insight from metabolomics. J Pharm Pharmacol. 2023;75(6):837–45. https://doi.org/10.1093/jpp/rgad025.
Ouyang S, Li H, Lou L, Huang Q, Zhang Z, Mo J, et al. Inhibition of STAT3-ferroptosis negative regulatory axis suppresses tumor growth and alleviates chemoresistance in gastric cancer. Redox Biol. 2022;52:102317. https://doi.org/10.1016/j.redox.2022.102317.
Zhang T, Sun B, Zhong C, Xu K, Wang Z, Hofman P, et al. Targeting histone deacetylase enhances the therapeutic effect of Erastin-induced ferroptosis in EGFR-activating mutant lung adenocarcinoma. Transl lung. Cancer Res. 2021;10(4):1857–72. https://doi.org/10.21037/tlcr-21-303.
Feng Y, Huang J, Wang F, Lin Z, Luo H, Li Q, et al. Methylcrotonyl-CoA carboxylase subunit 1 (MCCA) regulates multidrug resistance in multiple myeloma. Life Sci. 2023;333:122157. https://doi.org/10.1016/j.lfs.2023.122157.
Zeng Y, Jiang H, Zhang X, Xu J, Wu X, Xu Q, et al. Canagliflozin reduces chemoresistance in hepatocellular carcinoma through PKM2-c-Myc complex-mediated glutamine starvation. Free Radic Biol Med. 2023;208:571–86. https://doi.org/10.1016/j.freeradbiomed.2023.09.006.
Qu S, Qi S, Zhang H, Li Z, Wang K, Zhu T, et al. Albumin-bound paclitaxel augment temozolomide treatment sensitivity of glioblastoma cells by disrupting DNA damage repair and promoting ferroptosis. J Exp Clin Cancer Res. 2023;42(1):285. https://doi.org/10.1186/s13046-023-02843-6.
Wang JG, Li DL, Fan R, Yan MJ. Zerumbone combined with gefitinib alleviates lung cancer cell growth through the AKT/STAT3/SLC7A11 axis. Neoplasma. 2023;70(1):58–70. https://doi.org/10.4149/neo_2022_220418N423.
Acknowledgements
Not available.
Funding
This work was supported by the Innovation Fund of National Clinical Research Center for Orthopedics, Sports Medicine, and Rehabilitation (Grant No. 2021-NCRC-CXJJ-PY-13), Young Elite Scientists Sponsorship Program by CAST (Grant No. 2021-QNRC2-A06), the Major Project of Science and Technology in Henan Province (222102310428), the Joint Construction Project of Medical Science and Technique in Henan Province (LHGJ20200243), and the Project of TCM research in Henan Province (20-21ZY2234, 2023ZY2136).
Author information
Authors and Affiliations
Contributions
J.L. and H.J. spearheaded and supervised all the experiments. N.Y., M.L. and Y.Z. provided research ideas and designed the experiments. F.Z. conducted the experiments. Y.L. and J.Z. provided the technical support. Y.H. in charge of the formal analysis. F.Z. and Y.H. prepared the manuscript. All data were generated in-house laboratory, and no delivery to an outside paper mill. All authors agree to be accountable for all aspects of work ensuring integrity and accuracy.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not available.
Consent for publication
Not available.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
WB results.
Additional file 2:
Table S1. Primer sequences for q-PCR.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, F., Hao, Y., Yang, N. et al. Oridonin-induced ferroptosis and apoptosis: a dual approach to suppress the growth of osteosarcoma cells. BMC Cancer 24, 198 (2024). https://doi.org/10.1186/s12885-024-11951-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-024-11951-1