Abstract
Background
PTEN is the most frequently lost tumor suppressor in primary prostate cancer (PCa) and its loss is associated with aggressive disease. However, the transcriptional changes associated with PTEN loss in PCa have not been described in detail. In this study, we highlight the transcriptional changes associated with PTEN loss in PCa.
Methods
Using a meta-analysis approach, we leveraged two large PCa cohorts with experimentally validated PTEN and ERG status by Immunohistochemistry (IHC), to derive a transcriptomic signature of PTEN loss, while also accounting for potential confounders due to ERG rearrangements. This signature was expanded to lncRNAs using the TCGA quantifications from the FC-R2 expression atlas.
Results
The signatures indicate a strong activation of both innate and adaptive immune systems upon PTEN loss, as well as an expected activation of cell-cycle genes. Moreover, we made use of our recently developed FC-R2 expression atlas to expand this signature to include many non-coding RNAs recently annotated by the FANTOM consortium. Highlighting potential novel lncRNAs associated with PTEN loss and PCa progression.
Conclusion
We created a PCa specific signature of the transcriptional landscape of PTEN loss that comprises both the coding and an extensive non-coding counterpart, highlighting potential new players in PCa progression. We also show that contrary to what is observed in other cancers, PTEN loss in PCa leads to increased activation of the immune system. These findings can help the development of new biomarkers and help guide therapy choices.
Similar content being viewed by others
Background
Previous molecular studies have explored the genomic heterogeneity of prostate cancer (PCa) revealing distinct molecular subsets characterized by common genome alterations [1,2,3]. Among these molecular alterations, loss of the tumor suppressor gene phosphatase and tensin homolog (PTEN) – which is implicated in the negative-regulation of the PI3K-AKT-mTOR pathway – has been identified as one of the most common genomic drivers of primary PCa [4, 5]. Since alterations in the PI3K pathway are present in more than 30% of human cancers, the identification of an expression signature associated with PTEN loss has been investigated in different tumor contexts, including breast, bladder, lung, and PCa [6, 7].
Assessment of PTEN status by fluorescence in situ hybridization and immunohistochemistry (IHC) in large clinical PCa cohorts have shown a consistent association with adverse pathological features such as high Gleason score, extra-prostatic extension, as well as prognostic value for biochemical recurrence and cancer-related death [4, 8]. IHC-based assessment of PTEN status has been shown to correlate tightly with genomic alterations of the PTEN locus and captures not only loss of the gene, but also mutation and epigenetic changes that lead to PTEN functional inactivation [4, 9, 10] and the potential clinical utility of PTEN IHC as a valuable prognostic marker has been demonstrated previously [11,12,13,14].
Though PTEN is involved in a myriad of cellular processes spanning cellular proliferation to tumor microenvironment interactions [5], the transcriptional landscape related to PTEN expression has not yet been explored in-depth, and the role of long non-coding RNAs (lncRNAs) remains elusive [15]. These observations, added to the evidence that subtle PTEN downregulation can lead to cancer susceptibility [16], demonstrate the important role of PTEN in cancer biology but also highlight the need for additional studies.
Similarly, gene rearrangements of the ETS transcription factor, ERG, with the androgen-regulated gene Transmembrane Serine Protease 2 (TMPRSS2) are present in ~ 50% of PCa from patients of European descent. TMPRSS2-ERG fusion (herein denoted as ERG+ for fusion present and ERG− for the absence of fusion) has been shown to activate the PI3K-kinase pathway similarly to PTEN loss [17], leading to increased proliferation and invasion. Importantly, tumors harboring TMPRSS2-ERG rearrangements show enrichment for PTEN loss [17, 18]. The co-occurrence of these two genomic alterations makes it challenging to dissect the contributions of each to the transcriptomic landscape.
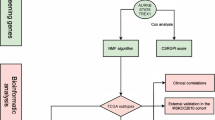
The goal of this study was to elucidate the transcriptional landscape of PTEN loss in PCa through the analysis of two large and very well clinically curated cohorts, for which PTEN and ERG status was assessed by clinical-grade IHC: The Natural History (NH) cohort, in which patients that underwent radical prostatectomy for clinically localized PCa did not receive neoadjuvant therapy or adjuvant hormonal therapy prior to documented distant metastases [19]; and the Health Professionals Follow-up Study (HPFS) cohort in which the patients were followed for over 25 years [20]. Based on IHC-assessed PTEN status for these cohorts, we built a PTEN-loss signature highly concordant across the independent datasets, in both presence and absence of TMPRSS2-ERG fusion. Overall, this PTEN-loss signature was associated with cellular processes associated with aggressive tumor behavior (e.g., increased motility and proliferation) and, surprisingly, with increases in gene sets related to the immune response. In addition, through our recently developed FANTOM-CAT/recount2 (FC-R2) resource [21] and copy-number-variation data, we expanded this signature beyond coding genes and report the non-coding RNA repertory resulting from PTEN loss.
Methods
Data collection and immunostaining
All expression data used in this work were gathered from public domain databases. In this work, we made use of three cohorts: FC-R2 TCGA, Natural History (NH), and Health Professionals Follow-up Study (HPFS). Information about each cohort is summarized in Table 1. Information about PTEN status by immunohistochemistry for the HPFS cohort was readily available and therefore obtained from the public domain. For NH cohort samples, IHC staining for PTEN and ERG was performed using a previously validated protocol [22]. Last, for TCGA we used the Copy Number Variation (CNV) called by the GISTIC algorithm to define PTEN status and the expectation-maximization algorithm to define ERG status.
Meta-analysis of NH and HPFS cohorts
Normalized microarray expression sets for the Natural History and HPFS cohorts were obtained from the Gene Expression Omnibus (GEO) [23]. We performed a meta-analysis approach using a Bayesian hierarchical multi-level model (BHM) for cross-study detection of differential gene expression implemented in the Bioconductor package XDE [24] on microarray-based cohorts to obtain a PTEN-null signature from PTEN IHC validated samples. The model was fitted using the Δgp model with empirical starting values and 1000 bootstraps were performed. All remaining parameters were set to default values. This analysis was also performed stratifying the samples by ERG status to evaluate the impact of the ERG rearrangement in the signature.
Differential expression analysis in the TCGA cohort
Raw coverage was obtained from the FC-R2 expression atlas [21] and divided by the average read length to obtain read counts. Only primary tumor samples with a PTEN GISTIC score of − 2 and 0 were used in this analysis. Low count genes (< 5 CPM) were filtered and the remaining genes were normalized with the trimmed mean of M-Values method [25]. A generalized linear model approach coupled with empirical Bayes moderation of standard errors and voom precision weights [26, 27] was used to detect differentially expressed genes in the TCGA cohort. Adjusted p-values controlling for multiple hypothesis testing were performed using the Benjamini-Hochberg method [28] and genes with false discovery rate (FDR) equal or less than 0.01 were reported.
Gene set enrichment analysis (GSEA)
The results from the meta-analysis performed in the NH and HPFS cohort were ranked by the weighted size effect (average of the posterior probability of concordant differential expression multiplied by the Bayesian effect size of each cohort). The results from the TCGA cohort were ranked by t-statistics. Ranked lists were tested for gene set enrichment. Gene set enrichment analysis (GSEA) was performed using a Monte Carlo adaptive multilevel splitting approach, implemented in the fgsea [29] package. A collection of gene sets (Hallmarks, REACTOME, and GO Biological Processes) were obtained from the Broad Institute MSigDB database. The androgen response gene set was obtained from Scheaffer et al. [30]. Gene sets with less than 15 and more than 1500 genes were removed from the analysis, except for the GO biological processes whose max size was set to 300 to avoid overly generic gene sets. The enriched pathways were collapsed to maintain only independent ones using the function collapsePathways from fgsea.
Results
Meta-analysis of natural history and health professionals follow-up study cohorts
We sought to obtain a consensus signature of PTEN loss that could be reproduced across independent cohorts. We utilized a meta-analysis approach leveraging a multi-level model for cross-study detection of differential gene expression (DGE). We fitted a Bayesian hierarchical model (BHM) for analysis of differential expression across multiple studies that allowed us to aggregate data from two previously described tissue microarray-based cohorts where PTEN and ERG status was determined by IHC (Table 1 and Fig. 1) and we derived a PTEN-loss signature (Fig. 2). In this analysis, we observed 813 genes for which the differential expression was highly concordant (Bayesian Effect Size (BES) ≥ 1, posterior probability of concordant differential expression (PPCDE) ≥ 0.95) (Table S1).
Cross-study meta-analysis of differential gene expression. Genes in the same loci as PTEN such as RLN1 and ATAD1 were found downregulated. PTEN-null vs PTEN-intact meta-analysis of HPFS/PHS and NH cohorts with Bayesian Hierarchical Model for DGE using XDE showing the top 25 most concordant differentially up- and down-regulated genes. PTEN status was based on IHC assays
The consequences of PTEN loss on cell cycle regulation and tumor cell invasion have been extensively reported previously [4, 31, 32]. Accordingly, beyond PTEN itself, the top DEG genes in our signature reflected this profile (Fig. 2 and Table S1). Dermatopontin (DPT) (BES = − 2.59, PPCDE = 1) and Alanyl membrane aminopeptidase (ANPEP) (BES = − 2.53, PPCDE = 1) were found downregulated upon PTEN loss. Leucine-Rich Repeat Neuronal 1 (LRRN1) was among the genes up-regulated upon PTEN loss (BES = 3.36, PPCDE = 1). These and other genes found differentially expressed upon PTEN loss have all been shown to be associated with a more aggressive phenotype in several cancer types [5].
Notably, we found ERG among the top upregulated genes in the signature (Fig. 2). As expected [18, 33, 34], ERG rearrangement was more common among cases with PTEN loss compared to intact PTEN in all cohorts (Fisher exact test, p ≤ 0.001). Given this enrichment, it was not surprising that ERG was among the most up-regulated genes in the BHM signature, as well as PLA2G7, which is among the most highly overexpressed genes in ERG-rearranged PCa compared to those lacking ERG rearrangements [35]. The presence of ERG and ERG-regulated transcripts in the PTEN-loss signature suggested that this signature might be confounded by enrichment of ERG rearranged tumors among the tumors with PTEN loss.
Since ERG rearrangements represent a major driver event in PCa and PTEN loss is enriched in ERG-rearranged tumors, we next investigated the role of ERG in our PTEN-loss signature. To this end, we repeated the Bayesian hierarchical model for the analysis of differential expression by stratifying the samples by ERG status. In the background with ERG rearrangement, we observed a similar signature to the previous overall PTEN-loss signature, but without the aforementioned ERG-associated genes (Supplementary figure S1 and Supplementary Table S2).
Extending the PTEN-loss signature
To validate our PTEN loss signatures in an independent cohort, we next examined the TCGA PRAD cohort [36], where PTEN status was estimated by genomic copy number (CN) assessment, which was closely aligned with PTEN gene expression (Figure S2). We recently developed a comprehensive expression atlas based on the FANTOM-CAT annotations. This meta-assembly is currently the broadest collection of the human transcriptome [21, 37]. These gene models include many novel lncRNAs, such as enhancers and promoters, that were annotated by the FANTOM consortium based on transcriptomic and epigenomic data, allowing the signature to be further expanded beyond the coding repertoire. We used TCGA expression data from the FC-R2 expression atlas [21] to perform DGE analysis stratified by the PTEN status as derived from CN analysis. We also performed the same analysis in a stratified manner as in the HPFS and NH cohorts, using the ERG expression with expectation maximization (EM) algorithm to define ERG status given the bimodal nature of ERG expression in PCa.
We observed 521 differentially expressed genes (DEG) when comparing PTEN-null and PTEN-wild-type samples (FDR ≤ 0.01, LogFC ≥1), of which 257 were coding genes and 264 were non-coding genes (Supplementary Table S3). When stratifying the samples by ERG status, we obtained 435 and 364 DEG in the background with and without ERG rearrangement (Supplementary Tables S4 and S5), respectively, with similar proportions of coding and non-coding genes. Using hypergeometric confidence intervals, we evaluated the concordance between the TCGA and the meta-analysis signatures. The results were found to be significantly concordant (Figure S3), confirming that CN is a reasonable proxy to IHC-staining in TCGA. Despite differences in technology, PTEN-status call, and statistical analysis, the high concordance between the signatures suggests that they are robust and reproducible, which allowed us to expand this signature to genes that are not encompassed in microarray, especially long non-coding RNAs.
Therefore, in this analysis, we were able to identify a variety of differentially expressed lncRNAs that have been already reported to be involved in PCa development and progression such as PCA3, PCGEM1, SCHLAP1, KRTAP5-AS1, Mir-596 [38,39,40,41,42,43,44,45,46,47] (Supplementary Tables S3, S4 and S5). PCA3 is a prostate-specific lncRNA overexpressed in PCa tissue. Similarly, lncRNA PCGEM1 expression is increased and highly specific in PCa where it promotes cell growth and it has been associated with high-risk PCa patients [42, 43]. On the other hand, to the best of our knowledge, KRTAP5-AS1 expression has not been previously associated with PCa.
In addition, among highly ranked differentially expressed lncRNAs were the lncRNAs SChLAP1 and its uncharacterized antisense neighbor AC009478.1. SchLAP1 is overexpressed in a subset of PCa where it antagonizes the tumor-suppressive function of the SWI/SNF complex and can independently predict poor outcomes [46, 47]. Besides, we observed a strong correlation between SchLAP1 and AC009478.1 expression in TCGA data only for PCa and bladder cancer (R = 0.94 and 0.85, respectively, with p < 2.2e-26, Figure S4), suggesting a possible, still unknown role also for this latter lncRNA in such tumor types.
A substantial proportion of the 264 lncRNAs differentially expressed upon PTEN-loss have not been previously reported in PCa, and 134 were only annotated in the FANTOM-CAT meta-assembly (Table 2). The FANTOM consortium has recently characterized hundreds of lncRNAs via molecular phenotyping [48], however, none of those associated with PTEN-loss was included in their study, and therefore they still lack an assigned function. Interestingly, it was shown that the expression levels of genes in the same topological domain are highly correlated only in tissue types in which these genes play a functional role [48]. For this reason, we characterized our novel PTEN-loss lncRNAs by analyzing the expression correlation with nearby genes across all cancer types in TCGA.
Among the FANTOM-CAT exclusive genes with the highest fold change in close proximity with coding genes, CATG00000038715 and CATG00000079217 were down-regulated, while CATG00000117664 was up-regulated (Figure S5). Notably, such genes were mostly expressed in PCa as opposed to other cancer types in TCGA (Fig. 3). CATG00000038715 is near CYP4F2 and CYP4F11, encoding members of the cytochrome P450 enzyme superfamily, and the expression levels of CATG00000038715 and CYP4F2 are most highly correlated in PCa (R = 0.91, p < 2.2e-16) suggesting specificity for this cancer type (Figure S6). CATG00000079217 is close to the coding gene FBXL7, an F-box gene that is a component of the E3 ubiquitin ligase complex. These genes showed only a weak correlation (R = 0.14, p < 7.4e-4), however, CATG00000079217 expression was notably higher in PCa and breast cancer than in other tumors, and it was moderately correlated with several PCa biomarkers (e.g. KLK2, KLK3, STEAP2, PCGEM1, SLC45A3) [42, 43, 49,50,51,52,53] (R = 0.37–0.57, p < 2.2e-16) in TCGA. Finally, CATG00000117664 is located near GPR158, a G protein-coupled receptor highly expressed in the brain. The expression between GPR158 and CATG00000117664 was significantly correlated (R = 0.54, p < 2.2e-16), and highly specific to PCa [54] (Figure S6).
PTEN loss induces the innate and adaptive immune system
We performed Gene Set Enrichment Analysis (GSEA) using fgsea [29] and tested both the BHM- and TCGA-generated molecular signatures for enrichment in three collections of the Molecular Signature Database (MSigDB) [55, 56]: HALLMARKS, REACTOME, and GO Biological Processes. Results were similar in both signatures, with positive enrichment of proliferation and cell cycle-related gene sets (e.g. MYC1 targets, MTORC1 signaling, cell cycle checkpoints, and DNA repair) and both innate and adaptive immune system associated gene sets (e.g. Neutrophil degranulation, MHC antigen presentation, interferon-alpha, and gamma) (Figs. 4 and 5 and Supplementary Tables S6, S7, S8, S9, S10, S11, S12, S13, S14, S15, S16, S17, S18, S19 and S20). The positive enrichment of MHC antigen presentation, interferon-alpha, and -gamma in PTEN-null tumors is consistent with our previous study showing that the absolute density of T-cells is increased in PCa with PTEN loss [57].
Top enriched gene sets enriched in the TCGA and BHM cohorts. The cohorts stratified by ERG status and overall shows a predominance of enriched cell cycle/proliferation and immune-response gene sets upon PTEN loss. Heatmap of mean-centered log2 signed p-values (normalized enrichment score multiplied by -log10 of p-value) showing the top 10 enriched gene sets of each collection (ranked by signed p-value)
Expression of immune-related genes stratified by PTEN status. The top 20 genes were selected based on the leading edge of the GSEA of the adaptive and innate immune system gene sets from REACTOME. Significances based on t-test between PTEN-null and PTEN-intact using log2 CPM + 1 value. Significance cutoffs: * = ≤ 0.05; ** ≤ 0.01; *** ≤ 0.001; **** ≤ 0.0001
Since PTEN-null tumors are known to have decreased androgen output, which is a strong suppressor of inflammatory immune cells [30, 58, 59], we hypothesized that this decrease in androgen levels could activate an immune response. We, therefore, performed a GSEA analysis using a collection of androgen-regulated genes from Schaeffer et al. [30] to test if the PTEN-null signature was enriched in this gene set. Both the TCGA- and BHM-signature were shown to be positively enriched in genes that were shown to be repressed upon dihydrotestosterone treatment (NES =1.39–155, FDR ≤ 0.05) (Figure S7).
Discussion
With an estimated prevalence of up to 50%, PTEN loss is recognized as one of the major driving events in PCa [60]. PTEN antagonizes PI3K-AKT/PKB and is a key modulator of the AKT-mTOR signaling pathways which are important in regulating cell growth and proliferation. Accordingly, PTEN loss is consistently associated with more aggressive disease features and poor outcomes. Saal and collaborators previously generated a transcriptomic signature of PTEN loss in breast cancer [6]. While this signature was correlated with worse patient outcomes in breast and other independent cancer datasets, including PCa, the signature unsurprisingly fails to capture key characteristics of PCa such as ERG-rearrangement [6, 11]. Significantly, a comprehensive transcriptomic signature reflecting the landscape of PTEN loss in PCa has not been described to date.
Immunohistochemistry assay is a clinically utilized technique to determine the status of the PTEN gene, with high sensitivity and specificity for protein levels, which are reduced when genomic deletions occur [22] (Fig. 1). Therefore, we analyzed transcriptome data from two large PCa cohorts – the Health Professional Follow-up Study and the Natural History study – for which IHC-based PTEN and ERG status was available (n = 390 and 207, respectively), deriving a PTEN-loss gene expression signature specific to PCa (Fig. 2 and Supplementary Table S1). Genes that are associated with increased proliferation and invasion in several cancer types, such as DPT, ANPEP, and LRRN1, were among the most concordant DEG in this signature. DPT has been shown to inhibit cell proliferation through MYC repression and to be down-regulated in both oral and thyroid cancer [61, 62]. It has also been shown to control cell adhesion and invasiveness, with low expression leading to the worst prognosis [62, 63]. ANPEP is known to play an important role in cell motility, invasion, and metastasis progression [63, 64], and lower expression of this gene has been associated with the worst prognosis [65]. LRRN1 is a direct transcriptional target of MYCN and an enhancer of the EGFR and IGRF signaling pathway [66]. Higher levels of LRRN1 expression promote tumor cell proliferation, inhibiting cell apoptosis, and play an important role in preserving pluripotency-related proteins through AKT phosphorylation [66,67,68], leading to a poor clinical outcome in gastric and brain cancer.
Notably, ERG was shown to be upregulated in our signature, which led us to perform a stratified analysis to avoid capturing signals driven mostly by ERG overexpression. In this latter analysis, we were not able to detect significant differences by PTEN status in the ERG−samples for the HPFS and NH cohorts, which were quantified by gene expression microarrays. Conversely, when analyzing the TCGA cohort, we were able to detect significant changes by PTEN status in the ERG− samples (Supplementary Tables S3, S4 and S5). However, given the known limitations of gene expression microarrays performed on formalin-fixed material, such as the limited dynamic range of expression values [69], observations in the HPFS and NH datasets could have been limited by the technology employed. Nevertheless, concordance between the BHM- and TCGA- cohorts were similar in both the overall and the ERG+ background comparisons (Supplementary Figure S3).
When we expanded our analyses to the non-coding transcriptome using the TCGA cohort, we identified several lncRNAs that have been already associated with PCa progression. For instance, among these lncRNAs, were PCA3 and KRTAP5-AS1. PCA3 acts by a variety of mechanisms such as down-regulation of the oncogene PRUNE2 and by acting as a miRNA sponge for mir-1261, which down-regulate the PRKD3 gene, leading to increase proliferation and migration [38, 39]. Conversely, knockdown of PCA3 can lead to partial reversion of epithelial-mesenchymal transition (EMT) [40] which can lead to increased cell invasion, motility, and survival [41]. Although KRTAP5-AS1 has not been associated with PCa, it has recently shown that it can act as a miRNA sponge for miRNAs such as mir-596, which targets CLDN4, an oncogene enhancing the invasion capacity of cancer cells promoting EMT [41, 44]. Thereby overexpression of KRTAP5-AS1 can potentially lead to increased levels of CLDN4 [45]. Mir-596 has also been shown to be overexpressed in response to androgen signaling and associated with anti-androgen therapy resistance [45].
Moreover, many lncRNAs exclusively annotated in the FANTOM-CAT [37] were associated with PTEN-loss and were shown to be expressed mostly in PCa (Fig. 3). Since these genes are novel without an elucidated function, we analyzed their potential roles by investigating coding genes located in the same genomic loci, under the premise of “guilty-by-association”. The genes encoding for CYP4F2, FBXL7, and GPR158, respectively, are positioned in the same loci as 3 of the top DE lncRNAs only known in the FANTOM-CAT (CATG00000038715, CATG00000079217, and CATG00000117664, Figure S5). CYP4F2 is involved in the process of inactivating and degrading leukotriene B4 (LTB4). LTB4 is a key gene in the inflammatory response that is produced in leukocytes in response to inflammatory mediators and can induce the adhesion and activation of leukocytes on the endothelium [70]. FBXL7 regulates mitotic arrest by degradation of AURKA, which is known to promote inflammatory response and activation of NF-κB [71, 72]. Likewise, increase expression of GPR158 is reported to stimulate cell proliferation in PCa cell lines, and it is linked to neuroendocrine differentiation [73].
We consistently observed a strong enrichment in immune response genes and gene sets upon PTEN loss (Fig. 4 and Supplementary Tables S6, S7, S8, S9, S10, S11, S12, S13, S14, S15, S16, S17, S18, S19 and S20). Immune-associated genes (i.e. GP2 and PLA2G2A) were found amongst the top up-regulated genes in our signature (Fig. 2). Positive enrichment of Interferon-alpha- and gamma-response genes (FDR ≤ 0.01) further suggests that a strong immuno-responsive environment, with both innate and adaptive systems activated, is developed in PTEN-null tumors (Fig. 5). The positive enrichment of MHC class II antigen presentation, neutrophil degranulation, vesicle-mediated transport, and FC receptor pathway-related genes suggests that PTEN-null tumors may be immunogenic (Fig. 4). This finding was particularly surprising given that PTEN is itself a key positive regulator of the innate immune response, controlling the import of IRF3, which is responsible for IFN production. Accordingly, disruption of PTEN expression has previously been reported to lead to decreased innate immune response [74]. Conversely, it has also been hypothesized that the increased genomic instability caused by, or associated with, PTEN loss can increase immunogenicity in the tumor microenvironment (TME) [75]. This finding is of particular interest given that immune-responsive tumors can be good candidates for immunotherapy-based approaches.
Remarkably, despite the loss of PTEN being associated with higher expression of the immune checkpoint gene programmed death ligand-1 (PD-L1) in several cancer types [76, 77], this is not true in PCa [78]. It has been shown that PCa employ different combinations of immune evasion mechanisms such as immunological ignorance, upregulated cytotoxic T lymphocyte-associated protein 4, and upregulated decoy receptor 3 [79]. So far, current immunotherapeutic interventions, such as PD-1 blockade, in PCa have not been successful. One of the possible reasons is the lack of PD-L1 expression [78]. Therefore, alternative targets must be considered for immunotherapy in PCa. One alternative target is the checkpoint molecule B7-H3 (CD276), whose expression has already been associated with PCa progression and worse prognosis [80] and has been suggested as a target for immunotherapy [81, 82]. CD276 was one of the most concordant up-regulated genes in our signature (Fig. 2) suggesting that its expression is associated with PTEN loss. Interestingly, B7-H3 expression may be down-regulated by androgens [83].
The effects of androgen on the immune system have already been extensively studied and reviewed [58]. Androgens are known to suppress inflammatory immune cells and impair the development and function of B- and T-cells [59]. We, therefore, hypothesized that the decreased levels of androgen in PTEN-null TME could lead to an unsuppressed immune system. By testing our signature for enrichment in androgen-related genes (AR) derived from Schaeffer et al. [30], we observed that upon PTEN-loss, androgen-sensitive genes that are typically suppressed by DHT are positively enriched, indicating that androgen levels or androgen response in PTEN-null tumors may be lower than in their PTEN-intact counterparts (Figure S7). This decrease in AR-signaling has been described in PTEN-null tumors, in which activation of the PI3K pathway inhibits AR activity [84]. Furthermore, AR inhibition activates AKT signaling by inhibiting AKT phosphatase levels further boosting cell proliferation [84], which has also been noted in this study (Fig. 3). Finally, in the non-coding repertoire, both PCA3 and PCGEM1 are modulated by androgen [85, 86] and were down-regulated upon PTEN loss which tracks with the observed decreased androgen response in PTEN-null tumors (Figure S5 and S7).
Altogether, we have generated a highly concordant gene signature for PTEN loss in PCa across three independent datasets. We show that this signature was highly enriched in proliferation and cell cycle genes, leading to a more aggressive phenotype upon PTEN loss, which is concordant with the literature. We have also highlighted some lncRNAs whose expression shows high specificity in PCa. Unfortunately, the roles of these lncRNAs are currently unknown and further functional studies are warranted, we have noted that they are in proximity to genes involved in immune response. We have shown that PTEN loss is associated with an increase in both innate and adaptive immune responses. Although the literature shows that PTEN loss usually leads to immuno-suppression, we find evidence that this finding may be reversed in PCa. This observation has potential implications in the context of precision medicine since immune responsive tumors are more likely to respond to immunotherapies. Therefore, PTEN-null tumors might benefit more from this approach than PTEN-intact tumors. Potentially, PTEN status can guide immunotherapy combination with other approaches such as androgen ablation.
Conclusion
Using the FC-R2 resource, we were able to highlight many lncRNAs that may be associated with PCa progression. Although functional characterization of these lncRNAs is beyond the scope of this study, we have shown that these novel lncRNAs are highly specific to PCa and track with several coding mRNAs and lncRNAs already reported to be involved in PCa development and progression, most notably, genes involved in the immune response. By providing a PCa-specific signature for PTEN loss and highlighting potential new players, we hope to empower further studies on the mechanisms leading to the development and progression of PCa that can aid in the development of potential biomarkers, drug targets, and guide therapies choice.
Availability of data and materials
The datasets analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository under accession numbers: GSE79957, GSE79021, and GSE62872. The TCGA cohort with FANTOM-CAT annotations is available at https://jhubiostatistics.shinyapps.io/recount/. All the code used in this study is available at https://github.com/eddieimada/PTEN_analysis.
Abbreviations
- PCa:
-
Primary prostate cancer
- PTEN:
-
Phosphatase and tensin homolog
- IHC:
-
Immunohistochemistry
- lncRNAs:
-
Long non-coding RNAs
- ERG:
-
ETS transcription factor
- TMPRSS2:
-
Transmembrane Serine Protease 2
- NH:
-
Natural History
- HPFS:
-
Health Professionals Follow-up Study
- FC-R2:
-
FANTOM-CAT/recount2
- CNV:
-
Copy Number Variation
- GEO:
-
Gene Expression Omnibus
- FDR:
-
False discovery rate
- GSEA:
-
Gene set enrichment analysis
- BHM:
-
Bayesian hierarchical model
- BES:
-
Bayesian Effect Size
- PPCDE:
-
Posterior probability of concordant differential expression
- CN:
-
Copy number
- DEG:
-
Differentially expressed genes
- MSigDB:
-
Molecular Signature Database
- EMT:
-
Epithelial-mesenchymal transition
- TME:
-
Tumor microenvironment
- AR:
-
Androgen-related genes
References
Cancer Genome Atlas Research Network, TCGA. The molecular taxonomy of primary prostate Cancer. Cell. 2015;163(4):1011–25.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. https://doi.org/10.1016/j.ccr.2010.05.026.
Baca S, Garraway L. The genomic landscape of prostate cancer. Front Endocrinol (Lausanne). 2012;3:69.
Jamaspishvili T, Berman DM, Ross AE, Scher HI, De Marzo AM, Squire JA, et al. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol. 2018;15(4):222–34. https://doi.org/10.1038/nrurol.2018.9.
Lee Y-R, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol. 2018;19(9):1–16.
Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She Q-BBQ-B, Maurer M, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci. 2007;104(18):7564–9. https://doi.org/10.1073/pnas.0702507104.
Ong CW, Maxwell P, Alvi MA, McQuaid S, Waugh D, Mills I, et al. A gene signature associated with PTEN activation defines good prognosis intermediate risk prostate cancer cases. J Pathol Clin Res. 2018;4(2):103–13. https://doi.org/10.1002/cjp2.94.
Morais CL, Han JS, Gordetsky J, Nagar MS, Anderson AE, Lee S, et al. Utility of PTEN and ERG immunostaining for distinguishing high-grade PIN from intraductal carcinoma of the prostate on needle biopsy. Am J Surg Pathol. 2015;39(2):169–78. https://doi.org/10.1097/PAS.0000000000000348.
Lotan TL, Wei W, Ludkovski O, Morais CL, Guedes LB, Jamaspishvili T, et al. Analytic validation of a clinical-grade PTEN immunohistochemistry assay in prostate cancer by comparison with PTEN FISH. Nat Genet. 2016;29(8):904–14.
Lotan TL, Heumann A, Rico SD, Hicks J, Lecksell K, Koop C, et al. PTEN loss detection in prostate cancer: comparison of PTEN immunohistochemistry and PTEN FISH in a large retrospective prostatectomy cohort. Oncotarget. 2017;8(39):65566–76. https://doi.org/10.18632/oncotarget.19217.
Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009 Aug 1;22(8):1083–93. https://doi.org/10.1038/modpathol.2009.69.
Leapman MS, Nguyen HG, Cowan JE, Xue L, Stohr B, Simko J, et al. Comparing prognostic utility of a single-marker immunohistochemistry approach with commercial gene expression profiling following radical prostatectomy. Eur Urol. 2018;74(5):668–75. https://doi.org/10.1016/j.eururo.2018.08.020.
Ahearn TU, Pettersson A, Ebot EM, Gerke T, Graff RE, Morais CL, et al. A Prospective Investigation of PTEN Loss and ERG Expression in Lethal Prostate Cancer. J Natl Cancer Inst. 2016;108(2). https://doi.org/10.1093/jnci/djv346.
Lotan TL, Tomlins SA, Bismar TA, Van der Kwast TH, Grignon D, Egevad L, et al. Report from the International Society of Urological Pathology (ISUP) consultation conference on molecular pathology of urogenital cancers. I. Molecular biomarkers in prostate Cancer. Am J Surg Pathol. 2020;44(7):e15–29. https://doi.org/10.1097/PAS.0000000000001450.
Misawa A, Takayama KI, Inoue S. Long non-coding RNAs and prostate cancer. Cancer Sci. 2017;108(11):2107–14. https://doi.org/10.1111/cas.13352.
Alimonti A, Carracedo A, Clohessy JG, Trotman LC, Nardella C, Egia A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42(5):454–8. https://doi.org/10.1038/ng.556.
King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41(5):524–6. https://doi.org/10.1038/ng.371.
Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41(5):619–24. https://doi.org/10.1038/ng.370.
Ross AE, Johnson MH, Yousefi K, Davicioni E, Netto GJ, Marchionni L, et al. Tissue-based genomics augments post-prostatectomy risk stratification in a natural history cohort of intermediate- and high-risk men. Eur Urol. 2016;69(1):157–65. https://doi.org/10.1016/j.eururo.2015.05.042.
Penney KL, Sinnott JA, Tyekucheva S, Gerke T, Shui IM, Kraft P, et al. Association of prostate cancer risk variants with gene expression in normal and tumor tissue. Cancer Epidemiol Biomark Prev. 2015;24(1):255–60. https://doi.org/10.1158/1055-9965.EPI-14-0694-T.
Imada EL, Sanchez DF, Collado-Torres L, Wilks C, Matam T, Dinalankara W, et al. Recounting the FANTOM CAGE--Associated Transcriptome. Genome Res. 2020;30(7):gr--254656.
Lotan TL, Gurel B, Sutcliffe S, Esopi D, Liu W, Xu J, et al. PTEN protein loss by immunostaining: analytic validation and prognostic Indicator for a high risk surgical cohort of prostate Cancer patients. Clin Cancer Res. 2011;17(20):6563–73. https://doi.org/10.1158/1078-0432.CCR-11-1244.
Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10. https://doi.org/10.1093/nar/30.1.207.
Scharpf RB, Tjelmeland H, Parmigiani G, Nobel AB. A Bayesian model for cross-study differential gene expression. J Am Stat Assoc. 2009;104(488):1295–310. https://doi.org/10.1198/jasa.2009.ap07611.
Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. https://doi.org/10.1186/gb-2010-11-3-r25.
Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3.
Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;Series B(1):289–300.
Sergushichev AA. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. BioRxiv. 2016;60012. https://doi.org/10.1101/060012.
Schaeffer EM, Marchionni L, Huang Z, Simons B, Blackman A, Yu W, et al. Androgen-induced programs for prostate epithelial growth and invasion arise in embryogenesis and are reactivated in cancer. Oncogene. 2008;27(57):7180–91. https://doi.org/10.1038/onc.2008.327.
Leinonen KA, Saramaki OR, Furusato B, Kimura T, Takahashi H, Egawa S, et al. Loss of PTEN is associated with aggressive behavior in ERG-positive prostate Cancer. Cancer Epidemiol Biomark Prev. 2013;22(12):2333–44. https://doi.org/10.1158/1055-9965.EPI-13-0333-T.
Yoshimoto M, Ludkovski O, DeGrace D, Williams JL, Evans A, Sircar K, et al. PTEN genomic deletions that characterize aggressive prostate cancer originate close to segmental duplications. Genes Chromosom Cancer. 2012;51(2):149–60. https://doi.org/10.1002/gcc.20939.
Mehra R, Salami SS, Lonigro R, Bhalla R, Siddiqui J, Cao X, et al. Association of ERG/PTEN status with biochemical recurrence after radical prostatectomy for clinically localized prostate cancer. Med Oncol. 2018;35(12):152. https://doi.org/10.1007/s12032-018-1212-6.
Krohn A, Freudenthaler F, Harasimowicz S, Kluth M, Fuchs S, Burkhardt L, et al. Heterogeneity and chronology of PTEN deletion and ERG fusion in prostate cancer. Mod Pathol. 2014;27(12):1612–20. https://doi.org/10.1038/modpathol.2014.70.
Massoner P, Kugler KG, Unterberger K, Kuner R, Mueller LAJ, Fälth M, et al. Characterization of transcriptional changes in ERG rearrangement-positive prostate cancer identifies the regulation of metabolic sensors such as neuropeptide Y. PLoS One. 2013;8(2):e55207. https://doi.org/10.1371/journal.pone.0055207.
Network CGAR, Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–20.
Hon C-C, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJLL, Gough J, et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543(7644):199–204. https://doi.org/10.1038/nature21374.
Salameh A, Lee AK, Cardó-Vila M, Nunes DN, Efstathiou E, Staquicini FI, et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad Sci. 2015;112(27):8403–8. https://doi.org/10.1073/pnas.1507882112.
He J-HH, Li B-XX, Han Z-PP, Zou M-XX, Wang L, Lv Y-BB, et al. Snail-activated long non-coding RNA PCA3 up-regulates PRKD3 expression by miR-1261 sponging, thereby promotes invasion and migration of prostate cancer cells. Tumor Biol. 2016;37(12):16163–76. https://doi.org/10.1007/s13277-016-5450-y.
Lemos AEGEGEG, Ferreira LB, Batoreu NM, de Freitas PP, Bonamino MH, Gimba ERP. PCA3 long noncoding RNA modulates the expression of key cancer-related genes in LNCaP prostate cancer cells. Tumor Biol. 2016;37(8):11339–48. https://doi.org/10.1007/s13277-016-5012-3.
Agarwal R, D’Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res. 2005;65(16):7378–85. https://doi.org/10.1158/0008-5472.CAN-05-1036.
Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L, et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci. 2000;97(22):12216–21. https://doi.org/10.1073/pnas.97.22.12216.
Petrovics G, Zhang W, Makarem M, Street JP, Connelly R, Sun L, et al. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004;23(2):605–11. https://doi.org/10.1038/sj.onc.1207069.
Lin X, Shang X, Manorek G, Howell SB. Regulation of the epithelial-mesenchymal transition by claudin-3 and claudin-4. PLoS One. 2013;8(6):e67496. https://doi.org/10.1371/journal.pone.0067496.
Song YX, Sun JX, Zhao JH, Yang YC, Shi JX, Wu ZH, et al. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8(1):289. https://doi.org/10.1038/s41467-017-00304-1.
Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45(11):1392–403. https://doi.org/10.1038/ng.2771.
Mehra R, Udager AM, Ahearn TU, Cao X, Feng FY, Loda M, et al. Overexpression of the long non-coding RNA SChLAP1 independently predicts lethal prostate cancer. Eur Urol. 2016;70(4):549–52. https://doi.org/10.1016/j.eururo.2015.12.003.
Ramilowski JA, Yip CW, Agrawal S, Chang JC, Ciani Y, Kulakovskiy IV, et al. Functional annotation of human long noncoding RNAs via molecular phenotyping. Genome Res. 2020;30(7):1060–72. https://doi.org/10.1101/gr.254219.119.
Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8(7):497–511. https://doi.org/10.1038/nrc2402.
Nam RK, Zhang WW, Klotz LH, Trachtenberg J, Jewett MAS, Sweet J, et al. Variants of the hK2 protein gene (KLK2) are associated with serum hK2 levels and predict the presence of prostate cancer at biopsy. Clin Cancer Res. 2006;12(21):6452–8. https://doi.org/10.1158/1078-0432.CCR-06-1485.
Cicek MS, Liu X, Casey G, Witte JS. Role of androgen metabolism genes CYP1B1, PSA/KLK3, and CYP11$α$ in prostate cancer risk and aggressiveness. Cancer Epidemiol Prev Biomarkers. 2005;14(9):2173–7. https://doi.org/10.1158/1055-9965.EPI-05-0215.
Whiteland H, Spencer-Harty S, Morgan C, Kynaston H, Thomas DH, Bose P, et al. A role for STEAP2 in prostate cancer progression. Clin Exp Metastasis. 2014;31(8):909–20.
Perner S, Rupp NJ, Braun M, Rubin MA, Moch H, Dietel M, et al. Loss of SLC45A3 protein (prostein) expression in prostate cancer is associated with SLC45A3-ERG gene rearrangement and an unfavorable clinical course. Int J Cancer. 2013;132(4):807–12. https://doi.org/10.1002/ijc.27733.
Patel N, Itakura T, Jeong S, Liao C-PP, Roy-Burman P, Zandi E, et al. Expression and functional role of orphan receptor GPR158 in prostate cancer growth and progression. Robson CN, editor. PLoS One. 2015;10(2):e0117758.
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011 Jun 15;27(12):1739–40. https://doi.org/10.1093/bioinformatics/btr260.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. https://doi.org/10.1073/pnas.0506580102.
Kaur HB, Guedes LB, Lu J, Maldonado L, Reitz L, Barber JR, et al. Association of tumor-infiltrating T-cell density with molecular subtype, racial ancestry and clinical outcomes in prostate cancer. Mod Pathol. 2018;31(10):1539–52. https://doi.org/10.1038/s41379-018-0083-x.
Trigunaite A, Dimo J, Jørgensen TN. Suppressive effects of androgens on the immune system. Cell Immunol. 2015;294(2):87–94. https://doi.org/10.1016/j.cellimm.2015.02.004.
Ylitalo EB, Thysell E, Jernberg E, Lundholm M, Crnalic S, Egevad L, et al. Subgroups of castration-resistant prostate Cancer bone metastases defined through an inverse relationship between androgen receptor activity and immune response. Eur Urol. 2017;71(5):776–87. https://doi.org/10.1016/j.eururo.2016.07.033.
Wise HM, Hermida MA, Leslie NR. Prostate cancer, PI3K, PTEN and prognosis. Clin Sci. 2017;131(3):197–210. https://doi.org/10.1042/CS20160026.
Guo Y, Li H, Guan H, Ke W, Liang W, Xiao H, et al. Dermatopontin inhibits papillary thyroid cancer cell proliferation through MYC repression. Mol Cell Endocrinol. 2019;480:122–32. https://doi.org/10.1016/j.mce.2018.10.021.
Yamatoji M, Kasamatsu A, Kouzu Y, Koike H, Sakamoto Y, Ogawara K, et al. Dermatopontin: a potential predictor for metastasis of human oral cancer. Int J Cancer. 2012;130(12):2903–11. https://doi.org/10.1002/ijc.26328.
Ishii K, Usui S, Sugimura Y, Yoshida S, Hioki T, Tatematsu M, et al. Aminopeptidase N regulated by zinc in human prostate participates in tumor cell invasion. Int J Cancer. 2001;92(1):49–54. https://doi.org/10.1002/1097-0215(200102)9999:9999<::AID-IJC1161>3.0.CO;2-S.
Hashida H, Takabayashi A, Kanai M, Adachi M, Kondo K, Kohno N, et al. Aminopeptidase N is involved in cell motility and angiogenesis: its clinical significance in human colon cancer. Gastroenterology. 2002;122(2):376–86. https://doi.org/10.1053/gast.2002.31095.
Sørensen KD, Abildgaard MO, Haldrup C, Ulhøi BP, Kristensen H, Strand S, et al. Prognostic significance of aberrantly silenced ANPEP expression in prostate cancer. Br J Cancer. 2013;108(2):420–8. https://doi.org/10.1038/bjc.2012.549.
Hossain S, Takatori A, Nakamura Y, Suenaga Y, Kamijo T, Nakagawara A. LRRN1 enhances EGF-mediated MYCN induction in neuroblastoma and accelerates tumor growth in vivo. Cancer Res. 2012;72(17):4587–96. https://doi.org/10.1158/0008-5472.CAN-12-0943.
Hossain MS, Ozaki T, Wang H, Nakagawa A, Takenobu H, Ohira M, et al. N-MYC promotes cell proliferation through a direct transactivation of neuronal leucine-rich repeat protein-1 (NLRR1) gene in neuroblastoma. Oncogene. 2008;27(46):6075–82. https://doi.org/10.1038/onc.2008.200.
Liao C-H, Wang Y-H, Chang W-W, Yang B-C, Wu T-J, Liu W-L, et al. Leucine-rich repeat neuronal protein 1 regulates differentiation of embryonic stem cells by post-translational modifications of pluripotency factors. Stem Cells. 2018;36(10):1514–24. https://doi.org/10.1002/stem.2862.
Wilhelm BT, Landry J-R. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48(3):249–57. https://doi.org/10.1016/j.ymeth.2009.03.016.
Hardwick JP. Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochem Pharmacol. 2008;75(12):2263–75. https://doi.org/10.1016/j.bcp.2008.03.004.
Coon TA, Glasser JR, Mallampalli RK, Chen BB. Novel E3 ligase component FBXL7 ubiquitinates and degrades Aurora a, causing mitotic arrest. Cell Cycle. 2012;11(4):721–9. https://doi.org/10.4161/cc.11.4.19171.
Katsha A, Soutto M, Sehdev V, Peng D, Washington MK, Piazuelo MB, et al. Aurora kinase A promotes inflammation and tumorigenesis in mice and human gastric neoplasia. Gastroenterology. 2013;145(6):1312–22.e1–8.
Fenner A. Orphan receptor GPR158 finds a home in prostate cancer growth and progression. Nat Rev Urol. 2015;12(4):182. https://doi.org/10.1038/nrurol.2015.44.
Li S, Zhu M, Pan R, Fang T, Cao Y-YY, Chen S, et al. The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat Immunol. 2016;17(3):241–9. https://doi.org/10.1038/ni.3311.
Vidotto T, Melo CM, Castelli E, Koti M, Dos Reis RB, Squire JA. Emerging role of PTEN loss in evasion of the immune response to tumours. Br J Cancer. 2020;122(12):1732–43. https://doi.org/10.1038/s41416-020-0834-6.
Lastwika KJ, Wilson W, Li QK, Norris J, Xu H, Ghazarian SR, et al. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung Cancer. Cancer Res. 2016;76(2):227–38. https://doi.org/10.1158/0008-5472.CAN-14-3362.
Berghoff AS, Kiesel B, Widhalm G, Rajky O, Ricken G, Wöhrer A, et al. Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro-Oncology. 2015;17(8):1064–75. https://doi.org/10.1093/neuonc/nou307.
Martin AM, Nirschl TR, Nirschl CJ, Francica BJ, Kochel CM, van Bokhoven A, et al. Paucity of PD-L1 expression in prostate cancer: innate and adaptive immune resistance. Prostate Cancer Prostatic Dis. 2015;18(4):325–32. https://doi.org/10.1038/pcan.2015.39.
Bou-Dargham MJ, Sha L, Sang Q-XA, Zhang J. Immune landscape of human prostate cancer: immune evasion mechanisms and biomarkers for personalized immunotherapy. BMC Cancer. 2020;20(1):572. https://doi.org/10.1186/s12885-020-07058-y.
Yuan H, Wei X, Zhang G, Li C, Zhang X, Hou J. B7-H3 over expression in prostate cancer promotes tumor cell progression. J Urol. 2011;186(3):1093–9. https://doi.org/10.1016/j.juro.2011.04.103.
Papanicolau-Sengos A, Yang Y, Pabla S, Lenzo FL, Kato S, Kurzrock R, et al. Identification of targets for prostate cancer immunotherapy. Prostate. 2019;79(5):498–505. https://doi.org/10.1002/pros.23756.
Yang S, Wei W, Zhao Q. B7-H3, a checkpoint molecule, as a target for cancer immunotherapy. Int J Biol Sci. 2020;16(11):1767–73. https://doi.org/10.7150/ijbs.41105.
Benzon B, Zhao SG, Haffner MC, Takhar M, Erho N, Yousefi K, et al. Correlation of B7-H3 with androgen receptor, immune pathways and poor outcome in prostate cancer: an expression-based analysis. Prostate Cancer Prostatic Dis. 2017;20(1):28–35. https://doi.org/10.1038/pcan.2016.49.
Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19(5):575–86. https://doi.org/10.1016/j.ccr.2011.04.008.
Gezer U, Tiryakioglu D, Bilgin E, Dalay N, Holdenrieder S. Androgen stimulation of PCA3 and miR-141 and their release from prostate cancer cells. Cell J. 2015;16(4):488–93. https://doi.org/10.22074/cellj.2015.494.
Parolia A, Crea F, Xue H, Wang Y, Mo F, Ramnarine VR, et al. The long non-coding RNA PCGEM1 is regulated by androgen receptor activity in vivo. Mol Cancer. 2015;14(1):46. https://doi.org/10.1186/s12943-015-0314-4.
Acknowledgments
Not applicable.
Funding
This publication was made possible through support from the National Institutes of Health–National Cancer Institute (NIH-NCI) grants U01CA196390, and R01CA200859; and the U.S. Department of Defense (DoD) Office of the Congressionally Directed Medical Research Programs (CDMRP) award W81XWH-16-1-0739 and W81XWH-16-1-0737; and Fundação de Amparo a Pesquisa do Estado de Minas Gerais award BDS-00493-16. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
L.M. and T.L. conceived the idea; L.M., E.L.I., and T.L. designed the study; E.L.I., D.F.S., W.D., T.L., and L.M. performed the analysis; E.L.I., D.F.S., T.L., T.V., G.R.F., and L.M. interpreted the results; T.L., E.M.E., S.T., L.A.M., M.L., and E.M.S. provided data and tools; E.L.I., D.F.S., T.L., and L.M. wrote the manuscript; all authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Tables.
Excel spreadsheet containing the raw results of all bioinformatics analysis.
Additional file 2: Supplementary Figures.
PDF file containing the raw results of all bioinformatics analysis. Figure S1. Cross-study of differential gene expression in PTEN-null vs PTEN-intact in ERG+ samples. Meta-analysis of HPFS/PHS and NH cohorts with Bayesian Hierarchical Model for DGE using XDE showing the top 25 most concordant differentially up- and down-regulated genes. PTEN status were based on IHC assays. Figure S2. PTEN expression levels stratified by CNV. Figure shows PTEN expression levels distribution by copy number variation (CNV), called by GISTIC algorithm. Figure S3. Correspondence-at-the-top (CAT) plot between TCGA CNV-based calls and the Bayesian Hierarchical Model approach (BHM). Agreement of genes ranked by t-statistics (TCGA) and average Bayesian Effect Size (BHM). Lines represent agreement between tested cohorts for PTEN-intact vs PTEN-null. Black-to-light grey shades represent the decreasing probability of agreeing by chance based on the hypergeometric distribution, with intervals ranging from 0.999999 (light grey) to 0.95 (dark grey). Lines outside this range represent agreement in different cohorts with a higher agreement than expected by chance. Figure S4. Expression of AC009478.1 is shown to be highly specific to PRAD, BLCA, to a lesser extent in UECA and BRCA. Figure shows raw expression values of SchLAP1 and AC009478.1 across cancer types. Pearson correlations and p-values are shown in red. Figure S5. Expression of FANTOM-CAT lncRNAs genes (top) and close coding genes (bottom) stratified by PTEN status. Significances based on t-test between PTEN-null and PTEN-intact using log2 CPM + 1 value. Significance cutoffs: * 0.05; **≤0.01; ***≤0.0001. Figure S6. Person correlation gene CATG00000038715 and CYP4F2 across cancer types. CATG00000038715 and CYP4F2 expression are shown to be highly correlated in PCa. Moreover, CATG00000038715 expression is shown to be highly specific to PCa. With exception of leukemia cells, none of the other tumors expressed high levels of CATG00000038715. Figure S7. Gene set enrichment for Androgen repressed genes. Gene set enrichment analysis of gene signature showing positive enrichment of genes repressed by dihydrotestosterone after 6 h of exposure obtained from Schaeffer et al. Enrichment for BHM-signature is shown in panel A and TCGA-signature in panel B.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Imada, E.L., Sanchez, D.F., Dinalankara, W. et al. Transcriptional landscape of PTEN loss in primary prostate cancer. BMC Cancer 21, 856 (2021). https://doi.org/10.1186/s12885-021-08593-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-021-08593-y