Abstract
Background
Septo-optic dysplasia (SOD), also known as de-Morsier syndrome, is a rare disorder characterized by any combination of optic nerve hypoplasia, pituitary gland hypoplasia, and midline abnormalities of the brain including absence of the septum pellucidum and corpus callosum dysgenesis. The variable presentation of SOD includes visual, neurologic, and/or hypothalamic-pituitary endocrine defects. The unclear aetiology of a large proportion of SOD cases underscores the importance of identifying novel SOD-associated genes.
Case presentation
To identify the disease-causing gene in a male infant with neonatal hypoglycaemia, dysmorphic features, and hypoplasia of the optic nerve and corpus callosum, we designed a targeted next-generation sequencing panel for brain morphogenesis defects. We identified a novel hemizygous deletion, c.6355 + 4_6355 + 5delAG, in intron 38 of the FLNA gene that the patient had inherited from his mother. cDNA studies showed that this variant results in the production of 3 aberrant FLNA transcripts, the most abundant of which results in retention of intron 38 of FLNA.
Conclusions
We report for the first time a case of early-onset SOD associated with a mutation in the FLNA gene. This finding broadens the spectrum of genetic causes of this rare disorder and expands the phenotypic spectrum of the FLNA gene.
Similar content being viewed by others
Background
Septo-optic dysplasia (SOD), also known as de-Morsier syndrome (OMIM # 182230), is defined as any combination of the following: (1) optic nerve hypoplasia (ONH); (2) midline neuro-radiological abnormalities such as agenesis of the corpus callosum and absence of the septum pellucidum; and (3) pituitary hypoplasia with consequent panhypopituitarism [1]. Approximately 30% of all cases are characterized by the presence of all 3 alterations, while hypopituitarism and absent septum pellucidum are observed in 62 and 60% of cases, respectively [1]. SOD is a rare congenital anomaly with a reported incidence of 1/10,000 and is equally prevalent in males and females [1].
SOD represents a clinical spectrum rather than a specific entity [2]. Affected patients may present during infancy with systemic alterations including neonatal hypoglycaemia, jaundice, seizures, failure to thrive, developmental delay and microgenitalia. Several ophthalmic manifestations are also described, including visual impairment, nystagmus, strabismus, and, occasionally, refractive errors.
The majority of cases are sporadic. In rare cases a familial history can be identified. Studies of familial cases point to a mutation in a developmental gene (HESX1, SOX2, SOX3) [3, 4]. In this case report, we present the first evidence implicating a mutation in the FLNA gene in early-onset SOD.
Case presentation
Our patient is a boy born in 2015, the second child of unrelated, healthy white parents. He has a healthy older sister who was born in 2011. He was born at 376/7 weeks gestation and delivered by vaginal birth (weight, 2.4 kg; z-score, − 1,65; length, 45 cm; z- score, − 2.45; head circumference, 31 cm; z-score, − 1.84; Apgar scores, 8 at 1 min and 8 at 5 min). Five hours after birth he was admitted to the neonatal unit with hypoglycaemia (point-of-care, 29 mg/dl) and received intravenous glucose infusion for 72 h. The results of cerebral ultrasound and laboratory tests for congenital cytomegalovirus infection were normal.
An initial evaluation at 6 months of age revealed global developmental delay. The patient’s build, nourishment, and stature were normal for his age (z-score, + 0.64). Examination revealed microcephaly (z-score, − 1.56), dysmorphic features (thin upper lip, ogival palate, low hair implantation, long philtrum, extensive palpebral fissures), limb anomalies (implantation anomalies of the right toes and the plantar pads on both feet), microphallus, and undescended testes.
The patient presented generalized hypotonia; he was unable to achieve head control and showed horizontal jerk nystagmus, ocular motility disturbances, and visual impairment. There was no history of seizures. Cranial magnetic resonance imaging (MRI) showed delayed myelination, hypoplasia of the corpus callosum (Fig. 1), and pituitary hypoplasia. The flash visual-evoked potential response was absent.
a Sagittal T1-weighted image showing hypoplasia of the corpus callosum (white arrow). b Axial T1-weighted (left) and axial T2-weighted (right) images showing hypoplasia of the corpus callosum and altered, poorly-defined signal in the bilateral subcortical parietal white matter (white arrows). c Coronal T2-weighted image showing hypoplasia of the optic chiasm (white arrow)
Two months later, the patient achieved head control, but was unable to sit unaided. The results of metabolic tests (lactate, pyruvate, creatine kinase, amino acids, organic acids, thiamine, riboflavin, free fatty acids, LCFA, biotinidase, carnitine, glutaric acid, 4-OH butyric acid, purines, and pyrimidines) were normal.
During the first 2 years of life the patient’s growth rate was normal but decreased progressively, with z-scores of + 5.79 (first year) and + 0.85 (second year). After 2 years of age (when growth hormone [GH] begins to play a more important role) the patient’s growth rate dropped markedly between 24 and 30 months (z-score, − 2.1) and between 30 and 36 months (z-score, − 5.05).
Analysis of endocrine status revealed that at baseline (without stimulus) the patient’s hormone levels were within the normal range for the thyroid (TSH, T4), growth (IGF1, IGFBP3) and gonadal (FSH, LH) axes.
At 2 years of age the patient was unable to maintain eye contact and had not developed language skills. He was unable to sit unaided until the age of 3. At this point, brain MRI showed bilateral optic nerve hypoplasia and confirmed hypoplasia of the corpus callosum (Fig. 1). An echocardiogram performed to evaluate congenital heart defects revealed interventricular septum hypertrophy without obstruction.
Methods
Next-generation sequencing analysis
Targeted enrichment was performed using in-solution hybridization technology (Sure Select XT; Agilent Technologies; Santa Clara, California) and the MiSeq platform (Illumina; San Diego, California) was used for subsequent sequencing. A custom Sure Select probe library was designed to capture the exons and exon-intron boundaries of 166 genes (list available on request). This list included genes associated with all forms of brain morphogenesis defects identified in searches of the Online Mendelian Inheritance in Man (OMIM) database and recently published studies. To ensure optimal representation of all regions of interest, we designed different subgroups of probes according to guanine-cytosine content and location with respect to repetitive sequences [5]. Sequence capture, enrichment, and elution were performed according to the manufacturer’s instructions. Captured fragments were sequenced in pair-end 100-base mode using the MiSeq platform. Image analysis and processing of fluorescence intensity in sequences (“base calling”) was performed using Real Time Analysis (RTA) software v.1.8.70 (Illumina), and the FastQC v0.10.1 program [6] was used for data quality control. Reads were aligned to the reference genome GRCh37 with BWA v0.7.9a [7]. NGSrich v0.7.5 software [8] was used for quality control prior to variant detection, and BEDTools 2.17.0 [9] and Picard 1.114 [10] for the intermediate steps. VarScan v.2.3.6 [11] and SAMtools v0.1.19 [12] software were used for variant detection and SnpEff v4.3 for variant annotation [13]. Prioritization was based on the frequency of variants (MAF < 0.01) in 1000 genomes, dbSNP 137, Exome Aggregation Consortium ExAC, the exome variant server (EVS), the Genome Aggregation Database (gnomAD), and our in-house population database. Possible variant pathogenicity was assessed using CONDEL [14], GERP++ [15], and Human Splicing Finder v.HSF3.0 [16].
To rule out the presence of other possible variants that could potentially mimic the patient’s phenotype, we performed a wider scope panel (Neuroexome), which sequenced a total of 1860 genes (list available on request) implicated in rare metabolic and neurological diseases. We detected no variants that could be potentially associated with our patient’s phenotype.
mRNA expression studies
Total RNA was extracted from T-cells and reverse-transcribed using M-MLV Reverse Transcriptase (Invitrogen, San Diego, CA, USA). The resulting cDNA was amplified by PCR using primer pairs (forward, 5′- CAAGGTGGACATCAACACAG-3′; reverse, 5′- GTCACCTGGGCTGTCATAT-3′) corresponding to sequences located in exons 38 and 40 of the FLNA gene. PCR fragments were separated by agarose gel electrophoresis and sequenced by Sanger sequencing.
Results
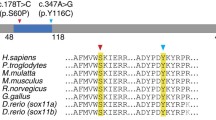
Next-generation sequencing detected a rare intronic variant c.6355 + 4_6355 + 5delAG in hemizygous state in the FLNA gene (reference sequence NM_001456.3). This variant was not present in any public variant databases, nor in any of the 1084 chromosomes from our in-house database. The variant consists of the deletion of 2 nucleotides in positions + 4 and + 5 of intron 38. Given its location in a region that is potentially sensitive to splicing, in silico analysis was performed using Human Splicing Finder (HSF v3.0) software to evaluate the possible effect of the variant on FLNA mRNA splicing. The results of the in silico analysis indicate that 2 powerful exonic acceptor splice sites created at the beginning of intron 38 of FLNA likely hinder correct splicing (Fig. 2). This explains the retention of intron 38 (or part thereof) in the 3 mutant transcripts.
The patient’s variant was inherited, and it was identified as a de novo mutation in the patient’s mother. The patient’s sister is a heterozygous carrier of the variant.
cDNA studies were conducted to determine whether the variant affected splicing. RT-PCR amplification revealed the total absence of wild type cDNA in the patient (A band) and 3 mutated cDNAs, all of which appear to give rise to a premature stop codon. In the most abundant mutant transcript (B band) intron 38 is retained while in the longest transcript (D band) introns 38 and 39 are retained (Fig. 3). In the smallest cDNA (C band) approximately half of intron 38 is present but exon 39 is absent. cDNA analysis of the patient’s mother revealed the presence of wild type mRNA, as well as the same 3 mutated transcripts found in the patient, albeit to a lesser extent. cDNA PCR was performed under different conditions to determine whether there was any variation in the PCR products obtained. In all cases similar results were obtained.
a Electrophoresis of cDNAs from the FLNA genes of a healthy control, the patient’s mother (heterozygous for the FLNA variant), and the patient (hemizygous). b Different FLNA splicing isoforms found in the patient: the A-band corresponds to wild type cDNA, the B-band includes intron 38; the C-band includes a portion of intron 38 but not exon 39; and the D-band includes introns 38 and 39
Discussion & Conclusions
SOD-associated mutations have been identified in genes that encode transcription factors essential for normal forebrain/pituitary development, such as HESX1, SOX2, SOX3, OTX2, TCF7L1 or TAX1BP3 implicated in regulation of the Wnt/β-catenin signaling pathway [4, 17,18,19,20,21,22,23]. Mutations in genes implicated in Kallmann syndrome have also been recently described in SOD patients. These include fibroblast growth factor receptor 1 (FGFR1), the ligand FGF8, and PROKR2 [24,25,26], all of which are involved in the maturation of gonadotrophin-releasing hormone (GnRH) neurons and/or their migration from the olfactory placode to the ventral forebrain [27, 28]. Nonetheless, the aetiology of SOD remains unclear in the majority of patients.
The X-linked FLNA gene encodes filamin A, a non-muscle actin-binding protein that serves as a molecular scaffold to transduce various receptor and intracellular signals throughout the actin cytoskeleton. It is required for locomotion of many cell types. It is expressed at high levels in the developing cortex, where it contributes to neuronal migration to the cortex, and is also essential for embryogenesis [29,30,31,32,33,34,35,36,37]. As described for many other X-linked genes, there are significant phenotypic differences between males and females with FLNA mutations. Heterozygous truncating mutations in FLNA result in periventricular nodular heterotopia (PVNH) as the principal presenting feature, a disorder in which the migration of neurons to the cerebral cortex is disrupted and neurons persist as nodules lining the ventricular surface. The majority of patients with FLNA-associated PVNH are female, since truncating variants are usually lethal in males [38]. Surviving males with truncating FLNA variants usually have distal or mosaic mutations, whereby some residual protein function is retained [39,40,41,42,43,44]. Females with FLNA-associated PVNH present with seizures and, occasionally, subtle learning disabilities [39], as well as other clinical signs including vascular and cardiac defects and congenital intestinal pseudo-obstruction (CIPO), which is characterized by anomalous intestinal smooth muscle layering. Some FLNA-associated PVNH patients show variable soft connective tissue involvement as well as X-linked cardiac valvular dystrophy (XCVD), giving rise to a phenotype known as Ehlers-Danlos syndrome (EDS) with periventricular heterotopia [45,46,47,48,49,50]. In a subset of male patients, the clinical presentation is characterized by PVNH accompanied by cardiovascular malformations and/or CIPO [51, 52]. The mechanisms underlying this phenotypic variability remain unclear.
Localized gain-of-function variants in FLNA lead to otopalatodigital (OPD) spectrum disorders, characterized primarily by skeletal dysplasia including otopalatodigital syndrome types I (OPD1) and II (OPD2), Melnick-Needles syndrome (MNS), frontometaphyseal dysplasia (FMD1) and terminal osseous dysplasia (TODPD), affecting both males and females (except for TODPD which has only been described in females) [53,54,55,56,57]. Several of these mutations are recurrent, and all are clustered in 4 regions of the gene: the actin-binding domain and rod domain repeats 3, 10, and 14/15. The patterns of mutation, X-chromosome inactivation, and phenotypic manifestations in this class of mutations indicate gain-of-function effects, implicating filamin A in signalling pathways that mediate organogenesis in multiple systems during embryonic development.
Our patient was diagnosed with SOD based on the presence of 2 out of 3 of the classic triad features (ONH, pituitary hormone abnormalities, and midline brain defects). He exhibits bilateral optic nerve hypoplasia, ocular motility disturbances, and midline brain defects with neonatal hypoglycaemia, but no clear endocrinological defects to date.
The patient carries a hemizygous splicing mutation inherited from his mother, in whom this variant arose as a consequence of a de novo event. This explains the lack of other cases in the patient’s family as well as the deleterious nature of the mutation. The results of our in-silico variant evaluation and mRNA studies indicate that the mutation affects FLNA splicing. PCR amplification revealed three mutant FLNA transcripts and the total absence of wild type cDNA. In the most abundant mutant transcript (B band) intron 38 is retained and a premature stop codon emerges in exon 39. In the longest transcript (D band), in which introns 38 and 39 are retained, a stop codon emerges in intron 38. In the smallest cDNA (C band), in which approximately half of intron 38 is present but exon 39 is skipped, a stop codon appears in exon 40. cDNA analysis of the patient’s mother revealed the presence of wild type mRNA, as well as the 3 same mutated transcripts found in the patient, albeit to a lesser extent, which probably explains the absence of disease in both the mother and sister. The survival of the patient suggests that the novel transcripts generated by this variant retain some residual protein function. Our patient also presents interventricular septum hypertrophy and limb anomalies, also characteristic of patients with FLNA mutations. Taken together, these findings strongly suggest that our patient’s variant is implicated in the clinical picture. Nonetheless, further functional studies will need to be performed in order to better elucidate the effect of this mutation.
In summary, we describe for the first time a case of SOD caused by an underlying FLNA mutation. This finding broadens the spectrum of genetic causes of this complex pathology and expands the phenotypic spectrum of FLNA gene mutations. Early diagnosis will be useful to provide prospective parents with genetic counselling regarding future pregnancies.
Availability of data and materials
The principal data generated and/or analyzed in this study are included in the published article. The corresponding datasets are available from the corresponding author on request.
Abbreviations
- ACTH:
-
Adrenocorticotropic hormone
- cDNA:
-
Complementary DNA
- CIPO:
-
Congenital intestinal pseudo-obstruction
- EDS:
-
Ehlers-Danlos syndrome
- LCFA:
-
Long chain fatty acid
- MRI:
-
Magnetic resonance imaging
- ONH:
-
Optic nerve hypoplasia
- OPD:
-
Otopalatodigital
- PCR:
-
Polymerase chain reaction
- PVNH:
-
Periventricular nodular heterotopia
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- SD:
-
Standard deviation
- SOD:
-
Septo-optic dysplasia
- TODPD:
-
Terminal osseous dysplasia with pigmentary defects
- XCVD:
-
X-linked cardiac valvular dystrophy
References
Kelberman D, Dattani MT. Genetics of septo-optic dysplasia. Pituitary. 2007;10:393–407.
Kelberman D, Dattani MT. Septo-optic dysplasia - novel insights into the aetiology. Horm Res. 2008;69:257–65.
Kelberman D, de Castro SC, Huang S, et al. SOX2 plays a critical role in the pituitary, forebrain and eye during human embryonic development. J Clin Endocrinol Metab. 2008;93:1865–73.
McCabe MJ, Alatzoglou KS, Dattani MT. Septo-optic dysplasia and other midline defects: the role of transcription factors: HESX1 and beyond. Best Pract Res Clin Endocrinol Metab. 2011;25(1):115–24.
Agilent SureDesign [Internet]. [cited 2018 Oct 11]. Available from: https://earray.chem.agilent.com/suredesign/
Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data [Internet]. [cited 2018 Oct 11]. Available from: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25(14):1754–60.
NGSrich [Internet]. SourceForge. [cited 2018 Oct 11]. Available from: https://sourceforge.net/projects/ngsrich/
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2.
Picard Tools - By Broad Institute [Internet]. [cited 2018 Oct 11]. Available from: http://broadinstitute.github.io/picard/
Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25(17):2283–5.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9.
Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w 1118 ; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92.
González-Pérez A, López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score. Condel Am J Hum Genet. 2011;88(4):440–9.
Cooper GM. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15(7):901–13.
Desmet F-O, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67.
Dattani MT, Martinez-Barbera JP, Thomas PQ, et al. Mutations in the homeobox gene HESX1/Hesx1 associated with septo-optic dysplasia in human and mouse. Nat Genet. 1998;19:125–33.
Kelberman D, Rizzoti K, Avilion A, et al. Mutations within Sox2/SOX2 are associated with abnormalities in the hypothalamo-pituitary-gonadal axis in mice and humans. J Clin Invest. 2006;116:2442–55.
Woods KS, Cundall M, Turton J, et al. Over- and underdosage of SOX3 is associated with infundibular hypoplasia and hypopituitarism. Am J Hum Genet. 2005;76:833–49.
Dateki S, Kosaka K, Hasegawa K, et al. Heterozygous orthodenticle homeobox 2 mutations are associated with variable pituitary phenotype. J Clin Endocr Metab. 2010;95:756–64.
Webb EA, Dattani MT. Septo-optic dysplasia. Eur J Hum Genet. 2010;18(4):393-7.
Gaston-Massuet C, McCabe MJ, Scagliotti V, Young RM, Carreno G, Gregory LC, et al. Transcription factor 7-like 1 is involved in hypothalamo-pituitary axis development in mice and humans. Proc Natl Acad Sci U S A. 2016;113(5):E548–57.
Reinstein E, Orvin K, Tayeb-Fligelman E, Stiebel-Kalish H, Tzur S, Pimienta AL, et al. Mutations in TAX1BP3 cause dilated cardiomyopathy with septo-optic dysplasia. Hum Mutat. 2015;36(4):439–42.
McCabe MJ, Gaston-Massuet C, Tziaferi V, et al. Novel FGF8 mutations associated with recessive holoprosencephaly, craniofacial defects, and hypothalamo-pituitary dysfunction. J Clin Endocrinol Metab. 2011;96:E1709–18.
McCabe MJ, Gaston-Massuet C, Gregory LC, et al. Variations in PROKR2, but not PROK2, are associated with hypopituitarism and septo-opticdysplasia. J Clin Endocrinol Metab. 2013;98:E547–57.
Raivio T, Avbelj M, McCabe MJ, et al. Genetic overlap in Kallmann syndrome, combined pituitary hormone deficiency, and septo-opticdysplasia. J Clin Endocrinol Metab. 2012;97:E694–9.
Pitteloud N, Zhang C, Pignatelli D, et al. Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. PNAS USA. 2007;104:17447–52.
Falardeau J, Chung WC, Beenken A, et al. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest. 2008;118:2822–31.
Lian G, Lu J, Hu J, et al. Filamin a regulates neural progenitor proliferation and cortical size through Wee1-dependent Cdk1 phosphorylation. J Neurosci. 2012;32:7672–84.
Huelsmann S, Rintanen N, Sethi R, Brown NH, Ylänne J. Evidence for the mechanosensor function of filamin in tissue development. Sci Rep. 2016;6:32798.
Lad Y, Kiema T, Jiang P, et al. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. EMBO J. 2007;26:3993–4004.
Liu J, Das M, Yang J, et al. Structural mechanism of integrin inactivation by filamin. Nat Struct Mol Biol. 2015;22:383–9.
Nakamura F, Song M, Hartwig JH, Stossel TP. Documentation and localization of force-mediated filamin A domain perturbations in moving cells. Nat Commun. 2014;5:4656.
Popowicz GM, Schleicher M, Noegel AA, Holak TA. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem Sci. 2006;31:411–9.
Fox JW, Lamperti ED, Ekşioğlu YZ, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–25.
Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–8.
Robertson SP, Twigg SR, Sutherland-Smith AJ, et al. Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet. 2003;33:487–91.
Unger S, Mainberger A, Spitz C. Filamin A mutation is one cause of FG syndrome. Am J Med Genet A. 2007;143a:1876–9.
Parrini E, Ramazzotti A, Dobyns WB, et al. Periventricular heterotopia: phenotypic heterogeneity and correlation with Filamin a mutations. Brain. 2006;129:1892–906.
Sheen VL, Dixon PH, Fox JW, et al. Mutations in the X-linked filamin 1 gene cause periventricular nodular heterotopia in males as well as in females. Hum Mol Genet. 2001;10:1775–83.
Guerrini R, Mei D, Sisodiya S, et al. Germline and mosaic mutations of FLN1 in men with periventricular heterotopia. Neurology. 2004;63:51–6.
Hehr U, Hehr A, Uyanik G, Phelan E, Winkler J, Reardon W. A filamin a splice mutation resulting in a syndrome of facial dysmorphism, periventricular nodular heterotopia, and severe constipation reminiscent of cerebro-fronto-facial syndrome. J Med Genet. 2006;43:541–4.
Berrou E, Adam F, Lebret M, et al. Gain-of-function mutation in Filamin a potentiates platelet integrin αIIbβ3 activation. Arterioscler Thromb Vasc Biol. 2017;37:1087–97.
Oegema R, Hulst JM, Theuns-Valks SD, et al. Novel no-stop FLNA mutation causes multi-organ involvement in males. Am J Med Genet A. 2013;161A:2376–84.
Ritelli M, Morlino S, Giacopuzzi E, et al. Ehlers-Danlos syndrome with lethal cardiac valvular dystrophy in males carrying a novel splice mutation in FLNA. Am J Med Genet A. 2017;173:169–76.
Hommel AL, Jewett T, Mortenson M, Caress JB. Juvenile muscular atrophy of the distal upper extremities associated with x-linked periventricular heterotopia with features of Ehlers-Danlos syndrome. Muscle Nerve. 2016;54(4):794–7.
Reinstein E, Frentz S, Morgan T, et al. Vascular and connective tissue anomalies associated with X-linked periventricular heterotopia due to mutations in Filamin a. Eur J Hum Genet. 2013;21:494–502.
Sheen VL, Walsh CA. Periventricular heterotopia: new insights into Ehlers-Danlos syndrome. Clin Med Res. 2005;3:229–33.
Sheen VL, Jansen A, Chen MH, et al. Filamin A mutations cause periventricular heterotopia with Ehlers-Danlos syndrome. Neurology. 2005;64:254–62.
Antonucci A, Fronzoni L, Cogliandro L, et al. Chronic intestinal pseudo-obstruction. World J Gastroenterol. 2008;14:2953–61.
De Giorgio R, Cogliandro RF, Barbara G, Corinaldesi R, Stanghellini V. Chronic intestinal pseudo-obstruction: clinical features, diagnosis, and therapy Gastroenterol. Clin North Am. 2011;40:787–807.
Stefanova M, Meinecke P, Gal A, Bolz H. A novel 9 bp deletion in the filamin a gene causes an otopalatodigital-spectrum disorder with a variable, intermediate phenotype. Am J Med Genet A. 2005;132A:386–90.
Hidalgo-Bravo A, Pompa-Mera EN, Kofman-Alfaro S, Gonzalez-Bonilla CR, Zenteno JC. A novel filamin a D203Y mutation in a female patient with otopalatodigital type 1 syndrome and extremely skewed X chromosome inactivation. Am J Med Genet A. 2005;136:190–3.
Zenker M, Nährlich L, Sticht H, Reis A, Horn D. Genotype-epigenotype-phenotype correlations in females with frontometaphyseal dysplasia. Am J Med Genet A. 2006;140:1069–73.
Kondoh T, Okamoto N, Norimatsu N, Uetani M, Nishimura G, Moriuchi H. A Japanese case of Oto-palato-digital syndrome type II: an apparent lack of phenotype-genotype correlation. J Hum Genet. 2007;52:370–3.
Mariño-Enríquez A, Lapunzina P, Robertson SP, Rodríguez JI. Otopalatodigital syndrome type 2 in two siblings with a novel filamin A 629G>T mutation: clinical, pathological, and molecular findings. Am J Med Genet A. 2007;143A(10):1120–5.
Sun Y, Almomani R, Aten E, et al. Terminal osseous dysplasia is caused by a single recurrent mutation in the FLNA gene. Am J Hum Genet. 2010;87:146–53.
Acknowledgements
The authors are indebted to the patient’s family for their collaboration.
Funding
This work was supported by Instituto de Salud Carlos III Spain (FIS-PI13/02177 and JR13/0019).
Author information
Authors and Affiliations
Contributions
MSPP: performed clinical evaluations, conducted molecular tests, and drafted the clinical portion of the manuscript. AF: performed mRNA experiments and Sanger cDNA sequencing. EML, MTM, MJCP, MLC: performed clinical evaluations and collected clinical data. AFM: performed genetic analyses, drafted the genetic portion of the manuscript, edited the final manuscript, and coordinated the study. The submitted version of the manuscript has been approved by all authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All authors declare that they obtained written consent from the parents/legal guardians for all genetic studies.
Consent for publication
Written consent to publication of clinical, radiological, and biological data was obtained from the parents/legal guardians of the patient.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Fernández-Marmiesse, A., Pérez-Poyato, M.S., Fontalba, A. et al. Septo-optic dysplasia caused by a novel FLNA splice site mutation: a case report. BMC Med Genet 20, 112 (2019). https://doi.org/10.1186/s12881-019-0844-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-019-0844-5