Abstract
Background
Rubinstein-Taybi syndrome (RSTS; OMIM #180849, #613684) is a rare autosomal dominant genetic condition characterized by broad thumbs and halluces, facial dysmorphism, short stature and variable degree of intellectual disability. RSTS is associated with mutations in CREBBP and EP300 genes in 50–60% and 5–8% of cases, respectively. The majority of cases are de novo heterozygous mutations.
Case presentation
Here we describe a familial RSTS case, associated with a novel EP300 mutation. The proband was a 9 years old female, with mild learning difficulties. Her mother, who also had learning difficulties, was found to have short and broad thumbs. MLPA and panel-based NGS of CREBBP and EP300 were performed. A novel heterozygous frameshift mutation in exon 31 of the EP300 gene (c.7222_7223del; p.(Gln2408Glufs*39)) was found in both.
Conclusions
This case represents the first case of inherited EP300-RSTS. The location of the frameshift deletion not affecting HAT domain and PHD finger, could explain the mild phenotype and the well-preserved intelligence. These patients are mildly affected, and this case highlights the possible missed diagnosis. We would recommend molecular testing of apparently healthy parents, and in the case of inherited mutations, of all adult first degree relatives at risk.
Similar content being viewed by others
Background
Rubinstein-Taybi syndrome (RSTS; OMIM #180849, #613684) is a rare (1:125000) complex neurodevelopmental disorder characterized by broad thumbs and halluces, facial abnormalities (downslanted palpebral fissures, low hanging columella, high palate, grimacing smile, and talon cusps), postnatal growth delay and intellectual disability (ID) [2, 10]. In addition, RSTS patients have a slightly increased predisposition to cancer [9]. RSTS is an autosomal dominant condition and the vast majority of cases occur sporadically due to de novo heterozygous mutations, being vertical transmission extremely rare [7]. So far, Bartsch et al. [3] described only five familial cases of RSTS, consistent with autosomal dominant inheritance, and recently another possible case has been described in India [15].
Two genes have been implicated with RSTS: CREBBP, located on chromosome 16p13.3, encoding a CREB-binding protein (CBP), and EP300, which maps to 22q13.2 and encodes E1A-associated protein p300. They are very similar and function as transcriptional coactivators in the regulation of gene expression mediating many of the same signalling pathways. CBP and p300 are highly conserved and have potent histone acetyltransferase (HAT) activity. Consequently, there is a direct link between loss of acetyltransferase activity and RSTS suggesting an aberrant chromatin regulation [17, 18]. Despite their high sequence similarity (>70%), CBP and p300 have differences in their respective functions [1]. RSTS is caused in approximately 50–60% of cases by mutations of the CREBBP gene (RSTS 1, OMIM #180849), and by EP300 gene mutations (RSTS 2, OMIM #613684) in 5–8%. To date, 246 disease causing mutations in the CREBBP gene have been reported to cause RSTS [7]. In contrast, only 23 RSTS patients with EP300 mutations, and a total of 34 mutations, have been described in this gene [10, 14]. The mutations described in both range from point mutations to whole gene deletions and chromosome rearrangements, and spread across the entire length of the genes.
Several reasons are being postulated for the lower detection rate of EP300 mutations: a lower mutation rate; underdiagnosis at similar mutation rates following the generally milder phenotype in cases of RSTS with EP300 mutation; or misdiagnosis in cases with severe phenotype resembling other syndromes with congenital malformations such as Cornelia de Lange syndrome (CdLS) which interestingly is caused by alterations in other chromatin associated proteins [1, 9, 13, 17].
In this case report we describe an inherited RSTS case associated with a novel EP300 mutation, representing the first familial case found in Spain.
Case presentation
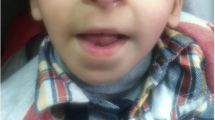
Proband was a 9-year-old girl, referred to the Genetics Clinic by the neuropediatrician who had been following her for mild learning difficulties. RSTS was suspected after seeing her mother, who also had learning difficulties and had short broad thumbs. Our patient was born at term via Cesarean section (pelvi-fetal disproportion) after an uneventful pregnancy. Both, weight and length at birth were low: 2.3 kg (−2.38 SD) and 46 cm (−2.07 SD), respectively. Occipitofrontal head circumference (OHC) at birth was not recorded. The girl’s development was within normal range in the first years of life but increasing learning difficulties were noted at primary school. She is at her expected grade with educational support. On physical examination the proband’s weight was 21.4 kg (−1.74 SD), her length 119 cm (−3.15 SD on Spain 2000 growth chart), and her OHC 48.7 cm (−2.9 SD). She had a thickened and low hanging columella. Her hands and feet (thumbs and halluces) were quite normal (Fig. 1. Phenotype table (Additional file 1)).
Images showing classical RSTS features of patient including thickened and low hanging columella (a, b), and detail of mother’s talon cusp at an upper incisor (c). Photographs showing normal thumbs and halluces of the proband (d, e), short and broad but not angulated thumbs of her mother (f, g) and her grandmother (h, i)
The mother (42 y.o.) had short stature (141 cm (−3.87 SD)) and proportionately small OHC (49 cm (−4 SD)). She had struggled at school but managed to get the Secondary School Certificate. Nowadays she is working as a check-out in a supermarket. Her general health is good. She is a single mother of two children: the proband and a healthy 7 year old boy. She had slightly posteriorly rotated ears, a prominent nose with high nasal bridge, low hanging columella, and talon cusp at an upper incisor. Her thumbs were short and broad but not angulated and her halluces were normal (Fig. 1. Phenotype table (Additional file 1)). Proband’s maternal aunt and grandmother also had short stature and proportionate OHC but normal intelligence and normal hands and feet. The grandmother’s length was 142.1 cm (−3.5 SD) and her OHC 52.5 cm (−1.9 SD). Maternal grandfather died aged 50 from liver disease.
Previous assessments of the proband included a hand X-ray for bone age, which was according to chronological age, and endocrine tests with normal levels of growth hormone in two stimulation tests. A brain MRI scan showed a pineal cyst (9 × 9 × 5 mm) and no other anomalies.
Clinical data, samples and photographs were obtained after written informed consent. This work has been approved by the Committee for Ethics in Clinical Research in La Rioja (CEICLAR).
Blood samples from the proband, her mother and her grandmother were collected in EDTA tubes. DNA was extracted using QIAamp DNA Mini Kit (QIAGEN) following the manufacture’s protocol. MLPA of CREBBP and EP300 was performed (P313 and P333 Kit, MRC-Holland). Panel-based next generation sequencing (NGS) of CREBBP and EP300 genes was carried out. Briefly, libraries encompassing exons and introns of CREBBP and EP300 genes were prepared using the SureSelectXT2 Custom kit (Agilent) and sequenced to generate 150 bp single reads. The resulting reads were mapped to the human genome hg19 using BWA (version 0.7.12). Sequence variants were called using the Genome Analysis Toolkit (GATK) version 3.3 and called variants were annotated with Annovar. ExAC browser of Broad Institute, 1000 Genomes database and dbSNP138, as well as, the Human Gene Mutation Database (HGMD), Leiden Open Variation Database (LOVD) and ClinVar databases were checked to assess the presence/absence of detected alterations in variations repositories.
Results and discussion
MLPA did not reveal any deletion within the CREBBP or EP300 genes. Only one variant was identified in the panel-based NGS of CREBBP and EP300 genes: a heterozygous mutation in exon 31 of the EP300 gene (RefSeq NM_001429.3: c.7222_7223del; p.(Gln2408Glufs*39)). As in the proband, this variant was also found in the heterozygous state in the mother as is shown by the number of reads (238 reads with deletion of a total of 486, for the proband; and 219/485 for her mother). This finding was confirmed by Sanger sequencing. This variant was not found in 100 healthy controls and it is not present in 1000G, ExAC and dbSNP, indicating that this variant is not common in population. To the best of our knowledge this variant has not been previously described, and it is not included in ClinVar, HGMD or LOVD. The variant has now been included in ClinVar database (SCV000266471) and in LOVD (individual #00064625). The deletion generates a frameshift that leads to loss of the original stop codon and results in a prolonged protein 31 aminoacids longer. This variation is considered to be pathogenic/likely pathogenic, according to ACMG interpretation: null variant in a gene where LOF is a known mechanism of disease, absent in population databases, protein length changing variant, and patient’s phenotype highly specific for gene [12].
According to the literature, point mutations are widespread along the whole EP300 gene without any remarkable hot spot, being frameshift mutations the most common found, as is our case [10, 14]. Likewise, genotype-phenotype correlations indicated that patients with larger deletions did not always have a more severe phenotype than those with smaller deletions or point mutations. On the other hand, it seems that mutations that do not alter HAT domain could explain non-classical RSTS cases [3, 18]. In this sense, the location of the frameshift deletion described in this report, very close to the 5′-end of EP300 gene, not affecting HAT domain and PHD finger, could explain the mild phenotype and the well-preserved intelligence.
Addressing the lower frequency of EP300 mutations (EP300 mutations are 10 times less frequent than CREBBP mutations), it may be possible that EP300 gene had a lower mutation rate than CREBBP, nevertheless, many more polymorphisms have been found in this gene, including some that lead to amino acid changes [13]. Hence, EP300 mutations might account for a higher frequency of RSTS patients than previously thought. However, because of phenotypic variability such patients are not ascertained and not studied.
Molecular analysis was performed in mother and grandmother and it was positive in the mother and negative in the grandmother, confirming the inherited character of this mutation, suggesting three possible options: 1) the mutation appeared de novo in the mother and was passed on to her daughter, 2) the grandmother could have germline mosaicism, that could not be corroborated or 3) the grandfather, who presented broad thumbs, could also be a carrier of the mutation. Thereby, despite the inherited confirmation of this RSTS case, it has not been possible to determine the exact point where the mutation appeared.
Familial RSTS is extremely rare, in this regards, between 1000 and 2000 cases of RSTS have been reported but only 11 cases of familial RSTS have been described [3–6, 8, 11], and are most likely linked to somatic mosaicism. Moreover, in all the cases in which the molecular cause was determined, it was associated with the CREBBP gene. Recently, Tamhankar et al., reported a patient with mutation in EP300 gene inherited from a healthy mother [15]. Also Negri et al., and Wincent et al., [10, 16] found a missense variation in both, a patient and his healthy mother and father, respectively, but both were considered as not pathogenic. Although Tamhankar et al. suggested the possibility of low penetrance, these variants should be considered not pathogenic, since they have been detected in healthy parents. Moreover, only one EP300 missense mutation has been previously observed in RSTS patients [10], indicating that this type of change is not probable as causative of RSTS.
Affected children tend to have more pronounced features than their affected parent in familial RSTS [3]. Conversely, in our case it was the mother who showed the characteristic thumbs, facial features and dental anomalies, giving rise to the suspicion of RSTS diagnosis. On the other hand, the familial short stature with proportionately small HC turned out not to segregate with the EP300 mutation and may therefore be associated with other genetic factors not studied.
This study broadens the number of phenotypic EP300-patients described to 24 providing additional knowledge about the phenotype of these RSTS-2 cases. In general, this cohort presents mild presentation of the phenotype, and also few phenotypic peculiarities, what is attested by the age at diagnosis, usually higher in EP300 patients [9, 10, 14]. Our observations are in accordance with this, since the proband was diagnosed at the age of 9, and her mother was not diagnosed, until this study.
According to the literature, these patients usually present a mild presentation of the syndrome. It is frequently reported in these RSTS-2 patients classic facial RSTS features (prominent nose with low-hanging nasal septum, downward slanting palpebral fissures, etc.); variable degree of intellectual disability, ranging from mild to moderate (no patient has a severe impairment); mild skeletal phenotype, even if there are cases described with normal thumbs and toes, it is common to find broad thumbs and big toes although with no radial deviation of them [1, 10, 16]. In our case, mother and proband showed mild learning difficulties, but the mother was able to lead a normal life. Both showed facial characteristics of RSTS although they were more evident in the mother. Also, the mother had broad but not angulated thumbs, whereas the child had normal thumbs and halluces.
Conclusion
In summary, this study presents the first case of inherited EP300-RSTS. This report underscores the possible lack of diagnosis in these patients with non-classic presentations, as is the case of patient’s mother. Therefore, variable clinical expression exists in RSTS and normal functioning individuals may carry disease-associated mutations, even in a non-mosaic state. These findings have implications for genetic counselling and we would recommend molecular testing of apparently healthy parents, and in the case of inherited mutations, of all adult first degree relatives at risk. Thus, it is important to highlight that since most individuals with RSTS due to EP300 mutation are mildly affected, this entity is likely underdiagnosed and special attention should be paid.
Abbreviations
- ACMG:
-
American College of Medical Genetics and Genomics
- CBP:
-
CREB-binding protein
- CdLS:
-
Cornelia de Lange syndrome
- CEICLAR:
-
Committee for ethics in clinical research in La Rioja
- GATK:
-
Genome analysis toolkit
- HAT:
-
Histone acetyltransferase
- HGMD:
-
Human gene mutation database
- ID:
-
Intellectual disability
- LOVD:
-
Leiden open variation database
- MRI:
-
Magnetic resonance imaging
- NGS:
-
Next generation sequencing
- OHC:
-
Occipitofrontal head circumference
- PHD:
-
Plant homeodomain
- RSTS:
-
Rubinstein-Taybi syndrome
References
Bartholdi D, Roelfsema JH, Papadia F, Breuning MH, Niedrist D, Hennekam RC, et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: delineation of the phenotype of the first patients carrying mutations in EP300. J Med Genet. 2007;44(5):327–33.
Bartsch O, Labonté J, Albrecht B, Wieczorek D, Lechno S, Zechner U, et al. Two patients with EP300 mutations and facial dysmorphism different from the classic Rubinstein -Taybi syndrome. Am J Med Genet. 2010;152A(1):181–4.
Bartsch O, Kress W, Kempf O, Lechno S, Haaf T, Zechner U. Inheritance and variable expression in Rubinstein-Taybi syndrome. Am J Med Genet A. 2010;152A(9):2254–61.
Chiang PW, Lee NC, Chien N, Hwu WL, Spector E, Tsai AC. Somatic and germ-line mosaicism in Rubinstein-Taybi syndrome. Am J Med Genet A. 2009;149A(7):1463–7.
Cotsirilos P, Taylor JC, Matalon R. Dominant inheritance of a syndrome similar to Rubinstein-Taybi. Am J Med Genet. 1987;26(1):85–93.
Hennekam RC, Lommen EJ, Strengers JL, Van Spijker HG, Jansen-Kokx TM. Rubinstein-Taybi syndrome in a mother and son. Eur J Pediatr. 1989;148(5):439–41.
Kamenarova K, Simeonov E, Tzveova R, Dacheva D, Penkov M, Kremensky I, et al. Identification of a novel de novo mutation of CREBBP in a patient with Rubinstein-Taybi syndrome by targeted next-generation sequencing: a case report. Hum Pathol. 2016;47(1):144–9.
Marion RW, Garcia DM, Karasik JB. Apparent dominant transmission of the Rubinstein-Taybi syndrome. Am J Med Genet. 1993;46(3):284–7.
Negri G, Milani D, Colapietro P, Forzano F, Della Monica M, Rusconi D, et al. Clinical and molecular characterization of Rubinstein-Taybi syndrome patients carrying distinct novel mutations of the EP300 gene. Clin Genet. 2015;87(2):148–54.
Negri G, Magini P, Milani D, Colapietro P, Rusconi D, Scarano E, et al. From Whole Gene Deletion to Point Mutations of EP300-Positive Rubinstein-Taybi Patients: New Insights into the Mutational Spectrum and Peculiar Clinical Hallmarks. Hum Mutat. 2016;37(2):175–83.
Petrij F, Dorsman JC, Dauwerse HG, Giles RH, Peeters T, Hennekam RC, et al. Rubinstein-Taybi syndrome caused by a De Novo reciprocal translocation t(2;16)(q36.3;p13.3). Am J Med Genet. 2000;92(1):47–52.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24.
Roelfsema JH, White SJ, Ariyürek Y, Bartholdi D, Niedrist D, Papadia F, et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am J Hum Genet. 2005;76(4):572–80.
Solomon BD, Bodian DL, Khromykh A, Mora GG, Lanpher BC, Iyer RK, et al. Expanding the phenotypic spectrum in EP300-related Rubinstein-Taybi syndrome. Am J Med Genet A. 2015;167A(5):1111–6.
Tamhankar PM, Merchant R, Shah A. Rubinstein Taybi Syndrome in an Indian Child due to EP300 Gene Mutation. Indian J Pediatr. 2016;83(5):473–4.
Wincent J, Luthman A, van Belzen M, van der Lans C, Albert J, Nordgren A, et al. CREBBP and EP300 mutational spectrum and clinical presentations in a cohort of Swedish patients with Rubinstein-Taybi syndrome. Mol Genet Genomic Med. 2015;4(1):39–45.
Woods SA, Robinson HB, Kohler LJ, Agamanolis D, Sterbenz G, Khalifa M. Exome sequencing identifies a novel EP300 frame shift mutation in a patient with features that overlap Cornelia de Lange syndrome. Am J Med Genet A. 2014;164A(1):251–8.
Zimmermann N, Acosta AM, Kohlhase J, Bartsch O. Confirmation of EP300 gene mutations as a rare cause of Rubinstein-Taybi syndrome. Eur J Hum Genet. 2007;15(8):837–42.
Acknowledgments
We are grateful to the patient and her family for their cooperation and their participation in the study.
Funding
This report did not require any funding source.
Availability of data and materials
The datasets during and/or analysed during the current study available from the corresponding author on reasonable request.
Authors’ contributions
Conception or design of the work: ML, EDG. Data collection: VS, PCC. Data analysis and interpretation: ML, CCA, PS. Drafting the article: ML, EDG, VS. Critical revision of the article: CCA, PS, PCC. Final approval of the version to be published: ML, CCA, PS, VS, PCC, EDG.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Consent to publish from the participant (or legal parent or guardian for children) to report individual patient data has been obtained.
Ethics approval and consent to participate
This work has been approved by the Committee for Ethics in Clinical Research in La Rioja (CEICLAR). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent from all individual participants included in the study and parental consent for their child to participate were obtained.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1:
Phenotype table. Phenotype found in patient and her mother. (XLS 26 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
López, M., Seidel, V., Santibáñez, P. et al. First case report of inherited Rubinstein-Taybi syndrome associated with a novel EP300 variant. BMC Med Genet 17, 97 (2016). https://doi.org/10.1186/s12881-016-0361-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-016-0361-8