Abstract
Background
Peutz–Jeghers syndrome (PJS) is a rare autosomal dominant inherited disorder characterized by gastrointestinal (GI) hamartomatous polyps, mucocutaneous hyperpigmentation, and an increased risk of cancer. Mutations in the serine–threonine kinase 11 gene (SKT11) are the major cause of PJS.
Case presentation
Blood samples were collected from six PJS families including eight patients. Mutation screening of STK11 gene was performed in these six families by Sanger sequencing and multiplex ligation-dependent probe amplification (MLPA) assay. Three novel mutations (c.721G > C, c.645_726del82, and del(exon2–5)) and three recurrent mutations (c.752G > A, c.545 T > C and del(exon1)) in STK11 were detected in six Chinese PJS families. Genotype-phenotype correlations suggested that truncating mutations trend to result in severe complications.
Conclusion
These findings broaden the mutation spectrum of the STK11 gene and would help clinicians and genetic counselors provide better clinical surveillance for PJS patients, especially for ones carrying truncating mutation.
Similar content being viewed by others
Background
Peutz–Jeghers syndrome (PJS) is a rare autosomal dominant inherited disorder characterized by gastrointestinal (GI) hamartomatous polyps, mucocutaneous hyperpigmentation of the lips, buccal mucosa, and digits. PJS polyps often lead to severe complications, such as intussusceptions and intestinal obstruction. Beyond these symptoms, PJS patients also have an increased risk of cancer at multiple sites, including the GI tract, breast, ovary, testis, and lung [1]. The cumulative lifetime risk is 20, 43, 71, and 89 % at ages 40, 50, 60 and 65 years, respectively [2].
Germline mutations in the serine–threonine kinase 11 (STK11) gene on chromosome 19p13.3 were identified as a cause of PJS in 1998 [3, 4]. The gene, 23 kb in size, consists of nine coding exon and one non-coding exon and encodes a 433-amino acid protein, which consists of three domains: the N-terminal non-catalytic domain, the catalytic kinase domain, and the C-terminal non-catalytic regulatory domain. This protein which also acts as a tumor suppressor factor is involved in cell growth, cell polarity, and energy metabolism and plays a pleiotropic role in tumorigenesis [5]. Recent studies showed that mutations in STK11 can be found in 57–88 % of PJS cases, including point mutations and large genomic deletions/insertions/duplications [6–9].
In this study, we report three novel mutations and three recurrent mutations in STK11, which were detected in six Chinese PJS families by Sanger sequencing and the multiplex ligation-dependent probe amplification (MLPA) assay.
Materials and methods
Patients
Eight PJS patients from six unrelated Chinese families were enrolled at the Genetics Clinic of the State Key Laboratory of Medical Genetics (SKLMG). Clinical diagnosis for PJS was based on no less than two of these three criteria: characteristic mucocutaneous pigmentation, hamartomatous polyps, and a family history. 100 healthy Chinese individuals (age range from 17 to 40 years, average 22 years) were recruited as controls. Informed consent for this investigation was obtained from all participating PJS patients and parents, and the principles outlined in the Declaration of Helsinki were followed.
Mutation screening and MLPA assay
Genomic DNA was extracted from peripheral blood by the phenol–chloroform method. The entire coding region and intron–exon boundaries of STK11 (RefSeq NM_000455) were analyzed by direct Sanger sequencing. Functional signification of novel mutations was predicted by two online software programs, PolyPhen-2 software (http://genetics.bwh.harvard.edu/pph2) and Swiss-Model software (http://swissmodel.expasy.org).
The MLPA test kit (SALSA P101-B1 STK11; MRC-Holland, Amsterdam, The Netherlands) was used to detect large genomic deletion/duplication of STK11 in samples with negative Sanger sequencing results. All operations were conducted using an ABI 3100 genetic analyzer (Applied Biosystems, Carlsbad, CA, USA) following the manufacturer’s recommendations. This experiment was done in triplicates.
Case presentation
A total of eight patients (2 females and 6 males) from six unrelated families were involved in this study (Table 1). Three of these families (family 3, 4, and 5) showed autosomal dominant pattern and the remaining families (family 1, 2, and 6) are sporadic (Fig. 1a). The patients’ age at STK11 gene testing ranged from 7 to 38 years old. Five probands in these families underwent the first loparotomy or polypectomy at an earlier age (from 7 to 22 years old), due to abdominal pain or hemafecia. Three out of these patients suffered from some severe complications, intussusceptions and intestinal obstruction. None of these patients enrolled in this study had developed cancer up to the test age except the patient 101 with colonic adenoma. This may be due to a young age (7–45 years old). Detail clinical information is showed in Table 1.
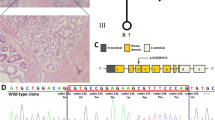
a Pedigrees of family 3, 4 and 5 with PJS showed a autosomal dominant pattern and family 1, 2, and 6 were sporadic. b Sanger sequencing showed four heterozygous mutations, c.721G > C, c.752G > A, c.545 T > C, and c.645_726del. c MLPA assay showed two heterozygous gross deletions, del(exon1) and del(exon2-5)
Results
Direct sequencing of STK11 gene revealed two recurrent mutation (c.525 T > C and c.752G > A) and two novel mutations (c.721G > C, c.645_726del) in four of six families (Table 1; Fig. 1b). Both of these two novel mutations occurred within the highly conserved kinase domain of STK11 (Fig. 2a, b). Mutation c.645_726del is de novo in sporadic family 6 and c.752G > A and c.545 T > C co-segregated with PJS in multiplex family 4 and 5, respectively. The unaffected parents in sporadic family 2 with c.721G > C were not be tested as DNA samples were not available. The novel missense mutations (c.721G > C) results in the substitution of amino acid in codon 241 (p.Ala241Pro) and was predicted to be “probably damaging” by PolyPhen-2 (Additional file 1: Figure S1). The novel 82-base deletion mutation (c.645_726del) in exon 5 of the STK11 gene would cause a frameshift change at codon 215, which would introduce a putative stop at codon 259 (p.Gly215GlyfsX45) with partial loss of the kinase domain and complete loss of the C-terminal of the a-helix (Fig. 2c).
a The structure of STK11 gene. It contains a large kinase domain which spans from exon1 to exon 8. b Evolutionary conservation of amino acid residues altered by c.721G > C (p.Ala241Pro) and c.645_726del (p.Gly215GlyfsX45) across different species. c The mutant protein (p.Gly215GlyfsX45) was predicted to result in partial loss of the kinase domain and complete loss of the C-terminal domain of the a-helix by Swiss-Model online software compared to the wild type. The blue Gly215 indicates the mutant site
Using MLPA analysis, two gross deletions del (exon1) and del(2–5) in the STK11 gene were detected in the remaining two families (Fig. 1c). Deletion of exon 1 detected in patient 101 has been previously reported and is de novo in sporadic family 1. The gross deletion of exon 2–5 found in patient 301 and patient 302 was not found in unaffected members in multiplex family 3, which, as well as novel mutations detected by direct sequencing (c.721G > C and c.645_726del), has not been previously reported in the Human Gene Mutation Database (HGMD), Leiden Open Variation Database (LOVD), NCBI-dbSNP, or Exome Variant Server (EVS). All six variants were not detected in the 100 unrelated normal controls.
Discussion
Currently, 396 mutations in the STK11 gene have been detected in patients with PJS or other disorders (HGMD Professional 2016.2), including missense/nonsense mutations (29.8 %), small deletions/insertions/indels (38.9 %), gross deletions/insertions/duplications (20.5 %), splice-site mutations (9.8 %), and complex rearrangements (1.0 %). Many mutations would influence kinase activity of the STK11 protein by preventing it from binding with MO25 and STRAD [10]. STK11 (+/−) mice in previous studies were found to develop gastrointestinal hamartomatous polyposis, suggesting that haploinsufficiency of STK11 is a mechanism of PJS [11].
In the present study, among the six mutations detected in six families with PJS, del(exon1) in family 1, c.752G > A in family 4, and c.545 T > C in family 5 have been previously reported [7, 12, 13]. In family 2, novel missense mutation c.721G > C was detected. Its mutant residue p.Ala241Pro is close to previously identified PJS-causing mutations c.724G > T (p.Gly242Trp) and c.751G > A (p.Gly251Ser), affecting highly conserved residues [12, 14]. The residue altered by p.Ala241Pro are located in the kinase core domain, in which mutations would lead to a loss of the kinase activity and disrupt the function of the STK11 protein, indicating that p.Ala241Pro is most likely disease-causing [15]. In addition, a prediction from PolyPhen-2 online software strongly suggested that it is a pathogenic mutation. Patient 601 in family 6 with typical PJS phenotype carried a novel frameshift mutation c.645_726del (p.Gly215GlyfsX45), which was predicted to generate a prematurely terminated protein with a partial loss of the kinase domain and a complete absence of the C–terminal. Beyond a loss of kinase activity, the truncated protein p.Gly215GlyfsX45 would impair STK11 polarizing activity and the STK11-mediated activation of the AMPK pathway [16]. Moreover, an absence of autophosphorylation and phosphorylation sites Thr336 and Ser428 would disrupt the cell growth suppressive capacity of STK11 protein [5]. In family 3, a novel gross deletion of exon 2–5 were identified in patient 301 and 302. This deletion would be pathogenic as deletions of exon 2–3 and exon 4–5 have been reported to cause a skip of partial exons within the kinase domain [3, 17], which would destroy the structure of STK11 protein and affect its stability by preventing the binding of Hsp90 and Cdc37 [18]. The mutations c.645_726del, del(exon1) and del(exon2–5), as well as previous indentified PJS-causing deletions [7], further demonstrates that haploinsufficiency of STK11 is a mechanism of PJS [11].
In our study, the probands in these six families exhibited the characteristic phenotypes of PJS, among which patient 101, 301, and 601 had complications intussusceptions and intestinal obstruction. As we know, the relationship between the types and sites of variants in STK11 and the phenotypes of PJS cases has been investigated in several studies. On one hand, individuals with missense mutations in STK11 typically have a later onset for PJS symptoms [19], but have cancer risks similar to the ones with truncating mutation [20]. In our study, the patients carrying truncating mutations (patient 101, 301, and 601) underwent the first polypectomy of GI polyps and related complications at earlier age (average age = 8.3, standard deviation = 1.2) than the ones with missense mutations (patient 401 and patient 501, average age = 14.5, standard deviation = 7.5). Moreover, we found that more individuals with truncating mutations underwent severe complications than ones with missense mutations, indicating that truncating mutations of STK11 may have a greater trend to result in severe complications in PJS patients. On the other hand, mutations in exon 3 and exon 6 have been associated with a higher cancer risk [20, 21]. In our study, since no cancer was found in most of these patients, a clear genotype-phenotype correction could not be established. However, colon adenoma detected in patient 101 once again verified a previous reported pathway of hamartoma-(adenoma)-carcinoma in PJS patients [22].
Timely surveillance and early treatment for PJS patients are crucial to improve their quality of life. Current surveillance protocols have two main purposes. One is to detect sizeable GI polyps which could cause severe complications and the other is to detect cancer at an early stage [23]. The potential relationship between truncating mutations of STK11 and severe complications discovered in this study may help PJS patients improve the quality of life by monitoring the development of GI polyps at an earlier age or at shorter intervals.
Conclusions
In summary, three novel mutations and three recurrent mutations in STK11 were identified in Chinese families with PJS, which further broaden the mutation spectrum of STK11. This potential relationship between truncating mutations of STK11 and a higher risk of severe complications would help clinicians and genetic counselors to provide better clinical services for PJS patients.
Abbreviations
- Cdc37:
-
Cell division cycle 37
- EVS:
-
Exome variant server
- GI:
-
Gastrointestinal
- HGMD:
-
Human gene mutation database
- Hsp90:
-
Heat-shock protein 90
- LOVD:
-
Leiden open variation database
- MLPA:
-
Multiplex ligation-dependent probe amplification
- MO25 (also known as CAB39):
-
The scaffolding protein calcium binding protein 39
- PJS:
-
Peutz-Jeghers syndrome
- STK11:
-
Serine-threonine kinase 11
- STRAD (also known as STRADA):
-
STE20-related kinase adaptor alpha
References
van Lier MG, Wagner A, Mathus-Vliegen EM, Kuipers EJ, Steyerberg EW, van Leerdam ME. High cancer risk in peutz-jeghers syndrome: a systematic review and surveillance recommendations. Am J Gastroenterol. 2010;105:1258–64. 1265.
Resta N, Pierannunzio D, Lenato GM, Stella A, Capocaccia R, Bagnulo R, et al. Cancer risk associated with STK11/LKB1 germline mutations in Peutz-Jeghers syndrome patients: results of an Italian multicenter study. Dig Liver Dis. 2013;45:606–11.
Jenne DE, Reimann H, Nezu J, Friedel W, Loff S, Jeschke R, et al. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet. 1998;18:38–43.
Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–7.
Korsse SE, Peppelenbosch MP, van Veelen W. Targeting LKB1 signaling in cancer. Biochim Biophys Acta. 1835;2013:194–210.
Weng MT, Ni YH, Su YN, Wong JM, Wei SC. Clinical and genetic analysis of Peutz-Jeghers syndrome patients in Taiwan. J Formos Med Assoc. 2010;109:354–61.
Orellana P, Lopez-Kostner F, Heine C, Suazo C, Pinto E, Church J, et al. Large deletions and splicing-site mutations in the STK11 gene in Peutz-Jeghers Chilean families. Clin Genet. 2013;83:365–9.
Volikos E, Robinson J, Aittomaki K, Mecklin JP, Jarvinen H, Westerman AM, et al. LKB1 exonic and whole gene deletions are a common cause of Peutz-Jeghers syndrome. J Med Genet. 2006;43:e18.
Aretz S, Stienen D, Uhlhaas S, Loff S, Back W, Pagenstecher C, et al. High proportion of large genomic STK11 deletions in Peutz-Jeghers syndrome. Hum Mutat. 2005;26:513–9.
Boudeau J, Baas AF, Deak M, Morrice NA, Kieloch A, Schutkowski M, et al. MO25alpha/beta interact with STRADalpha/beta enhancing their ability to bind, activate and localize LKB1 in the cytoplasm. EMBO J. 2003;22:5102–14.
Miyoshi H, Nakau M, Ishikawa TO, Seldin MF, Oshima M, Taketo MM. Gastrointestinal hamartomatous polyposis in Lkb1 heterozygous knockout mice. Cancer Res. 2002;62:2261–6.
Olschwang S, Boisson C, Thomas G. Peutz-Jeghers families unlinked to STK11/LKB1 gene mutations are highly predisposed to primitive biliary adenocarcinoma. J Med Genet. 2001;38:356–60.
Osoegawa A, Kometani T, Nosaki K, Ondo K, Hamatake M, Hirai F, et al. LKB1 mutations frequently detected in mucinous bronchioloalveolar carcinoma. Jpn J Clin Oncol. 2011;41:1132–7.
Resta N, Simone C, Mareni C, Montera M, Gentile M, Susca F, et al. STK11 mutations in Peutz-Jeghers syndrome and sporadic colon cancer. Cancer Res. 1998;58:4799–801.
Mehenni H, Gehrig C, Nezu J, Oku A, Shimane M, Rossier C, et al. Loss of LKB1 kinase activity in Peutz-Jeghers syndrome, and evidence for allelic and locus heterogeneity. Am J Hum Genet. 1998;63:1641–50.
Forcet C, Etienne-Manneville S, Gaude H, Fournier L, Debilly S, Salmi M, et al. Functional analysis of Peutz-Jeghers mutations reveals that the LKB1 C-terminal region exerts a crucial role in regulating both the AMPK pathway and the cell polarity. Hum Mol Genet. 2005;14:1283–92.
Resta N, Giorda R, Bagnulo R, Beri S, Della ME, Stella A, et al. Breakpoint determination of 15 large deletions in Peutz-Jeghers subjects. Hum Genet. 2010;128:373–82.
Boudeau J, Deak M, Lawlor MA, Morrice NA, Alessi DR. Heat-shock protein 90 and Cdc37 interact with LKB1 and regulate its stability. Biochem J. 2003;370:849–57.
Amos CI, Keitheri-Cheteri MB, Sabripour M, Wei C, McGarrity TJ, Seldin MF, et al. Genotype-phenotype correlations in Peutz-Jeghers syndrome. J Med Genet. 2004;41:327–33.
Lim W, Olschwang S, Keller JJ, Westerman AM, Menko FH, Boardman LA, et al. Relative frequency and morphology of cancers in STK11 mutation carriers. Gastroenterology. 2004;126:1788–94.
Mehenni H, Resta N, Park JG, Miyaki M, Guanti G, Costanza MC. Cancer risks in LKB1 germline mutation carriers. Gut. 2006;55:984–90.
Wang ZJ, Ellis I, Zauber P, Iwama T, Marchese C, Talbot I, et al. Allelic imbalance at the LKB1 (STK11) locus in tumours from patients with Peutz-Jeghers’ syndrome provides evidence for a hamartoma-(adenoma)-carcinoma sequence. J Pathol. 1999;188:9–13.
Beggs AD, Latchford AR, Vasen HF, Moslein G, Alonso A, Aretz S, et al. Peutz-Jeghers syndrome: a systematic review and recommendations for management. Gut. 2010;59:975–86.
Acknowledgements
We thank the patients, their families, their clinicians and volunteers for their participation in this study. This study was supported by grants from the National Key Basic Research Program of China (2012CB944600), the National Natural Science Foundation of China (81270706) and The National Key Technology R&D Program of China (2012BAI09B05).
Availability of data and materials
Most data generated and analysed during this study are included in this published article and its supplementary information files. More data are available from the corresponding author on reasonable request.
Authors’ contributions
HT, PY, HXL, YP, CC, DSL, and LQW collected clinical data of these PJS families. HT, YRH, LBM, and QP carried out molecular genetic studies for all the families and the controls. HT, XDW, and LBM did the bioinformatics analysis. HT, DSL, and LQW designed and supervised the study. HT drafted the manuscript and XDW and LQW revised it. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Informed consent for this investigation was obtained from all participating PJS patients and parents, and the principles outlined in the Declaration of Helsinki were followed. The project was carried out in agreement with the Ethical Committee of the State Key Laboratory of Medical Genetics (SKLMG).
Author information
Authors and Affiliations
Corresponding authors
Additional file
Additional file 1: Figure S1.
Polyphen-2 reports for the pathogenicity of the mutant amino acid residues in STK11. The amino acid substitution p.Ala241Pro was valued “PROBABLY DAMAGING” with the score 0.999 (HumDiv) and 0.971 (HumVar). (TIF 95 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Tan, H., Mei, L., Huang, Y. et al. Three novel mutations of STK11 gene in Chinese patients with Peutz–Jeghers syndrome. BMC Med Genet 17, 77 (2016). https://doi.org/10.1186/s12881-016-0339-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-016-0339-6