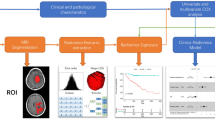
Abstract
Objective
To develop a nomogram based on tumor and peritumoral edema (PE) radiomics features extracted from preoperative multiparameter MRI for predicting brain invasion (BI) in atypical meningioma (AM).
Methods
In this retrospective study, according to the 2021 WHO classification criteria, a total of 469 patients with pathologically confirmed AM from three medical centres were enrolled and divided into training (n = 273), internal validation (n = 117) and external validation (n = 79) cohorts. BI was diagnosed based on the histopathological examination. Preoperative contrast-enhanced T1-weighted MR images (T1C) and T2-weighted MR images (T2) for extracting meningioma features and T2-fluid attenuated inversion recovery (FLAIR) sequences for extracting meningioma and PE features were obtained. The multiple logistic regression was applied to develop separate multiparameter radiomics models for comparison. A nomogram was developed by combining radiomics features and clinical risk factors, and the clinical usefulness of the nomogram was verified using decision curve analysis.
Results
Among the clinical factors, PE volume and PE/tumor volume ratio are the risk of BI in AM. The combined nomogram based on multiparameter MRI radiomics features of meningioma and PE and clinical indicators achieved the best performance in predicting BI in AM, with area under the curve values of 0.862 (95% CI, 0.819–0.905) in the training cohort, 0.834 (95% CI, 0.780–0.908) in the internal validation cohort and 0.867 (95% CI, 0.785–0.950) in the external validation cohort, respectively.
Conclusions
The nomogram based on tumor and PE radiomics features extracted from preoperative multiparameter MRI and clinical factors can predict the risk of BI in patients with AM.
Similar content being viewed by others
Introduction
Atypical meningioma (AM), one of 15 subtypes of meningioma, is classified as a WHO grade 2 tumor with a certain degree of aggressiveness, with an invasiveness between that of benign and malignant meningioma; it accounts for approximately 24.5% of all meningiomas [1], and it has a higher risk of postoperative recurrence than WHO grade 1 meningiomas [2,3,4]. Brain invasion (BI) refers to the presence of meningioma tissue in the adjacent brain tissue without a separate connective tissue layer and tumor cells infiltration into the brain parenchyma in irregular tongue-like projections without an intervening pia mater [2, 5]. BI was clarified in the 2016 revision of the CNS WHO 4 classification as an independent histological criterion for the diagnosis of AM [6]. The 2021 WHO CNS 5, the latest revision, emphasises BI as a pathological diagnostic criterion for AM and applies to any potential subtype [7].
BI in meningiomas has a distinct clinical significance and is independently associated with tumor progression, recurrence and poor prognosis [2, 8,9,10,11]. The presence of BI is closely related to the choice of surgical technique, such as the application of intraoperative navigation, expansion of surgical excision range, etc [12, 13]. . BI makes surgery far more difficult and may have negative implications on functional outcome. In addition, BI is a risk factor for preoperative epileptic seizure and postoperative bleeding [14,15,16]. Therefore, accurately identifying BI in meningioma is of important clinical significance. AM is more likely to recur and has a worse prognosis than WHO grade 1 meningioma [17]. Additionally, the prevalence of AM has increased to 20–35% following the use of BI as an independent diagnostic criterion for the condition [17, 18], necessitating studying AM as a distinct research object. Currently, the gold standard for the diagnosis of BI is the histopathological examination, but this is an invasive procedure that cannot capture the associated changes in real time. It is therefore important to develop practical means to prospectively and noninvasively determine BI in AM.
Several previous studies explored the correlation between BI and imaging features, such as peritumoral edema (PE), enhanced heterogeneity and irregular tumor shape, which were the independent risk predictors of BI [19,20,21,22,23]. Radiomics is an emerging image processing method developed in recent years that allows high-throughput extraction and quantitative analysis of radiomics features in images that cannot be identified by the naked eye [24, 25]. Although there are several radiomics studies on BI prediction of meningiomas [12, 26], there is a lack of radiomics studies on BI prediction of atypical meningiomas. Since the imaging features of atypical meningiomas are obviously different from those of meningiomas, it is necessary to develop a special radiomics study to establish a prediction model of BI for atypical meningiomas.
Therefore, using multicentre data, the aim of this study was to analyze the MRI-based radiomics features of AM tumors and PE, to develop a nomogram for predicting BI in AM patients, and to compare the predictive performance of different models.
Materials and methods
Patients
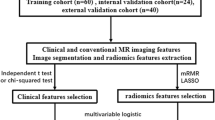
This retrospective study was approved by the Medical Ethics Committee of three medical centres, which waived the need for written informed consent from the patients. From the perspective of research design, we planed to include 3 radiomics scores (R-scores) and 6 clinical characteristics, to construct a logistic model. According to the 10 events per variable empirical principle, at least 90 positive and negative samples would be include into the training set.With a ratio of 7:3 for total sample, the total sample size should be greater than 258. From July 2016 to July 2022, 1382 patients with pathologically confirmed AM from medical centre 1 (Beijing Tiantan Hospital, Capital Medical University) were initially considered, and 390 patients with AM were finally included in the study. The detailed inclusion/exclusion criteria and enrolment process are shown in Fig. 1. In this study, the pathological samples were uniformly diagnosed with BI using light microscopy. BI was defined as the presence of adjacent intracerebral meningioma tissue without a separate connective tissue layer, with an appearance described as “an irregular tongue-like protrusion of tumor cells infiltrating the underlying brain parenchyma without an intervening pia mater” [2, 5], and the WHO classification was recorded; patients without BI were analysed by a combination of pathology reports and surgical records. Patients from centre 1 were randomly divided into a training cohort (n = 273) and an internal validation cohort (n = 117) at a ratio of 7:3 using the DeepWise Multimodal Research Platform version 2.5.1 (http://keyan.deepwise.com). A total of 79 AM patients from medical centre 2 (SanBo Brain Hospital, Capital Medical University) and centre 3 (Shaoxing Second Hospital) were collected as an external validation cohort. All data is based on one MRI which is the most recent MRI examination within 2 weeks before surgery. Representative images of multiparameter MRI and histopathology are shown in Fig. 2.
A 50-year-old female, presented with a tic of limbs, from centre 3. A mass is located in the right frontal lobe, which is diagnosed as atypical meningioma (AM) by pathological examination. (A) Axial T1-weighted MR images (T1C) show that the tumor and brain tissue are of equal intensity with unclear boundaries. (B, C) Axial T2-weighted MR images (T2) and T2-fluid attenuated inversion recovery images (FLAIR) show the equal intensity of the tumor and brain tissue, accompany by a large amount of peritumoral edema (PE), and the edema boundary is clearly displayed. (D) The T1C show that the tumor boundary is not smooth and the tumor-brain interface is blurred. (E) Pathological HE staining (10 × 10 magnification): the patient is diagnosed as AM with brain invasion (BI). Arrows show tumor tissue infiltrating into brain tissue
MR image acquisition and segmentation
Two radiologists with more than 10 years of experience in neuroimaging independently interpreted the tumor location (skull base/non-skull base) on preoperative MRI. Inconsistent results were resolved by consultation; neither radiologist had any knowledge of the pathology results nor was involved in the subsequent analysis. A diagnostic radiologist with more than 10 years of experience used the open source software 3D-slice (version 5.0.3, https://www.slicer.org) to manually delineate the volume of interest (VOI) of the tumor along the border on contrast-enhanced T1-weighted MR (T1C) images and T2-weighted MR (T2) images independently, and both the tumor and PE were outlined on T2-fluid attenuated inversion recovery (FLAIR) images. After recording the volumes of tumors on the T1C and T2 images, to ensure the reliability of the data, the average of the two was taken as tumor volume (VTumor), the volume outlined on the FLAIR images minus the volume of the tumor was considered as peritumoral edema volume (VPE), and VPE was divided by VTumor to obtain the peritumoral edema index (PEI = VPE/VTumor) [27, 28]. In our study, the clinical factors included clinical parameters (sex, age) and imaging parameters (tumor location, VTumor, VPE and PEI). The detailed scanning protocol and parameters are shown in Supplementary A.
Image preprocessing
After manual separation of the tumor and PE using 3D Slicer (version 5.0.3, http://www.slicer.org), to reduce the variation in images acquired by different MR scanners, we normalised the T1C, T2 and FLAIR sequence images using z score normalisation after manually segmenting the VOI to obtain a standard normal distribution of image intensities while resampling all voxels to 1.0 × 1.0 × 1.0 mm3. Feature discretization was performed using bin width: 25. To ensure the accuracy of the VOI, 40 patients were randomly selected from the training cohort, and the VOI on the FLAIR sequence images were outlined again by the former and another radiologist in the same way as described above. Then, the inter-/intraclass correlation coefficients (ICCs) were used to evaluate the consistency between the VOI of the 40 patients and those outlined by the previous doctors. Features were extracted separately according to the outlined VOI to determine and assess the reliability between the VOI outlined by the same outliner at different times and between the VOI outlined by different outliners. High reproducibility was indicated when the mean ICCs of all features was > 0.75.
Radiomics feature extraction and selection
We used the DeepWise Multimodal Research Platform version 2.5.1 (http://keyan.deepwise.com) with the Python PyRadiomics (version 3.0.1) and scikit-learn (version 0.22) packages as radiomics feature extraction and data analysis tools for this study. More details about feature extraction and selection are shown in Supplementary B.
Model construction and validation
Using the radiomics score (R-score) of the tumor and PE features obtained from multiparameter MRI after least absolute shrinkage and selection operator (LASSO) regression screening and calculation, T1C, T2, FLAIR single models were developed in the training cohort. In the process of parameter adjustment, the optimal hyper-parameters were selected by using fivefold cross-validation and and grid search for the training cohort. Then, the R-scores of the T1C, T2 and FLAIR features were analysed by univariate and multivariate logistic regression, meaningful R-scores were selected and used to develop the fused radiomics model. The above four radiomics models were then independently verified in the internal and external validation cohorts. The subject ROC curves of each model were plotted, and the area under the ROC curve (AUC), accuracy (ACC), sensitivity (SEN), specificity (SPEC), positive predictive value (PPV) and negative predictive value (NPV) were calculated. The predictive performance of each model was evaluated and compared to determine the optimal model for predicting BI in AM patients.
The clinical characteristics of all included patients were analysed by univariate analysis and multivariate logistic regression to identify clinical risk factors significantly associated with a prediction of BI and construct clinical prediction models in the training cohort. Furthermore, the identified clinical factors were introduced into multivariate logistic regression along with the multiparameter R-scores to construct a comprehensive prediction model. The clinical, fused radiomics and comprehensive models were then compared in terms of their predictive efficacy for BI in AM, and the models were independently validated in the internal and external validation cohorts.
Based on the results of the study, a clinical and radiomics nomogram of the optimal prediction model that was clinically meaningful was constructed, accurately predicting the likelihood of BI occurring in AM patients preoperatively. Figure 3 represents a radiomics flow chart of the study.
Statistical methods
Continuous variables are expressed as the mean ± standard deviation, and categorical variables are expressed as frequencies (percentages). ANOVA (continuous variables) and the chi-square test (categorical variables) were used for data processing. The two-independent sample t test was used for variables with normal distributions, while the Wilcoxon signed-rank test was used for variables with a skewed distribution. The DeLong test was used to compare the ROC curves among the different models. p < 0.05 was considered to indicate statistical significance. The Deepwise Multimodal Research Platform version 2.5.1 (https://keyan.deepwise.com) was used as statistical analysis tool for this study.
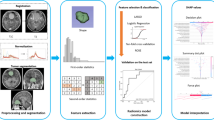
Flowchart of the radiomics study. First, data were collected, including contrast-enhanced T1-weighted MR images (T1C), T2-weighted MR images (T2) and T2-fluid attenuated inversion recovery (FLAIR) images, followed by manual tumor segmentation on the T1C and T2 sequences and manual tumor + PE segmentation on the FLAIR sequence. Second, the tumor and PE radiomics features were extracted, including first-order, shape, texture and filter features. Third, the least absolute shrinkage and selection operator (LASSO) was used to select the optimal features. Finally, T1C, T2, FLAIR and fused radiomics models were established to predict brain invasion in atypical meningioma patients, and their predictive performance was compared
Results
Clinical data
A total of 390 AM patients from centre 1 were enrolled in this study, including 225 (57.69%) patients with BI and 165 (42.31%) patients without BI. These patients were then divided into two cohorts at a ratio of 7:3, with 273 comprising the training cohort, including 157 patients with BI and 116 patients without BI, and 117 comprising the internal validation cohort, which included 68 patients with BI and 49 patients without BI. A total of 79 AM patients from centres 2 and 3 were used as the external validation cohort, including 37 patients with BI and 42 patients without BI. In the training cohort, patients with BI were statistically older (p = 0.046), but no such difference was observed in the internal and external validation cohorts. Sex and tumor location were not significantly different in the training and validation cohorts, and VTumor was statistically significant in the external validation cohorts and not significantly different in the training and internal validation cohorts. In the training, internal and external validation cohorts, VPE (p < 0.01) and PEI (p < 0.01) were significantly higher in the patients with BI than in those without BI. The baseline characteristics of the patients in the training and validation cohorts are shown in Table 1.
Clinical predictors of BI
Sex, VPE and PEI were included in the multivariate logistic regression analysis (Supplementary C), showing that VPE (p < 0.001, OR = 1.018, 95%CI: 1.008–1.027) and PEI (p = 0.035, OR = 1.333, 95%CI: 1.020–1.743) were significantly different between patients with and without BI and positively correlated with BI. After including VPE and PEI, the clinical model achieved AUCs of 0.818 (95%CI: 0.765–0.870), 0.796 (95%CI: 0.703–0.857) and 0.773 (95%CI: 0.666–0.881) were obtained in the training, internal, and external validation cohorts, respectively.
Selection of radiomics features
A total of 1409 T1C tumor features, 1409 T2 tumor features, and 1409 FLAIR tumor and PE features were extracted in this study. The ICCs of the outlines generated using the FLAIR images were excellent, with ICCs of 0.894 ± 0.175 and 0.850 ± 0.182, respectively. After Pearson correlation analysis and LASSO regression analysis, nine features from FLAIR, three features from T2 and two features from T1C images were selected. The heatmap of the Pearson correlation analysis for the selected features is shown in Supplementary D. As shown in the figure, the correlation between the selected radiomics features was low, and the feature redundancy was minimal. The analysis and weights of the selected radiomics features are shown in Table 2, which also shows that tumor and PE radiomics features based on the FLAIR sequence were predominant.
Construction and comparison of radiomics models
The R-scores of the T1C, T2, and FLAIR (T1C-score, T2-score and FLAIR-score) tumor and PE radiomics features were calculated using the corresponding formulae, and multivariate logistic regression was used to construct the T1C, T2, FLAIR and fused radiomics models separately. A comparison of the models is shown in Fig. 4(A-C). A comparison of the decision curves of the models is shown in Fig. 5(A-C).
The results showed that the AUC of the model built from the tumor and PE radiomics features extracted from the FLAIR sequence in predicting BI in AM was significantly higher than that of the models based on the T2 and T1C tumor features in the training cohort (FLAIR model AUC = 0.851, 95%CI: 0.806–0.897), internal validation cohort (AUC = 0.819, 95%CI: 0.742–0.898) and external validation cohort (AUC = 0.820, 95%CI: 0.728–0.913). Furthermore, there was a significant difference (DeLong test, p < 0.05) among the AUCs of the three models; ranked according to p value, FLAIR > T2 > T1C.
The T1C-score, T2-score, and FLAIR-score were included in univariate and multivariate logistic regression analyses (Supplementary E). The results of multiple logistic regression showed that the T2-score and FLAIR-score were significantly different between patients with and without BI (p = 0.005 and p < 0.001, respectively) and positively correlated with BI. The T1C-score was not significantly different between the two groups (p = 0.609); therefore, only the T2-score and FLAIR-score were analysed by multivariate logistic regression to construct a fused radiomics model. The AUC of the fused radiomics model was higher than that of the FLAIR model alone in the training cohort (AUC = 0.859, 95%CI: 0.815–0.902), internal validation cohort (AUC = 0.825, 95%CI: 0.752–0.904), and external validation cohort (AUC = 0.862, 95%CI: 0.777–0.946). In addition, the ACC, SPEC, PPV and NPV of the fused radiomics model were higher than those of the single radiomics models in both the training and validation cohorts.
Comparison of the ROC curves of the different models (A, B, C) ROC curves of the different radiomics models in the training, internal validation and external validation cohorts. The fused radiomics model showed the best diagnostic efficacy among them, with an AUC of 0.859 in the training cohort (A), 0.825 in the internal validation cohort (B) and 0.862 in the external validation cohort (C). (D, E, F) ROC curves of the clinical, fused radiomics and comprehensive models in the training, internal and external validation cohorts. The comprehensive model showed the best diagnostic efficacy among these three models, with an AUC of 0.862 in the training cohort (D), 0.834 in the internal validation cohort (E) and 0.867 in the external validation cohort (F)
Decision curve analysis for the different models (A, B, C) Decision curves of the T1C, T2, FLAIR and fused radiomics models in the training, internal validation and external validation cohorts. (D, E, F) Decision curves of the clinical, fused radiomics and comprehensive models in the training, internal validation and external validation cohorts
Comparison of clinical, fused radiomics and comprehensive models
Combining VPE, PEI, T2-score and FLAIR-score, a comprehensive model was constructed using multivariate logistic regression. The ROC curves of the six models are shown in Fig. 4, and the decision curves are shown in Fig. 5. The AUCs, ACCs, SENs, SPECs, PPVs and NPVs of the six models are shown in Table 3. The comparison of AUC among the clinical, radiomics and comprehensive models are shown in Fig. 6.
The results show that the comprehensive model was significantly superior to the clinical model (p = 0.030 in the training cohort, p = 0.021 in the external validation cohort) and had the highest AUC in predicting BI in AM patients, superior to that of the clinical model (p = 0.029 in the training cohort, p = 0.042 in the internal validation cohort and p = 0.020 in the external validation cohort) but not significantly different from that of the fused radiomics model (p > 0.05), which had the highest ACC, SEN, PPV, NPV and AUC in both the training and validation cohorts.
Development of the nomogram
A clinical and radiomics nomogram for preoperatively predicting the likelihood of BI in AM patients was constructed by combining VPE, PEI, T2-score and FLAIR-score, as seen in Fig. 7A, where each risk factor is labelled in quantitative form, and a total score is calculated based on the corresponding score for each risk factor for each AM patient to predict the risk of developing BI. The higher the total score is, the greater the risk of developing BI. Additionally, calibration curves were plotted for the training, internal and external validation cohorts to determine the predictive efficacy of the nomogram (Fig. 7B-D). The results show that the prediction curves are very close to the reference line, indicating strong predictive efficacy. Additionally, we show the confusion matrix of the comprehensive model (Fig. 7E-G), which reveals that the predicted false negatives and false positives are low in the training, internal validation and external validation cohorts.
Nomogram of the comprehensive model and its calibration curves and confusion matrixes (A) The comprehensive model was constructed to develop the nomogram. (B, C, D) Calibration curves of the comprehensive model-based nomogram in the training, internal validation and external validation cohorts. (E, F, G) Confusion matrixes of the comprehensive model in the training, internal validation and external validation cohorts
Discussion
In our study, the efficacy of MR radiomics features of AM tumor and PE in predicting the occurrence of BI in AM was compared between radiomics models built from different preoperative MRI sequences, a clinical model, a fused radiomics model and a comprehensive model constructed from the extracted MRI radiomics features and clinical features. The results showed that: (1) the AUC of the model built from the tumor and PE radiomics features extracted from FLAIR sequences in predicting BI in AM was significantly higher than that of the model based on the tumor features from the T2 and T1C sequences; (2) the radiomics features of PE play an important role in predicting BI; (3) PEI is an independent clinical risk factor in predicting BI in AM; and (4) the combined model (VPE, PEI, T2-score and FLAIR-score) showed the best performance in preoperatively predicting BI in AM patients in the training, internal validation and external validation cohorts.
Our study demonstrated that the incidence of BI in AM patients was 55.86%, which is higher than the 31.5% (95% CI 22.3–42.6%) before the revised criteria for AM were released [29,30,31,32] and in line with the findings from recent studies [8, 33]. The reason for this is that the new, revised criteria include BI as an independent, histological criterion for diagnosing AM. Formerly WHO grade 1 meningioma patients were included as having AM once the new BI-based criterion was applied to them, resulting in an increase of 1–10% in the incidence of AM [23, 34], as well as an increase in the proportion of BI in AM. In terms of clinical risk factors for BI in AM, our study found that patient sex, age, tumor location, and VTumor were not involved, consistent with the findings of several studies [8, 29]. In other studies, however, BI was more common in older, male meningioma patients and preferentially targeted areas other than the skull base [32, 35]. The possible reason for this inconsistency is that most of these other studies included all meningiomas and did not factor in the WHO classification. Our study showed that BI was associated with VPE and PEI, with the probability of BI increasing with increased VPE and PEI. Previous studies have suggested that PE is an independent predictor of BI [19, 20, 36], consistent with our study. Ong T et al. [20] studied BI in 60 meningioma patients and found that a larger PE may be associated with an increased incidence of BI. However, when controlling for tumor size, no statistically significant differences were found for PEI, in contrast to the results of our study.
Previous radiomics studies on predicting BI of meningiomas have primarily focused on analyzing the internal of the tumor and the tumor-brain interface [12, 26, 33, 36]. In this study, we developed a novel analysis by extracting the PE radiomics features and combining them with the internal features of the tumor. Nine features from FLAIR, three features from T2 and two features from T1C images were identified, demonstrating the clear preponderance of FLAIR sequence features. The results of the ROC analysis demonstrate that the AUC of the model constructed using tumor and PE radiomics features from the FLAIR sequence was significantly higher than that of the model based on the T2 and T1C tumor features in predicting BI in AM, which indicated the important role of both tumor and PE features in providing prediction-relevant information. One possible explanation for this finding is that when BI occurs in meningioma, changes also occur in the microenvironment surrounding the PE, and FLAIR sequences, which are sensitive to water in tissue, allow for clear delineation of edema boundaries. Another interesting result is that the FLAIR and T2 data proved to be more significant than the T1C data in predicting scores, this has also prompted us to pay more attention to the application of non-enhanced images in meningioma. In addition, the fused radiomics model combining FLAIR and T2 features outperformed both the single radiomics and clinical models. The nomogram constructed by combining VPE, PEI, T2-score and FLAIR-score demonstrated the best performance in predicting the occurrence of BI in AM patients preoperatively.
Li N et al. [33] conducted a study on 284 WHO grade 2 meningioma patients to predict BI based on tumor and tumor-brain border features.They found that combined clinical and conventional imaging indices had a slightly higher AUC than our study. However, the tumor-brain interface is not easy to delineate, and the inclusion of conventional imaging indicators was overrepresented, increasing the difficulty in implementing predictive models. Zhang J et al. [12] used T1C and T2 sequences to extract the radiomics features of meningioma. They developed a combined clinical-radiomics features nomogram for predicting BI in meningiomas, their study achieved an AUC of 0.857 in the training cohort and 0.819 in the validation cohort. However, it is worth noting that the majority of the study population in their research consisted of WHO grade 1 meningiomas, which may have introduced bias into the construction of the predictive models. Additionally, none of the results of the above studies had been externally validated. In our study, we included the largest number of AM cases compared to any published study, and we utilized a simple delineation for radiomics feature extraction and data from multiple centers. The AUC was 0.867 in the external validation cohort, indicating that our model is reliable and generalizable to other AM populations.
This study had several limitations. First, our study was retrospective, and there might be unavoidable selection bias. Second, the VOI was manually outlined using software, a time-consuming process that needs to be fully automated using better software to improve efficiency and avoid human error. Finally, as we retrospectively collected MR images from different centres, there were inevitable differences in equipment and scanning parameters. We standardised all the images to reduce the impact of these differences on the radiomics features, and good performance was obtained in the external validation cohort.
In conclusion, this study developed a predictive clinical and radiomics nomogram based on tumor and PE radiomics features extracted from multiparameter MRI, highlighting the important role that the radiomics features of PE play in predicting BI in AM patients. After further validation of data from multiple centers, the nomogram developed in this study could maximize the predictive accuracy for BI in AM patients in clinical practice and guide better clinical implementation of surgical protocols and personalized patient treatment, helping to improve the survival of AM patients.
Data availability
The datasets generated or analyzed during the study are available from the corresponding author on reasonable request.
Abbreviations
- ACC:
-
Accuracy
- AM:
-
Atypical Meningioma
- AUC:
-
Area Under the Curve
- BI:
-
Brain Invasion
- CNS:
-
Central Nervous System
- FLAIR:
-
T2-Fluid Attenuated Inversion Recovery
- ICCs:
-
Inter-/Intraclass Correlation Coefficients
- LASSO:
-
Least Absolute Shrinkage and Selection Operator
- NPV:
-
Negative Predictive Value
- PE:
-
Peritumoral Edema
- PEI:
-
Peritumoral Edema Index
- PPV:
-
Positive Predictive Value
- R-scores:
-
Radiomics Scores
- SEN:
-
Sensitivity
- SPEC:
-
Specificity
- T1C:
-
Contrast-enhanced T1-weighted MR image
- T2:
-
T2-weighted MR image
- VOI:
-
Volume Of Interest
- VTumor :
-
Tumor Volume
- VPE :
-
Peritumoral Edema Volume
- WHO:
-
World Health Organization
References
Bagshaw HP, Burt LM, Jensen RL, et al. Adjuvant radiotherapy for atypical meningiomas. J Neurosurg. 2017;126(6):1822–8.
Streckert E, Hess K, Sporns PB, et al. Clinical, radiological, and histopathological predictors for long-term prognosis after surgery for atypical meningiomas. Acta Neurochir (Wien). 2019;161(8):1647–56.
Garcia-Segura ME, Erickson AW, Jairath R, et al. Necrosis and Brain Invasion Predict Radio-Resistance and Tumor recurrence in atypical meningioma: a retrospective cohort study. Neurosurgery. 2020;88(1):E42–8.
Ressel A, Fichte S, Brodhun M, et al. WHO grade of intracranial meningiomas differs with respect to patient’s age, location, tumor size and peritumoral edema. J Neurooncol. 2019;145(2):277–86.
Perry A, Stafford SL, Scheithauer BW, et al. Meningioma grading: an analysis of histologic parameters. Am J Surg Pathol. 1997;21(12):1455–65.
Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–20.
Louis DN, Perry A, Wesseling P et al. The 2021 WHO classification of tumors of the Central Nervous System: a summary. Neuro Oncol 2021, 23(8):1231–51.
Brokinkel B, Hess K, Mawrin C. Brain invasion in meningiomas-clinical considerations and impact of neuropathological evaluation: a systematic review. Neuro Oncol. 2017;19(10):1298–307.
Spille DC, Hess K, Sauerland C, et al. Brain Invasion in meningiomas: incidence and correlations with clinical variables and prognosis. World Neurosurg. 2016;93:346–54.
Banan R, Abbetmeier-Basse M, Hong B, et al. The prognostic significance of clinicopathological features in meningiomas: microscopic brain invasion can predict patient outcome in otherwise benign meningiomas. Neuropathol Appl Neurobiol. 2021;47(6):724–35.
Picart T, Dumot C, Guyotat J, et al. Clinical and pathological impact of an optimal assessment of brain invasion for grade 2 meningioma diagnosis: lessons from a series of 291 cases. Neurosurg Rev. 2022;45(4):2797–809.
Zhang J, Yao K, Liu P et al. A radiomics model for preoperative prediction of brain invasion in meningioma non-invasively based on MRI: a multicentre study. EBioMedicine 2020, 58:102933.
Li HY, Ying YZ, Zheng D, et al. Is brain invasion sufficient as a stand-alone criterion for grading atypical meningioma? J Neurosurg. 2023;139(4):953–64.
Hess K, Spille DC, Adeli A, et al. Brain invasion and the risk of seizures in patients with meningioma. J Neurosurg. 2018;130(3):789–96.
Hinrichs FL, Brokinkel C, Adeli A, et al. Risk factors for preoperative seizures in intracranial meningiomas. J Neurosurg Sci. 2023;67(1):66–72.
Brokinkel B, Sicking J, Spille DC, et al. Letter to the editor. Brain invasion and the risk for postoperative hemorrhage and neurological deterioration after meningioma surgery. J Neurosurg. 2018;129(3):849–51.
Delgado-Lopez PD, Cubo-Delgado E, Gonzalez-Bernal JJ, et al. A practical overview on the Molecular Biology of Meningioma. Curr Neurol Neurosci Rep. 2020;20(12):62.
Zhi M, Girvigian MR, Miller MJ, et al. Long-term outcomes of newly diagnosed Resected atypical meningiomas and the role of Adjuvant Radiotherapy. World Neurosurg. 2019;122:e1153–61.
Adeli A, Hess K, Mawrin C, et al. Prediction of brain invasion in patients with meningiomas using preoperative magnetic resonance imaging. Oncotarget. 2018;9(89):35974–82.
Ong T, Bharatha A, Alsufayan R, et al. MRI predictors for brain invasion in meningiomas. Neuroradiol J. 2021;34(1):3–7.
Friconnet G, Baudouin M, Brinjikji W, et al. Advanced MRI shape analysis as a predictor of histologically aggressive supratentorial meningioma. J Neuroradiol. 2022;49(3):275–80.
Coroller TP, Bi WL, Huynh E, et al. Radiographic prediction of meningioma grade by semantic and radiomic features. PLoS ONE. 2017;12(11):e187908.
Jiang J, Yu J, Liu X, et al. The efficacy of preoperative MRI features in the diagnosis of meningioma WHO grade and brain invasion. Front Oncol. 2022;12:1100350.
Brunasso L, Bonosi L, Costanzo R et al. Updated systematic review on the role of Brain Invasion in Intracranial meningiomas: what, when. Why? Cancers (Basel) 2022, 14(17).
Park JE, Kim HS. Radiomics as a quantitative imaging biomarker: practical considerations and the current standpoint in Neuro-oncologic studies. Nucl Med Mol Imaging. 2018;52(2):99–108.
Xiao D, Zhao Z, Liu J, et al. Diagnosis of Invasive Meningioma based on brain-tumor interface Radiomics features on brain MR images: a Multicenter Study. Front Oncol. 2021;11:708040.
Loewenstern J, Aggarwal A, Pain M, et al. Peritumoral Edema Relative to meningioma size predicts functional outcomes after resection in older patients. Oper Neurosurg (Hagerstown). 2019;16(3):281–91.
Tamiya T, Ono Y, Matsumoto K, et al. Peritumoral brain edema in intracranial meningiomas: effects of radiological and histological factors. Neurosurgery. 2001;49(5):1046–51.
Pizem J, Velnar T, Prestor B, et al. Brain invasion assessability in meningiomas is related to meningioma size and grade, and can be improved by extensive sampling of the surgically removed meningioma specimen. Clin Neuropathol. 2014;33(5):354–63.
Barresi V, Lionti S, Caliri S, et al. Histopathological features to define atypical meningioma: what does really matter for prognosis? Brain Tumor Pathol. 2018;35(3):168–80.
Yoon H, Mehta MP, Perumal K, et al. Atypical meningioma: randomized trials are required to resolve contradictory retrospective results regarding the role of adjuvant radiotherapy. J Cancer Res Ther. 2015;11(1):59–66.
Nakasu S, Nakasu Y. Prognostic significance of brain invasion in meningiomas: systematic review and meta-analysis. Brain Tumor Pathol. 2021;38(2):81–95.
Li N, Mo Y, Huang C, et al. A clinical semantic and Radiomics Nomogram for Predicting Brain Invasion in WHO Grade II Meningioma based on Tumor and Tumor-to-brain interface features. Front Oncol. 2021;11:752158.
Rebchuk AD, Chaharyn BM, Alam A, et al. The impact of brain invasion criteria on the incidence and distribution of WHO grade 1, 2, and 3 meningiomas. Neuro-oncology (Charlottesville. Va). 2022;24(9):1524–32.
Timme M, Thomas C, Spille DC, et al. Brain invasion in meningiomas: does surgical sampling impact specimen characteristics and histology? Neurosurg Rev. 2020;43(2):793–800.
Joo L, Park JE, Park SY, et al. Extensive peritumoral edema and brain-to-tumor interface MRI features enable prediction of brain invasion in meningioma: development and validation. Neuro Oncol. 2021;23(2):324–33.
Acknowledgements
The authors would like to express their gratitude to Yi You who from the Department of Research Collaboration, Research & Development Center (R&D), Beijing Deepwise & League of Doctor of Philosophy (PHD) Technology Co., Ltd, Beijing, China for the technical support.
Funding
This study has received funding by the National Natural Science Foundation of China (No. 81772005); The collaborative innovative major special project supported by Beijing Municipal Science & Technology Commission (No. Z191100006619088); Medical and Health Research Projects of Health Commission of Zhejiang Province (No. 2024KY493).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. The conception and design of the study by JY, XC and SL. The MRI and clinical data were collected by JY, FZ and XL. The ROIs of tumors and PE were outlined by JY and DX. The model construction and data analysis were performed by XK, DS and CH. The first draft of the manuscript was written by JY and XK. The manuscript was edited and revised by CW, HX, SL and XC. All authors approve the final version of the manuscript for submission.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This is a retrospective study. It was approved by the Medical Ethics Committee of three medical centres (Beijing Tiantan Hospital, Capital Medical University; SanBo Brain Hospital, Capital Medical University; and Shaoxing Second Hospital), which waived the need for written informed consent from the patients.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Data overlap
The datasets were constructed based on the collection of this study and had not been published repeatedly in previous studies.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yu, J., Kong, X., Xie, D. et al. Multiparameter MRI-based radiomics nomogram for preoperative prediction of brain invasion in atypical meningioma:a multicentre study. BMC Med Imaging 24, 134 (2024). https://doi.org/10.1186/s12880-024-01294-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12880-024-01294-5