Abstract
Background
Solobacterium moorei, the only species in the genus Solobacterium, is a Gram-positive, non-spore-forming, strict anaerobic, short to long bacillus. It has rarely been documented to cause blood stream infections. Here we report the first case of bacteremia caused by S.moorei in China.
Case presentation
A 61-year-old male presented to Peking Union Medical College Hospital (Beijing) with thrombotic thrombocytopenic purpura (TTP) and several other underlying diseases. He also had persistent coma accompanied by intermittent convulsions, halitosis, and intermittent fever. Blood cultures taken when the patient had a high fever were positive, with the anaerobic bottle yielding an organism identified as S.moorei by 16S rRNA gene sequencing, whilst the aerobic bottle grew Streptococcus mitis. After replacement of venous pipeline, and empirical use of vancomycin and meropenem, the patient’s body temperature and white blood cell count returned to normal. Unfortunately, the patient died of severe TTP.
Conclusion
This is the first case report of S. moorei isolation from blood stream in China. 16S rRNA gene sequencing is the only method that can identify S. moorei. Blood cultures must be taken before administration of antibiotics, and anaerobic culture should be considered for such rare pathogens in patients with oral diseases and immune deficiency.
Similar content being viewed by others
Background
Solobacterium moorei (S. moorei) is a member of the indigenous human intestinal microflora, and was first isolated from human feces in 2000 [1]. The new genus Solobacterium, which belonged to the Clostridium cluster XVI, was created after 16S rRNA gene sequence analysis [2]. S.moorei is the only species in the genus, and shows close phylogenetic relationship with Bulleidia extructa, Holdemania filiformis, and Erysipelothrix rhusiopathiae [1, 2]. S.moorei has been reported to cause halitosis [3,4,5], other oral cavity diseases [6,7,8,9] and wound infections [10]. To our knowledge, there are only 4 reports on blood stream infection caused by S.moorei [11,12,13,14]. Herein, we present the first case of S. moorei bacteremia in China.
Case presentation
A 61-year-old man who previously worked in a cattle farm, and had a medical history of hypertension for more than 10 years, hyperlipidemia, type 2 diabetes, rectal cancer treated with 6 rounds of chemotherapy, and brucellosis, presented to a local primary hospital. On April 12,017, he had a fever (38.5 °C), cough, expectoration, dizziness, headache and fatigue. Pneumonia was suspected by Computed Tomography (CT) test at the local primary hospital. The symptoms did not improve after cefuroxime and moxifloxacin treatment for 3 days. On April 42,017, the patient developed seizures and unconsciousness, and was then transferred and admitted to the emergency department of Peking Union Medical College Hospital (PUMCH). Laboratory tests revealed that the patient’s white blood cell count was 5.67 × 109/L with 63% neutrophils, hemoglobin was 11.91 g/L, platelet count was 16.0 × 109/L, and procalcitonin (PCT) was < 0.5 ng/mL. The patient was treated with moxifloxacin for infection, and midazolam for sedation and recurrence of convulsions. He was also transfused with platelets and put on a ventilator.
On April 52,017, further lab examinations found that brucellosis Rose Bengal Test was positive, and thus minocycline and rifampicin administration were initiated for brucellosis treatment. The patient was also diagnosed with thrombotic thrombocytopenic purpura (TTP) syndrome with a very low platelet count of 16.0 × 109/L for which methylprednisolone and continuous plasma exchange were commenced to treat the patient. On April 132,017, sputum culture yielded extended-spectrum β-lactamase (ESBL) positive Klebsiella pneumoniae and ESBL negative Proteus mirabilis. Microbiology tests from other specimens (cerebrospinal fluid, bone marrow, femoral vein catheter blood and jugular vein catheter blood) were all negative from April 5 to April 132,017. The patient had serious halitosis as reported by the doctor in charge, but no pathogen was isolated from the oral secretion. On April 5 and 7, three sets of blood cultures were taken but all were negative for pathogens. On April 13, the patient developed a high fever, and another set of blood cultures was taken, and was positive after incubation for 25 h (aerobic) and 51 h (anaerobic) in the BacT/Alert automated blood culturing system. Further tests identified the cultured organisms as Streptococcus mitis (aerobic bottle) and S.moorei (anaerobic bottle). Treatment was adjusted accordingly, which included replacement of venous pipeline and empirical use of vancomycin and meropenem. The patient’s body temperature and white blood cell count returned to normal levels. Unfortunately, the patient had been in a coma since admission, and on April 21, the patient died of severe TTP.
Microbiology and molecular examination
S.moorei is a strict anaerobic organism which can only grow under anaerobic conditions. On blood agar it forms gray white, non-hemolytic colonies (around 0.5 mm in diameter) after 72 h of incubation at 37 °C. Gram staining morphology revealed short to long non-spore-forming Gram-positive bacilli. S.moorei grows slowly, produces relatively few positive biochemical reactions, and phenotypic variations appear to be commonly exhibited by different strains. Due to these challenges, S.moorei cannot be identified using any commercially available identification kits [15]. Moreover, the organism cannot be identified by Vitek (bioMérieux) or Bruker Matrix-Assisted Laser Desorption/ Ionization Time-of-Flight (MALDI-TOF) as the organism’s spectrum is unavailable in the respective databases.
Antimicrobial susceptibility of S.moorei was determined by the E-test gradient method according to the manufacturer’s instructions. Brucella blood agar supplemented with hemin and vitamin K was used as the primary plate, and Bacteroides fragilis ATCC 25285 was used as the quality control strain. The breakpoints for antimicrobial agents tested were according to Clinical and Laboratory Standards Institute (CLSI) guidelines 27rd informational supplement (M100-S27), Antimicrobial susceptibility results of S. moorei are shown in Table 1.
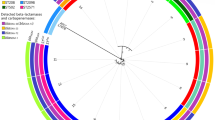
16S rRNA gene sequencing was performed to reliably identify the organism. The universal primers were 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′- TACGGCTACCTTGTTACGACTT − 3′), generating a sequence with 1403 base pairs. BLAST analysis matched the strain to S.moorei strain JCM 10645 with identity similarity of 99.9%. We also constructed a phylogenetic tree [16] of the clinical isolate 17B10385 in this study with S.moorei from GenBank and closely related genera [1, 12]. The phylogenetic tree analysis also confirmed our clinical isolate as S.moorei (Fig. 1). The 16S rRNA gene sequence of our S.moorei strain has been deposited in GenBank (accession number MK989992).
Phylogenetic tree showing the relationships of the blood culture isolate to S. moorei isolates and members of other related genera. The tree was constructed by using the neighbour-joining method and bootstrap values calculated from 1000 trees. The accession numbers shown are those in the GenBank database. ***: the clinical S.moorei isolate 17B10385 in this study
Discussion and conclusions
Early studies showed that S.moorei is phylogenetically related to Eubacterium. Eubacterium includes all anaerobic, non-spore-forming, Gram-positive bacilli except Propionibacterium, Lactobacillus and Bifidobacterium. The main glucose fermentation product for S.moorei is acetic acid, whilst for Propionibacterium, Lactobacillus, Bifidobacterium and Eubacterium, it is propionic acid, lactic acid, acetic acid, both butyric acids and formic acid [1]. Due to differences in culture conditions, G + C contents and fermentation products, S.moorei was assigned to a new species.
In the last decade, S. moorei has mainly been reported to cause oral diseases, including halitosis [3,4,5], endodontic infections [6, 7], peri-radicular lesions [8], refractory periodontitis [9], root canals [7], periodontal disease and dentoalveolar abscesses [2]. Studies show that S.moorei mainly colonizes the oral cavity of halitosis patients, with detection frequency in healthy volunteers much less than in halitosis patients. The distribution of S.moorei varies by race and gender, being more prevalent in African Americans, followed by Hispanics and Whites, and in males than females [3,4,5]. S.moorei adheres to oral epithelial cells through adhesins. Biofilm formation is a key step in the development of halitosis [17]. Furthermore, S.moorei produces beta galactosidase and hydrogen sulfide, which play important roles in halitosis [4].
In addition to oral diseases, S.moorei is reported to cause wound and blood stream infections, although this is relatively rare. To date, there are only four reports about S.moorei bacteremia [11,12,13,14], and one report on wound infection [10]. Table 2 summarizes S. moorei bacteremia and wound infections. Including the present case, there are only 9 cases of S. moorei bacteremia; 5 cases of S. moorei bacteremia were reported by Pedersen during a period of 7 years [14]. S. moorei deposited in the clone library was isolated from only 9 cases in a pool of 400 surgical wound infections. Clinical data indicates that patients with compromised immunity (such as malignant disease, intravenous drug abuser, diabetes and history of surgery) are more susceptible to S.moorei bacteremia across gender and age. S.moorei is susceptible to commonly used anti-anaerobic agents (e.g., penicillin, piperacillin-tazobactam, clindamycin, metronidazole, meropenem, moxifloxacin, tegacycline and vancomycin). For wound infections, surgical debridement and drainage is commonly required. In the present case, the high fever developed during treatment may be related to the entry of S.moorei and Streptococcus mitis into the bloodstream. After replacement of venous pipeline and empirical use of vancomycin and meropenem, the patient’s body temperature and the total leukocyte count returned back to normal.
S.moorei bloodstream infection may originate from oral infection, lung abscess, abdominal infection, and the habit of licking needle by intravenous drug users. The patient in this case had serious halitosis, which maybe the origin of the bacteremia. However, we failed to culture this pathogen from oral secretions, possibly due to the fact that oral secretions are not routinely cultured for anaerobic pathogens. Thus for patients with oral diseases such as halitosis, anaerobic cultures should be strongly recommended to identify suspicious pathogens.
The newly discovered S. moorei was isolated from patients with oral diseases and immune deficiency. Although rarely isolated, its pathogenicity in oral, wound and bloodstream infections is very clear. S. moorei exhibits susceptibility to common antibiotics used for anaerobic infections. Prior use of antibiotics before blood cultures are taken may yield negative results. Due to the special culture conditions and specialized identification method, the prevalence of S. moorei bacteremia may be underestimated. Laboratory staff and clinicians should pay more attention to such rare bacteria and their clinical significance. Optimization of blood culture procedures and utilization of 16S rRNA gene sequencing are powerful tools for rare pathogen identification from blood and other sterile body fluids.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CLSI:
-
Clinical and Laboratory Standards Institute
- CT:
-
Computed Tomography;
- ESBL:
-
Extended-Spectrum β-Lactamases
- MALDI-TOF:
-
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight
- MIC:
-
Minimal Inhibitory Concentration.
- PCT:
-
Procalcitonin;
- TTP:
-
Thrombotic thrombocytopenic purpura
References
Kageyama A, Benno Y. Phylogenic and phenotypic characterization of some Eubacterium-like isolates from human feces: description of Solobacterium moorei gen. Nov., Sp. Nov. Microbiol Immunol. 2000;44(4):223–7.
Downes J, Munson MA, Spratt DA, Kononen E, Tarkka E, Jousimies-Somer H, Wade WG. Characterisation of Eubacterium-like strains isolated from oral infections. J Med Microbiol. 2001;50(11):947–51.
Haraszthy VI, Zambon JJ, Sreenivasan PK, Zambon MM, Gerber D, Rego R, Parker C. Identification of oral bacterial species associated with halitosis. J Am Dent Assoc. 2007;138(8):1113–20.
Haraszthy VI, Gerber D, Clark B, Moses P, Parker C, Sreenivasan PK, Zambon JJ. Characterization and prevalence of Solobacterium moorei associated with oral halitosis. J Breath Res. 2008;2(1):017002.
Kazor CE, Mitchell PM, Lee AM, Stokes LN, Loesche WJ, Dewhirst FE, Paster BJ. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J Clin Microbiol. 2003;41(2):558–63.
Rocas IN, Hulsmann M, Siqueira JF Jr. Microorganisms in root canal-treated teeth from a German population. J Endod. 2008;34(8):926–31.
Rolph HJ, Lennon A, Riggio MP, Saunders WP, MacKenzie D, Coldero L, Bagg J. Molecular identification of microorganisms from endodontic infections. J Clin Microbiol. 2001;39(9):3282–9.
Schirrmeister JF, Liebenow AL, Pelz K, Wittmer A, Serr A, Hellwig E, Al-Ahmad A. New bacterial compositions in root-filled teeth with periradicular lesions. J Endod. 2009;35(2):169–74.
APV Colombo BSK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human Oral microbe identification microarray. J Periodontol. 2009;80(9):1421–32.
Zheng G, Summanen PH, Talan D, Bennion R, Rowlinson MC, Finegold SM. Phenotypic and molecular characterization of Solobacterium moorei isolates from patients with wound infection. J Clin Microbiol. 2010;48(3):873–6.
Detry G, Pierard D, Vandoorslaer K, Wauters G, Avesani V, Glupczynski Y. Septicemia due to Solobacterium moorei in a patient with multiple myeloma. Anaerobe. 2006;12(3):160–2.
Lau SK, Teng JL, Leung KW, Li NK, Ng KH, Chau KY, Que TL, Woo PC, Yuen KY. Bacteremia caused by Solobacterium moorei in a patient with acute proctitis and carcinoma of the cervix. J Clin Microbiol. 2006;44(8):3031–4.
CA Martin WRS, Borland CD, Karas JA. Femoral vein thrombophlebitis and septic pulmonary embolism due to a mixed anaerobic infection including Solobacterium moorei: a case report. J Med Case Rep. 2007;1:40.
Pedersen RM, Holt HM, Justesen US. Solobacterium moorei bacteremia: identification, antimicrobial susceptibility, and clinical characteristics. J Clin Microbiol. 2011;49(7):2766–8.
Blairon L, Maza ML, Wybo I, Pierard D, Dediste A, Vandenberg O. Vitek 2 ANC card versus BBL crystal anaerobe and RapID ANA II for identification of clinical anaerobic bacteria. Anaerobe. 2010;16(4):355–61.
Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 2013;30(5):1229–35.
Hiranmayi KV, Sirisha K, Ramoji Rao MV, Sudhakar P. Novel pathogens in periodontal microbiology. J Pharm Bioallied Sci. 2017;9(3):155–63.
Acknowledgements
Not applicable.
Funding
The study was financially supported by a National Major Science and Technology Project (grant number 2017ZX10103004), a Leading Innovation-oriented Biopharmaceutical Industry Cluster Project in Zheng-Luo-Xin National Innovation Demonstration Area (grant number181200211900), a CAMS Innovation Fund for Medical Sciences (grant number 2016-I2M-1-014), and a Beijing Science and Technology Project (Z181100001618015). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
WJL carried out experiments and wrote the manuscript; JY and YL were involved in the acquisition and analysis of data; MX and TK revised the manuscript critically for important intellectual content; YCX designed all the experiments, structured and wrote the paper and critically reviewed the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study protocol was approved by the Institutional Review Board of Peking Union Medical College Hospital (No. S-K615). Written informed consent was obtained from the patient’s family.
Consent for publication
Written informed consent was obtained from the patient’s family for publication of this case report and accompanying images. A copy of the written consent is available for review by the Editor of this journal.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, WJ., Xiao, M., Yi, J. et al. First case report of bacteremia caused by Solobacterium moorei in China, and literature review. BMC Infect Dis 19, 730 (2019). https://doi.org/10.1186/s12879-019-4359-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-019-4359-7