Abstract
Background
The vitamin D receptor (VDR) mediates the immunological function of vitamin D3, which activates macrophages, and vitamin D deficiency has been linked to tuberculosis risk. Single nucleotide polymorphisms (SNPs) in VDR may influence the function of vitamin D and susceptibility to tuberculosis.
Methods
This study included 217 patients with pulmonary tuberculosis (PTB) and 383 healthy subjects in a Tibetan Chinese population living in and near Xi’an. Association analyses of SNPs in VDR were performed with the SPSS 17.0 statistical packages, SNP stats software, Haploview software package (version 4.2), and the SHEsis software platform.
Results
Our results revealed a correlation between three SNPs (rs11574143, odds ratio [OR]: 1.47, 95 % confidence interval [CI]: 1.11 - 1.94, p = 0.006, p-adjust = 0.030; rs11574079, OR: 0.48, 95 % CI: 0.25 - 0.92, p = 0.023, p-adjust = 0.115; rs11168287, OR: 2.55, 95 % CI: 2.00 - 3.25, p = 1.730E-14, p-adjust = 0.865E-13) and PTB based on Chi-square tests. We observed the allele “A” of rs11574143 and rs11168287 increased the PTB risk and the allele “A” of rs11574079 provided a protective effect against PTB.
Conclusions
The goal of this study was the identification of putative associations between five SNPs (rs11574143, rs7975232, rs11574079, rs3819545 and rs11168287) in VDR and susceptibility to PTB. Our findings demonstrated associations between VDR polymorphisms and PTB development.
Similar content being viewed by others
Background
Pulmonary tuberculosis (PTB), which is, caused by the human pathogen Mycobacterium tuberculosis, is a major cause of morbidity and mortality in human populations worldwide. Susceptibility to disease upon M. tuberculosis infection is influenced by the agent, environmental and host genetic factors [1]. Each year more than 9 million people are infected by PTB and more than 1.7 million succumb to PTB annually [2]. Cell-mediated immunity is essential for the suppression of an M. tuberculosis infection as it is an intracellular parasite [3]. The fact that only 10 % of those infected with M. tuberculosis progress to clinical disease revealed that genetic factors, as well as environmental factors are involved in the pathophysiology of PTB [4]. In addition, the host genetic basis of PTB has been confirmed by twin studies that indicated a two times higher risk of disease in identical twins compared to non-identical twins [5]. There is a much higher risk of severe disease and death in children than in adults, therefore, the pediatric tuberculosis remains a public health emergency [6]. Many studies have shown that genetic factors play important roles in PTB disease development, and vitamin D receptor (VDR) gene has recently been found to be interesting candidate genes.
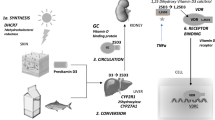
The active form of vitamin D, 1-25-dihydroxyvitamin D3, is an important hormone that modulates the activity of different defense and immune cells, including lymphocytes, monocytes, macrophages and epithelial cells [7]. Vitamin D restricts M. tuberculosis growth in macrophages through the production of the anti-microbial peptide, cathelicidin [8].Many studies showed an association between vitamin D deficiency and certain diseases such as systemic lupus erythematosus [9], diabetes mellitus [10], tuberculosis [11], etc. During a tuberculosis infection, vitamin D binds to VDR in macrophages, and this binging activates synthesis of the antimicrobial peptide cathelicidin, which restricts M. tuberculosis intracellular growth in macrophages [12], and eliminates Mycobacterium tuberculosis in phagolysosomes [13]. Thus, polymorphisms in VDR may contribute to PTB susceptibility.
In this study, we evaluated the roles of VDR gene polymorphisms and haplotypes on PTB susceptibility by conducting an extensive association analysis of a case–control study in a Tibetan Chinese population. We found considerable evidence for associations between three SNPs and PTB susceptibility.
Methods
Study participants
From October 2012 to September 2013, we recruited 217 patients to participate in an ongoing molecular epidemiological study at the Department of Respiratory Physicians of the Tangdu Hospital affiliated with The Fourth Military Medical University in Xi’an, China. The inclusion criteria in the case group were newly diagnosed pulmonary tuberculosis patients, Tibetan ethnic, have symptoms of PTB, positive sputum smear and chest radiography consistent with active disease. The exclusion criteria in case group were HIV positive, known to present diabetes mellitus and other severe diseases, and consuming immunosuppressive drugs.
From June 2012 to July 2013, we also recruited a random sample of 383 unrelated healthy individuals at the medical center of Tangdu Hospital to serve as controls. All of the study participants were Tibetan Chinese living in Xi’an and nearby. Controls subjects had no history PTB and no evidence of prior PTB noted on chest radiography. We enlisted them without regard to their age, gender, or disease stage.
SNP selection and genotyping
We selected five single nucleotide polymorphisms (SNPs) in VDR that had with minor allele frequencies (MAF) > 5 % and were associated with PTB in the HapMap Asian population. We extracted genomic DNA from peripheral blood samples using the GoldMag-Mini Whole Blood Genomic DNA Purification Kit (GoldMag Ltd. Xi'an, China) according to the manufacturer's protocol. Sequenom MassARRAY Assay Design 3.0 Software was used to design primers for amplification and extension reactions [14]. SNP genotyping using the standard protocol recommended by the manufacturer was performed by Sequenom MassARRAY RS1000 [14]. Finally, Sequenom Typer 4.0 Software was used to perform the data management and analysis [14, 15].
Statistical analysis
We used Microsoft Excel and the SPSS 17.0 statistical package (SPSS, Chicago, IL) to perform statistical analyses. All of the p-values presented in this study are two-sided, and p ≤ 0.05 was used as the threshold of statistical significance. Departure from Hardy-Weinberg Equilibrium (HWE) of each SNP frequency was assessed by an exact test on the control subjects. We calculated the genotype frequencies between the cases groups and control groups by Chi-square test [16].
Odds ratios (OR) and 95 % confidence intervals (CI) were calculated by unconditional logistic regression analyses [17]. We used the web-based software SNP stats to test the associations between SNPs and the risk for PTB in four genetic models (codominant, dominant, recessive, and additive) [18].
The software platform (http://sampsize.sourceforge.net/iface/s3.html) was used for evaluating the statistical power of this case-control study. We used the Haploview software package (version 4.2) and SHEsis software platform (http://www.nhgg.org/analysis/) for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci [19, 20].
Results
The PCR primers for the 5 selected SNPs, which were designed using the Sequenom MassARRAY Assay Design 3.0 Software, were listed in Table 1.
Basic information of candidate SNPs in this study was shown in Table 2. We also adjusted the p-value by Bonferroni correction. It listed the minor allele frequency (MAF) of cases and controls, we found there existed a correlation between three loci (rs11574143, OR: 1.47, 95 % CI: 1.11 - 1.94, p = 0.006, p-adjust = 0.030; rs11574079, OR: 0.48, 95 % CI: 0.25 - 0.92, p = 0.023, p-adjust = 0.115; rs11168287, OR: 2.55, 95 % CI: 2.00 - 3.25; p = 1.730E-14, p-adjust = 0.865 E-13) and PTB based on Chi-square tests. Rs7975232 and rs3819545 had no connection with PTB susceptibility. All of the tested SNPs conformed to HWE in the control group of this study (p > 0.05). Post-hoc power value we calculated of rs11574143, rs7975232, rs11574079, rs3819545, rs11168287 were 77.59 %, 45.41 %, 62.96 %, 4.13 %, 100 %, respectively.
The relationships between rs11574143, rs11574079, rs11168287 and PTB risk were listed in Table 3. We observed the allele “A” of rs11574143 and rs11168287 increased the PTB risk and the allele “A” of rs11574079 provided a protective effect against PTB.
For rs11574143, In the codominant model, the genotype “A/G” (OR =1.54; 95 % CI, 1.08 - 2.19; p = 0.024, p-adjust = 0.12) increased PTB risk by 1.54-fold; In the dominant model, the genotype “G/A” and “A/A” (OR = 1.59; 95 % CI, 1.13 - 2.23; p = 0.0077, p-adjust = 0.039) increased PTB risk by 1.59-flod; In the additive model, the allele “A” increased PTB risk by 1.48-fold (OR = 1.48; 95 % CI, 1.11 - 1.96; p = 0.0066, p-adjust = 0.033). We also observed another susceptibility SNP, rs11168287, p-value < 0.0001 (p-adjust < 0.0005) for all of the four models in SNP analyses. In the codominant model, the genotype “A/G” (OR = 2.38; 95 % CI, 1.57 - 3.60) and “A/A” (OR = 6.07; 95 % CI, 3.69 - 9.97) increased PTB risk by 2.38-fold and 6.07-fold respectively; In the dominant model, the genotype “A/G” and “A/A” increased more than 3-fold PTB risk (OR = 3.19; 95 % CI, 2.16 - 4.72); In the recessive model, the genotype “A/A” increased PTB risk by 3.60-fold (OR = 3.60; 95 % CI, 2.38 - 5.46). In the additive model, the allele “A” increased PTB risk by 2.46-fold (OR = 2.46; 95 % CI, 1.92 - 3.15).
The SNP rs11574079 correlated with a protective effect against PTB in the dominant model (OR: 0.48, 95 % CI: 0.24 - 0.92, p = 0.021, p-adjust =0.105) and in the additive model (OR: 0.47, 95 % CI: 0.25 - 0.91, p = 0.018, p-adjust = 0.09).
A block (rs4947492 and rs12718945) was detected in studied VDR SNPs by haplotype analyses (Fig. 1). The results of the association between the VDR haplotype and the risk of PTB were listed in Table 4. Altogether there were three haplotypes and only the haplotype “A/A” was found to increase risk of suffering from PTB by 1.45-fold (OR = 1.45; 95 % CI, 1.09 - 1.94; p = 0.01). We adjusted the p-value by Bonferroni correction, the haplotype “A/A” still existed a statistically significant difference (p = 0.02).
Discussion
Our comprehensive analysis of SNPs in VDR suggests that VDR genotypes and haplotypes associate with PTB risk. We found two PTB risk associated SNPs, rs11574143 and rs11168287, and one PTB protective SNP, rs11574079, in VDR. The other two VDR SNPs, rs7975232 and rs3819545, had did not associate with PTB susceptibility.
In previously published studies, there is no research reported that the association between VDR gene and PTB susceptibility. We for the first time studied the correlations between three SNPs (rs11574143, rs11168287 and rs11574079) in VDR gene and PTB susceptibility in a Tibetan Chinese population, rs3819545 showed no correlations with PTB susceptibility, and we got some valuable results. Further studies should involve some others regions and ethnic groups.
A few studies have analyzed the association between the VDR ApaI (rs7975232) polymorphism and TB susceptibility. A study conducted by Selvaraj et.al. in the Indian population shows that the ‘A/A’ genotype is associated with resistance to pulmonary TB in males, but not in females [21]. In a study carried out in pulmonary TB patients of south India, the Bb genotype of VDR BsmI was associated with susceptibility to TB, whereas A/A genotype of ApaI and B/B genotype of BsmI were associated with resistance to pulmonary TB [22, 23]. Our results are inconsistent with the results of some previously published studies, a possible explanation could be their ethnic differences.
Comparisons with other researches showed that population or various ethnicities give different results. Many studies have suggested associations between VDR SNPs and PTB in variety populations [22, 24, 25]. The different results shown by Central Indian and South Indian populations may be due to the diversity of these two Indian populations and their difference in origins [26]. A latest study was in accordance with those obtained by other authors for West African populations, but in total discrepancy with the results for Indian, Iranian and Chinese populations [27]. Several case-control studies, performed in populations with high TB incidence, had studied the correlation between the VDR gene polymorphisms and genetic susceptibility to PTB, but the results were inconclusive, presumably because the populations were ethnically and geographically different and genetically diverse [25, 28, 29]. Wei et al. reported no association was found between VDR polymorphisms (either allele or genotype) and susceptibility to pediatric PTB in the Chinese Han population [30]. Also, a study in Cambodia [31] and Tanzania [32] found no associations between VDR gene and PTB. These differing results could be caused by gene-environment interaction, gene-gene interaction, and gene-agent interactions. These reports suggest that the differing association results between VDR genotypes in different ethnic populations may result from gene-environment [33] and gene-gene interactions [34, 35].
There are other factors that may influence the results. It is known that before vitamin D enters the macrophage, inactive vitamin D in serum could be bound to vitamin D binding protein or in a free state. A recent study showed that VDR variant (rs7968585) can predict risk for hip fracture, cancer, myocardial infarction, and mortality among older Caucasian individuals with a low vitamin D status [36]. This association was only apparent in individuals with a low vitamin D status, highlighting the importance of taking into account gene-environment interactions. Vitamin D status appears to be involved in the activation of macrophages and restricts the intracellular development of mycobacteria [37]. This effect of vitamin D may be influenced by polymorphisms in the VDR gene. However, although the relationship between the internal environment and the host gene is unknown, it may explain the heterogeneity observed in the association studies and it should be addressed as an influencing factor. The polymorphic variants of VDR gene along with other gene and environmental factors may be responsible for an altered cell-mediated immunity against M. tuberculosis in a susceptible or resistant host. Although several investigators reported and suggested that the VDR polymorphism may be of immune-regulatory significance for PTB but it is not clear that the polymorphisms determine susceptibility to the development of clinical disease or susceptibility to infection. Further studies are required to determine how VDR polymorphisms influence susceptibility to infection or to clinical disease development. In order to understand the pathogenesis of pulmonary TB that is useful for prophylaxis and therapy. Ethnic-specific genetic association with TB susceptibility may guide TB therapy and prophylaxis in an ethnic-specific manner. A post hoc power analysis was conducted and the power analysis showed that except rs11168287, other SNPs failed to reach minimum level (80 %) for this case-control study. In this case, we could not conclude that the difference was not statistically significant, but need to increase sample size to verify. Future research should pay attention to this point.
Because we limited ethnicity of our subjects to Tibetan Chinese, and limited the locations of our subjects to Xi’an City and its surrounding area, there were no substantial population admixtures in our study populations. However, some limitations should be considered when interpreting our results. Our sample size was small and only included 217 PTB cases. Also, gene-environment and gene-gene interactions may effect VDR expression and activity. And the assessment of the environmental factors such as tobacco, diet and alcohol consumption and the possibility of inaccurate exposure should be considered. The susceptibility and resistance to PTB are the result of the interactions between environment, socioeconomic status and host genes, therefore, further studies are required to elucidate the complex associations between the VDR gene variants and the immune response against PTB. These data will lead to a better understanding of the immunological and genetic pathways in tuberculosis, and will provide new ideas for potential treatments and prophylaxes to reduce the prevalence and incidence of PTB caused by M. tuberculosis infections.
Conclusions
Our findings demonstrated that genetic variants of VDR play complex roles in the development of PTB and provided new evidence for associations between SNPs and haplotypes of VDR and the risk of PTB. This study offers new information on the relationship between VDR polymorphisms and PTB .Moreover, this study reveals the molecular markers associated with of PTB susceptibility and could therefore be used as diagnostic and prognostic markers for PTB patients in clinical study.
Abbreviations
VDR, vitamin D receptor; SNP, single nucleotide polymorphism; PTB, pulmonary tuberculosis; MAF, minor allele frequencies; HWE, Hardy-Weinberg Equilibrium; OR, odds ratio; CI, confidence interval
References
Bellamy R, Ruwende C, Corrah T, McAdam KP, Whittle HC, Hill AV. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N Engl J Med. 1998;338(10):640–4.
Thakur M. Global Tuberculosis Control: WHO report 2010. National Medical Journal of India. 2012;14(3):189–90.
Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19(1):93–129.
Casanova J-L, Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annu Rev Immunol. 2002;20(1):581–620.
Comstock G. Tuberculosis in twins: a re-analysis of the Prophit survey. Am Rev Respir Dis. 1978;117(4):621–4.
Sulaja BDM, Chauhan UK. rs 10735810 of Vitamin D Receptor (VDR) gene: association with pulmonary tuberculosis in children. Tuberculosis (TB). 2014;2014:04–21.
Bellamy R, Ruwende C, Corrah T, McAdam K, Thursz M, Whittle H, Hill A. Tuberculosis and chronic hepatitis B virus infection in Africans and variation in the vitamin D receptor gene. J Infect Dis. 1999;179(3):721–4.
Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, Ochoa MT, Schauber J, Wu K, Meinken C, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science (New York, NY). 2006;311(5768):1770–3.
Handono K, Marisa D, Kalim H. Association between the low levels of vitamin D and Treg function in systemic lupus erythematosus patients. Acta Med Indones. 2013;45:26–31.
Afzal S, Bojesen SE, Nordestgaard BG. Low 25-hydroxyvitamin D and risk of type 2 diabetes: a prospective cohort study and metaanalysis. Clin Chem. 2013;59(2):381–91.
Nnoaham KE, Clarke A. Low serum vitamin D levels and tuberculosis: a systematic review and meta-analysis. Int J Epidemiol. 2008;37(1):113–9.
Liu PT, Stenger S, Tang DH, Modlin RL. Cutting edge: vitamin D-mediated human antimicrobial activity against Mycobacterium tuberculosis is dependent on the induction of cathelicidin. J Immunol. 2007;179(4):2060–3.
Chun RF, Adams JS, Hewison M. Back to the future: a new look at ‘old’vitamin D. J Endocrinol. 2008;198(2):261–9.
Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009;2:12. 11-12.12. 16.
Thomas RK, Baker AC, DeBiasi RM, Winckler W, LaFramboise T, Lin WM, Wang M, Feng W, Zander T, MacConaill LE. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39(3):347–51.
Adamec C. Example of the use of the nonparametric test. Test x2 for comparison of 2 independent examples. Cesk Zdrav. 1964;12:613–9.
Bland JM, Altman DG. The odds ratio. BMJ. 2000;320(7247):1468.
Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–9.
Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5.
Yong Y, Lin H. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–8.
Selvaraj P, Chandra G, Jawahar MS, Rani MV, Rajeshwari DN, Narayanan PR. Regulatory role of vitamin D receptor gene variants of Bsm I, Apa I, Taq I, and Fok I polymorphisms on macrophage phagocytosis and lymphoproliferative response to mycobacterium tuberculosis antigen in pulmonary tuberculosis. J Clin Immunol. 2004;24(5):523–32.
Roth DE, Soto G, Arenas F, Bautista CT, Ortiz J, Rodriguez R, Cabrera L, Gilman RH. Association between vitamin D receptor gene polymorphisms and response to treatment of pulmonary tuberculosis. J Infect Dis. 2004;190(5):920–7.
Selvaraj P, Kurian SM, Chandra G, Reetha AM, Charles N, Narayanan PR. Vitamin D receptor gene variants of BsmI, ApaI, TaqI, and FokI polymorphisms in spinal tuberculosis. Clin Genet. 2004;65(1):73–6.
Banoei MM, Mirsaeidi MS, Houshmand M, Tabarsi P, Ebrahimi G, Zargari L, Kashani BH, Masjedi MR, Mansouri SD, Ramirez J. Vitamin D receptor homozygote mutant tt and bb are associated with susceptibility to pulmonary tuberculosis in the Iranian population. Int J Infect Dis. 2010;14(1):e84–85.
Kang TJ, Jin SH, Yeum CE, Lee SB, Kim CH, Lee SH, Kim KH, Shin ES, Chae GT. Vitamin D Receptor Gene TaqI, BsmI and FokI Polymorphisms in Korean Patients with Tuberculosis. Immune network. 2011;11(5):253–7.
Papiha SS. Genetic variation in India. Hum Biol. 1996;68(5):607–28.
Alexandra S-G, Georgiana DC, Nicoleta C, Daniela PM, Traian S, Veronica S. Apa I and Taq I polymorphisms of VDR (vitamin D receptor) gene in association with susceptibility to tuberculosis in the Romanian population. Rom Biotechnol Lett. 2013;18(1):7956–62.
Liu W, Cao WC, Zhang CY, Tian L, Wu XM, Habbema JD, Zhao QM, Zhang PH, Xin ZT, Li CZ, et al. VDR and NRAMP1 gene polymorphisms in susceptibility to pulmonary tuberculosis among the Chinese Han population: a case-control study. Int J Tuberc Lung Dis. 2004;8(4):428–34.
Bornman L, Campbell SJ, Fielding K, Bah B, Sillah J, Gustafson P, Manneh K, Lisse I, Allen A, Sirugo G, et al. Vitamin D receptor polymorphisms and susceptibility to tuberculosis in West Africa: a case-control and family study. J Infect Dis. 2004;190(9):1631–41.
WeiWei J, ZhaoNa L, Lin S, ShunYing Z, HuiMin L, AnXia J, YaJie G, ZaiFang J, ADong S. Vitamin D receptor gene polymorphisms and susceptibility to pediatric tuberculosis among the Chinese Han population. Chinese J Pract Pediatr. 2009;24(4):264–6.
Delgado JC, Baena A, Thim S, Goldfeld AE. Ethnic-specific genetic associations with pulmonary tuberculosis. J Infect Dis. 2002;186(10):1463–8.
Soborg C, Andersen AB, Range N, Malenganisho W, Friis H, Magnussen P, Temu MM, Changalucha J, Madsen HO, Garred P. Influence of candidate susceptibility genes on tuberculosis in a high endemic region. Mol Immunol. 2007;44(9):2213–20.
Wilkinson RJ, Llewelyn M, Toossi Z, Patel P, Pasvol G, Lalvani A, Wright D, Latif M, Davidson RN. Influence of vitamin D deficiency and vitamin D receptor polymorphisms on tuberculosis among Gujarati Asians in west London: a case-control study. Lancet (London, England). 2000;355(9204):618–21.
Velez DR, Hulme WF, Myers JL, Weinberg JB, Levesque MC, Stryjewski ME, Abbate E, Estevan R, Patillo SG, Gilbert JR, et al. NOS2A, TLR4, and IFNGR1 interactions influence pulmonary tuberculosis susceptibility in African-Americans. Hum Genet. 2009;126(5):643–53.
Ben-Selma W, Harizi H, Boukadida J. Association of TNF-alpha and IL-10 polymorphisms with tuberculosis in Tunisian populations. Microbes Infect/ Institut Pasteur. 2011;13(10):837–43.
Sulaja BDM. Rs 10735810 of Vitamin D Receptor (VDR) gene: association with pulmonary tuberculosis in children. J Infect Mol Biol. 2014;2(3):32–4.
Vidyarani M, Selvaraj P, Jawahar MS, Narayanan PR. 1, 25 Dihydroxyvitamin D3 modulated cytokine response in pulmonary tuberculosis. Cytokine. 2007;40(2):128–34.
Acknowledgements
We are grateful to all of the patients and individuals for their participation.
Funding
This work was supported by the Natural Science Foundation of Xizang (Tibet) Autonomous Region (No. 12KJZRYMY10) and the Major Science and Technology Research Projects of Xizang (Tibet) Autonomous Region (2015XZ01G17).
Availability of data and materials
Not applicable.
Authors’ contributions
QH designed the study, carried out the molecular genetic studies, participated in the statistical analyses, and drafted the manuscript. ZC and GL participated in the molecular genetic studies and the statistical analyses. FM, HZ and SX participated in the design of the study and performed the statistical analyses. YW conceived of the study, participated in the design of the study, coordinated the study and assisted in drafting the manuscript. TJ and LK also conceived of the study, participated in the design and coordination of the study, and funded the study. All of the authors read and approved the final manuscript.
Authors’ information
Qunying Hu and Zhengshuai Chen joint first authors.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All of the subjects were informed of the purpose and the experimental procedures of the study. The Human Research Committee of the Tangdu Hospital for Approval of Research Involving Human Subjects approved the use of human tissue in this study. We also obtained signed informed consent from each study participant.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Hu, Q., Chen, Z., Liang, G. et al. Vitamin D receptor gene associations with pulmonary tuberculosis in a Tibetan Chinese population. BMC Infect Dis 16, 469 (2016). https://doi.org/10.1186/s12879-016-1699-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-016-1699-4