Abstract
Background
Abdominal aortic aneurysm (AAA) is a common cardiovascular system disease with high mortality. The aim of this study was to identify potential genes for diagnosis and therapy in AAA.
Methods
We searched and downloaded mRNA expression data from the Gene Expression Omnibus (GEO) database to identify differentially expressed genes (DEGs) from AAA and normal individuals. Then, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis, transcriptional factors (TFs) network and protein-protein interaction (PPI) network were used to explore the function of genes. Additionally, immunohistochemical (IHC) staining was used to validate the expression of identified genes. Finally, the diagnostic value of identified genes was accessed by receiver operating characteristic (ROC) analysis in GEO database.
Results
A total of 1199 DEGs (188 up-regulated and 1011 down-regulated) were identified between AAA and normal individual. KEGG pathway analysis displayed that vascular smooth muscle contraction and pathways in cancer were significantly enriched signal pathway. The top 10 up-regulated and top 10 down-regulated DEGs were used to construct TFs and PPI networks. Some genes with high degrees such as NELL2, CCR7, MGAM, HBB, CSNK2A2, ZBTB16 and FOXO1 were identified to be related to AAA. The consequences of IHC staining showed that CCR7 and PDGFA were up-regulated in tissue samples of AAA. ROC analysis showed that NELL2, CCR7, MGAM, HBB, CSNK2A2, ZBTB16, FOXO1 and PDGFA had the potential diagnostic value for AAA.
Conclusions
The identified genes including NELL2, CCR7, MGAM, HBB, CSNK2A2, ZBTB16, FOXO1 and PDGFA might be involved in the pathology of AAA.
Similar content being viewed by others
Background
Abdominal aortic aneurysm (AAA), defined as the aortic diameter > 3.0 cm, is a cardiovascular system disease that is characterized by aortic dilation that exceeds the normal aortic diameter by more than 50%. AAA dilatation will lead to rupture of the aorta, which results in bleeding. Generally, it is asymptomatic until the rupture event occurs [1]. Additionally, AAA is common in adult patients, especially elderly men, and leads to severe complications [2,3,4]. Up to now, the etiology of AAA remains unclear. It is noted that some clinical risk factors including smoking history, advanced age, family history, hypertension, hyperlipidaemia, atherosclerosis, chronic obstructive pulmonary disease are remarkably related to AAA [3, 5,6,7]. It is also observed that the intricate interplay of apoptosis, inflammation and matrix degradation is involved in the development of this disorder [8,9,10]. Anyway, the pathophysiology of AAA is complex, but fundamentally aneurysm comes from the vessel wall structural integrity loss and the vessel wall weakening. It is pointed out that vascular smooth muscle cells are the critical cell type involved in the development of AAA [11].
In a word, AAA is a common and late onset disease, which can rupture with a high mortality if not treated. In some clinical practice, there is no effective treatment other than surgical approaches to repair AAA [12]. And endovascular aneurysm repair has improved detection and lower mortality rates of AAA [13,14,15,16,17]. However, morbidity and mortality after surgery are still common [18, 19]. Therefore, understanding the genetic architecture and pathological mechanism of the disease may provide valuable information for elucidation of pathogenic mechanisms and signal pathways in AAA and the discovery of potential biomarkers and drug targets in AAA diagnosis and non-surgical treatment therapy.
In this study, we tried to find differentially expressed genes (DEGs) in AAA by integrated analysis. Then, functional enrichment analysis including Gene Ontology (GO) and Kyoto Encyclopedia of Genes Genomes (KEGG) was used to investigate the biological function of DEGs followed by transcriptional factors (TFs) network an d protein-protein interaction (PPI) network construction of top 20 DEGs (10 up-regulated and 10 down-regulated). Immunohistochemical (IHC) staining was applied to validate the expression of candidate DEGs. Finally, receiver operating characteristic (ROC) analyses was applied to analyze diagnosis ability of identified DEGs. Our study may be helpful in understanding the pathogenic mechanism and finding valuable diagnosis biomarkers and therapy drug in AAA.
Methods
Datasets
In this study, we searched datasets from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) with the keywords abdominal aortic aneurysm [All Fields] AND (“gse”[Filter] AND “Homo sapiens”[Organism]). The study type was described as “expression profiling by array.” All selected datasets were genome-wide expression data of AAA group and/or normal group tissue samples. Those standardized or primary datasets were included in this study. Finally, a total of 3 datasets including GSE7084, GSE47472 and GSE57691 were screened, which was shown in Table 1.
Analysis of DEGs
Raw expression data of AAA patients in this study were downloaded. Limma and metaMA packages were used to identify the DEGs. And the inverse normal method was used to combine the p value in metaMA. The false discovery rate (FDR) was performed for multiple testing corrections of raw p value through the Benjamin and Hochberg method [20, 21]. The threshold of DEGs was set as FDR < 0.01.
Functional annotation analyses of DEGs
To obtain the biological function and signaling pathways of DEGs, the Metascape software was used for Gene Ontology (GO, http://www.geneontology.org/) annotation and Kyoto Encyclopedia of Genes Genomes (KEGG, http://www.genome. jp/kegg/pathway.html) pathway enrichment of DEGs. The threshold of GO function and KEGG pathway of DEGs was all set as FDR < 0.05.
PPI network construction
It is useful for understanding the molecule mechanism of AAA to study the interactions between proteins. In order to gain insights into the interaction between proteins encoded by DEGs and other proteins, the database of BioGRID (http://thebiogrid.org) was used to retrieve the predicted interactions between top 20 proteins encoded by DEGs (10 up-regulated and 10 down-regulated) and other proteins. The PPI network was generated by the Cytoscape Software (http://cytoscape.org/). A node in the PPI network denotes protein, and the edge denotes the interactions.
Analysis of potential TFs to target DEGs
TFs play a critical role in regulating gene expression. We downloaded the TFs in the human genome and the motifs of genomic binding sites from the TRANSFAC. Moreover, the 2 KB sequence in the upstream promoter region of DEGs was downloaded from UCSC (http://www.genome.ucsc.edu/cgi-bin/hgTables). Target sites of potential TFs were then distinguished. Finally, the transcriptional regulatory network was visualized by Cytoscape software.
Immunohistochemical (IHC) staining for CCR7 and PDGFA
In this study, a patient with AAA and a normal individual was enrolled for the IHC experiment. The 5 μm slides were incubated with anti CCR7 primary rabbit anti-human polyclone antibody (1:500 dilution; abcam) and anti PDGFA primary rabbit anti-human polyclone antibody (1:500 dilution; Invitrogen) followed incubated with peroxidase conjugated goat anti-rabbit secondary antibody (1:200 dilution; Vector). For color visualization, diaminobenzidine (DAB) substrate (Vector) was applied. The staining area was analyzed by the software of Image Pro-plus 6.0 (Media Cybernetics Corporation, arrendale, PA, USA), and quantified by the IHC staining score (intensity score × positive rate score). The negative (−), positive (+), positive (++), positive (+++) of intensity scores represented 0, 1, 2 and 3, respectively. The positive rate score including negative, 1–25%, 26–50%, 51–75% and 76–100% represented 0, 1, 2, 3 and 4, respectively. IHC staining score of 0, 1~ 4, 5~ 8 and 9~ 12 represented negative, slight positive, moderate positive and strong positive, respectively.
All patients provided written informed consent with the approval of the ethics committee of the First Affiliated Hospital of Wenzhou Medical University (2017147).
Receiver operating characteristic analyses
By using pROC package in R language we performed the receiver operating characteristic (ROC) analyses to assess the diagnostic value of DEGs (NELL2, CCR7, MGAM, HBB, CSNK2A2, ZBTB16, FOXO1 and PDGFA) in AAA. The area under the curve (AUC) was calculated and the ROC curve was generated.
Results
DEGs analysis
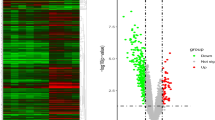
Raw expression profiles of AAA patients were downloaded from the data portal of the GEO database. A total of 1199 DEGs were identified as the threshold at FDR < 0.01, consisting of 188 up-regulated genes and 1011 down-regulated genes. The top 10 up- and down-regulated DEGs are listed in Table 2. The heat map of the top 50 DEGs is shown in Fig. 1.
The heat map of top 50 DEGs. The diagram presents the result of a two-way hierarchical clustering of the top 50 DEGs and samples. The clustering is constructed using the complete-linkage method together with the Euclidean distance. Each row represents a DEG and each column, a sample. The DEG clustering tree is shown on the right. The colour scale illustrates the relative level of DEG expression: red, below the reference channel; green, higher than the reference
Functional and pathway enrichment analyses of DEGs
To investigate the biological function of the identified DEGs in AAA, GO term and KEGG pathway enrichment analyses was performed. In GO term and KEGG pathway enrichment analyses, circulatory system development, muscle structure development and translational initiation were the most significant enrichment in biological process (Fig. 2); Oxidoreductase activity, electron carrier activity, protein domain specific binding were the most notable enrichment in molecular function (Fig. 3); Mitochondrial part, focal adhesion and intracellular ribonucleoprotein complex were the most significant enrichment in cellular component (Fig. 4). The top 10 GO terms of DEGs are shown in Table 3, and the KEGG enrichment signal pathways of DEGs shown in Table 4. The vascular smooth muscle contraction and pathways in cancer that were significantly related to AAA are shown in Fig. 5 and Fig. 6, respectively.
Establishment of TFs-target genes regulatory network
In order to study the TFs-target genes regulatory network for AAA, we utilized TRANSFAC to identify TFs regulating the top ten up-regulated or down-regulated DEGs. In the end, we obtained transcriptional regulatory networks comprised of 190 pairs of TFs-genes involving 40 TFs (Fig. 7). In this network, the top 7 downstream genes covered by most TFs were neural EGFL like 2 (NELL2, degree = 13), C-C motif chemokine receptor 7 (CCR7, degree = 9), maltase-glucoamylase (MGAM, degree = 8), hemoglobin subunit beta (HBB, degree = 8). Five hub TFs were HNF-4 (degree = 10), Oct-1 (degree = 10), Pax-4 (degree = 8), Evi-1 (degree = 6) and Nkx2–5 (degree = 6) (Table 5).
PPI network
To obtain the interaction between the proteins encoded by DEGs and other proteins, PPI network was explored and visualize by Cytoscape. PPI networks of the top 10 up-regulated and the top 10 down-regulated DEGs were presented in Fig. 8. As Fig. 8 shows, the network consisted of 539 nodes and 567 edges. The red and green diamonds indicate the up- and down-regulated genes in AAA, respectively. The blue ellipses present the proteins that interacted with those proteins encoded by DEGs. The top three proteins with a high degree were casein kinase 2 alpha 2 (CSNK2A2, degree = 184), zinc finger and BTB domain containing 16 (ZBTB16, degree = 113) and forkhead box O1 (FOXO1, degree = 53).
Validation of CCR7 and PDGFA in AAA
In order to validate the expression of CCR7 and platelet derived growth factor subunit A (PDGFA), we assessed the protein expression of CCR7 and PDGFA in AAA through the immunohistochemistry (Fig. 9 and Fig. 10). The result showed that CCR7 was obviously up-regulated in AAA compared with the control, which was consistent with the bioinformatic consequence. However, PDGFA was significantly up-regulated in AAA compared with the control, which was not in line with the bioinformatic result.
ROC curve analysis
In order to access the discriminatory ability of the NELL2, CCR7, MGAM, HBB, CSNK2A2, ZBTB16, FOXO1 and PDGFA among AAA tissues and adjacent non-tumor tissues generated from GEO database, ROC curve analyses were conducted and AUC were calculated. As Fig. 11 shown, the AUC of all these genes was more than 0.8. For AAA diagnosis, the sensitivity and specificity of these genes were very high.
Discussion
In spite of improvement to surgical techniques that have been made in AAA treatment, morbidity and mortality after operations are still common. AAA seriously influences the life quality of patients and brings a heavy burden on the family. Therefore, it is urgent to elucidate AAA pathogenesis mechanism for developing novel diagnose and therapeutic target.
TFs are key regulatory factors in gene expression. The construction of regulatory networks between TFs and target genes is helpful in understanding the biological regulatory mechanism in the development of AAA. In this study, we found NELL2, CCR7, MGAM and HBB were significantly expressed genes with the most degree under the regulation of TFs including HNF-4, Oct-1 and Pax-4. NELL2 is a neural tissue-enriched protein in mammal and it is a receptor for vascular endothelial growth factor-A, which plays an important role in angiogenesis. It is reported that the mRNA expression of NELL2 is up-regulated in benign prostate hyperplasia and prostate cancer [22]. In addition, NELL2 is regarded as the potential biomarker for bladder cancer [23]. CCR7 is a pro-inflammatory cytokine and is found in human atherosclerotic plaques [24]. It is found that expression of CCR7 is dramatically down-regulated in human carotid atherosclerotic plaques [25]. MGAM is found down-regulated and considered as a candidate serum biomarker in colorectal cancer [26]. Additionally, MGAM is a significantly mutated gene in lung adenocarcinoma [27]. HBB is suggested as a potential biomarker in the plasma sample of patients with AAA [28]. In this study, we found that NELL2, CCR7, MGAM and HBB were up-regulated in the AAA tissues, which played crucial roles in the carcinogenesis of AAA.
The interaction among proteins determines the characteristic of the cell, tissue and individual. The study of PPI is a useful way to find the potential drug target of AAA. Herein, we found three genes including CSNK2A2, ZBTB16 and FOXO1 were for the most degree in the PPI network. CSNK2A2 is found to be correlated with ovarian cancer patient survival. Furthermore, the down-regulation of CSNK2A2 will decrease the proliferation of ovarian cancer cells [29]. ZBTB16, also called PLZF, plays an important role in oncogenesis and is first identified in acute promyelocytic leukemia [30]. Based on a microarray study, ZBTB16 is found to be down-regulated in AAA [31]. FOXO1 is a transcription factor and plays roles in diverse physiological processes including Akt-dependent cell proliferation and apoptosis [32]. Additionally, FOXO1 is also involved in energy metabolism and autophagy [33]. In our study, we found down-regulated expression of CSNK2A2, ZBTB16 and FOXO1. It is worth mentioning that ZBTB16 and FOXO1 were also involved in the pathways in cancer according to the KEGG analysis. In addition, PDGFA was one of the top ten down-regulated genes and also involved in the pathways in cancer. PDGFA is expressed in vascular smooth muscle cells and has been involved in migration and proliferation of vascular smooth muscle cells [34]. Moreover, the importance of PDGFA in the arterial system has been demonstrated on account of that fact that the proliferation of arterial vascular smooth muscle cells was strongly stimulated by PDGFA [35]. Moreover, an in situ hybridization study has demonstrated mRNA for PDGFA in atherosclerotic plaques [36]. In this study, we found that PDGFA was down-regulated in AAA. However, the IHC result was not consistent with the bioinformatic analysis. The small sample size may account for the discrepancy. In a word, CSNK2A2, ZBTB16 and FOXO1 played a crucial role in the oncogenesis of AAA and could be considered as drug targets of AAA.
Apart from the cancer signal pathway, vascular smooth muscle contraction was another signal pathway identified that associated with AAA. Vascular smooth muscle cells have been shown to play an important synthetic role in vascular remodeling [37, 38]. It is pointed out that vascular smooth muscle cells are the main component of the aortic media and the dysfunction plays a key role in different arterial diseases, such as AAA [39]. In addition, vascular smooth muscle cell activation is the main hallmark of atherosclerosis, which is a risk factor of AAA [40,41,42,43]. Several down-regulated genes were significantly involved in the signal pathway, such as AGTR1, CALD1, EDNRA, MYH11, RAMP1, ROCK1 and ROCK2.
Angiotensin II receptor type 1 (AGTR1) is a cardiovascular risk gene. Jones, G. T et al. found that AGTR1 was remarkably associated with AAA [44]. In addition, the 1166A > C polymorphism in AGTR1 has been demonstrated to be associated with AAA [45, 46]. It is noted that AGTR1 blockers (ARBs) have been investigated for prevention or delay of aortic dilatation [47]. It is reported that the expression of caldesmon 1 (CALD1) is increased in aortas, which protects from aneurysm. This suggested that importance of CALD1 in maintaining vascular integrity in AAA. Endothelin receptor type A (EDNRA) is primarily located in the vascular smooth muscle cells and mediates vasoconstriction and proliferation [48]. It has been reported that EDNRA on chromosome 4q31 is related to intracranial aneurysm [49]. It is found that heterozygous mutation of myosin heavy chain 11 (MYH11) results in the early and severe decrease in the aortic wall elasticity [50]. Additionally, it has been demonstrated the relationship between MYH11 genetic and epigenetic and thoracic aortic aneurysms and dissections [51, 52]. Receptor activity modifying protein 1 (RAMP1) is a member of a family of calcitonin receptor modifying proteins and is thought to play an important role in regulating blood pressure by vascular relaxation. Tsujikawa K et al. found that ramp1-deficient mice exhibited elevated blood pressure [53]. It is pointed out that the mRNA levels of RAMP1 are decreased in AAA [31]. It is found that the expression of Rho associated coiled-coil containing protein kinase 1 (ROCK1) and Rho associated coiled-coil containing protein kinase 1 (ROCK2) was increased at the AAA lesion compared with control [54]. Thus it can be seen that AGTR1, CALD1, EDNRA, MYH11, RAMP1, ROCK1 and ROCK2 played an important role in vascular smooth muscle contraction, which was significantly associated with AAA.
In order to access the discriminatory ability of identified genes in the TFs and PPI network, eight genes including NELL2, CCR7, MGAM, HBB, CSNK2A2, ZBTB16, FOXO1 and PDGFA were applied to ROC curve analyses among AAA tissues and adjacent non-tumor tissues in GEO database. Our result showed that the AUC of all these genes was more than 0.8, especially HBB (AUC: 0.906), CSNK2A2 (AUC: 0.945), ZBTB16 (AUC: 0.950), FOXO1 (AUC: 0.940) and PDGFA (AUC: 0.930). This suggested that NELL2, CCR7, MGAM, HBB, CSNK2A2, ZBTB16, FOXO1 and PDGFA may have value in diagnosis of the development of AAA.
Conclusions
In summary, we found a series of DEGs in AAA. Among which, eight genes including NELL2, CCR7, MGAM, HBB, CSNK2A2, ZBTB16, FOXO1 and PDGFA could be used for the diagnosis biomarkers of AAA. Especially, CSNK2A2, ZBTB16 and FOXO1 could be considered as drug targets in the therapy of AAA. In addition, vascular smooth muscle contraction was an important signal pathway identified in this study, which played crucial roles in the aortic angiogenesis of AAA. There are limitations to our study. Firstly, the sample size in the IHC experiment is small and large numbers of tissue samples are needed to further validate the identified DEGs. Secondly, biological function of identified DEGs is not investigated, some in vivo or in vitro experiments are needed to further study the molecular mechanism of AAA. Thirdly, the sample size of normal individuals in the selected dataset is less than that of the patient group. In further studies, it is better to sample equal numbers of individuals in both groups in order to reduce the false positive/negative rate for up−/down-regulated DEGs detection.
Abbreviations
- AAA:
-
abdominal aortic aneurysm
- AGTR1:
-
angiotensin II receptor type 1
- ARBs:
-
AGTR1 blockers
- AUC:
-
area under the curve
- CALD1:
-
caldesmon 1
- CCR7:
-
C-C motif chemokine receptor 7
- CSNK2A2:
-
casein kinase 2 alpha 2
- DAB:
-
diaminobenzidine
- DEGs:
-
differentially expressed genes
- EDNRA:
-
endothelin receptor type A
- FDR:
-
false discovery rate
- FOXO1:
-
forkhead box O1
- GEO:
-
gene expression omnibus
- HBB:
-
hemoglobin subunit beta
- IHC:
-
immunohistochemical
- MGAM:
-
maltase-glucoamylase
- MYH11:
-
myosin heavy chain 11
- NELL2:
-
neural EGFL like 2
- PDGFA:
-
platelet derived growth factor subunit A
- PPI:
-
protein-protein interaction
- RAMP1:
-
receptor activity modifying protein 1
- ROC:
-
receiver operating characteristic
- ROCK1:
-
Rho associated coiled-coil containing protein kinase 1
- ROCK2:
-
Rho associated coiled-coil containing protein kinase 1
- TFs:
-
transcriptional factors
- ZBTB16:
-
zinc finger and BTB domain containing 16
References
Weintraub NL. Understanding abdominal aortic aneurysm. N Engl J Med. 2009;361(11):1114–6.
Hirsch AT, Haskal ZJ, Hertzer NR, Bakal CW, Creager MA, Halperin JL, Hiratzka LF, Murphy WR, Olin JW, Puschett JB, et al. Acc/Aha 2005 Practice Guidelines For The Management Of Patients With Peripheral Arterial Disease (Lower Extremity, Renal, Mesenteric, And Abdominal Aortic): A Collaborative Report From The American Association For Vascular Surgery/Society For Vascular Surgery, Society For Cardiovascular Angiography And Interventions, Society For Vascular Medicine And Biology, Society Of Interventional Radiology, And The Acc/Aha Task Force On Practice Guidelines (Writing Committee To Develop Guidelines For The Management Of Patients With Peripheral Arterial Disease): Endorsed By The American Association Of Cardiovascular And Pulmonary Rehabilitation; National Heart, Lung, And Blood Institute; Society For Vascular Nursing; Transatlantic Inter-Society Consensus; And Vascular Disease Foundation. Circulation. 2006;113(11):e463–654.
Lederle FA, Johnson GR, Wilson SE. Abdominal aortic aneurysm in women. J Vasc Surg. 2001;34(1):122–6.
Katz DJ, Stanley JC, Zelenock GB. Gender differences in abdominal aortic aneurysm prevalence, treatment, and outcome. J Vasc Surg. 1997;25(3):561–8.
Lederle FA, Johnson GR, Wilson SE, Chute EP, Littooy FN, Bandyk D, Krupski WC, Barone GW, Acher CW, Ballard DJ. Prevalence and associations of abdominal aortic aneurysm detected through screening. Aneurysm detection and management (ADAM) veterans affairs cooperative study group. Ann Intern Med. 1997;126(6):441–9.
Coady MA, Davies RR, Roberts M, Goldstein LJ, Rogalski MJ, Rizzo JA, Hammond GL, Kopf GS, Elefteriades JA. Familial patterns of thoracic aortic aneurysms. Archives of surgery (Chicago, Ill: 1960). 1999;134(4):361–7.
Lederle FA, Johnson GR, Wilson SE, Chute EP, Hye RJ, Makaroun MS, Barone GW, Bandyk D, Moneta GL, Makhoul RG. The aneurysm detection and management study screening program: validation cohort and final results. Aneurysm detection and management veterans affairs cooperative study investigators. Arch Intern Med. 2000;160(10):1425–30.
Bobryshev YV, Lord RS, Parsson H. Immunophenotypic analysis of the aortic aneurysm wall suggests that vascular dendritic cells are involved in immune responses. Cardiovascular surgery (London, England). 1998;6(3):240–9.
Ernst CB. Abdominal aortic aneurysm. N Engl J Med. 1993;328(16):1167–72.
Pearce WH, Koch AE. Cellular components and features of immune response in abdominal aortic aneurysms. Ann N Y Acad Sci. 1996;800:175–85.
Henderson EL, Geng YJ, Sukhova GK, Whittemore AD, Knox J, Libby P. Death of smooth muscle cells and expression of mediators of apoptosis by T lymphocytes in human abdominal aortic aneurysms. Circulation. 1999;99(1):96–104.
Kniemeyer HW, Kessler T, Reber PU, Ris HB, Hakki H, Widmer MK. Treatment of ruptured abdominal aortic aneurysm, a permanent challenge or a waste of resources? Prediction of outcome using a multi-organ-dysfunction score. Eur J Vasc Endovasc Surg. 2000;19(2):190–6.
Brewster DC, Cronenwett JL, Hallett JW Jr, Johnston KW, Krupski WC, Matsumura JS. Guidelines for the treatment of abdominal aortic aneurysms. Report of a subcommittee of the joint Council of the American Association for vascular surgery and Society for Vascular Surgery. J Vasc Surg. 2003;37(5):1106–17.
Cowan JA Jr, Dimick JB, Henke PK, Rectenwald J, Stanley JC, Upchurch GR Jr. Epidemiology of aortic aneurysm repair in the United States from 1993 to 2003. Ann N Y Acad Sci. 2006;1085:1–10.
Lederle FA, Wilson SE, Johnson GR, Reinke DB, Littooy FN, Acher CW, Ballard DJ, Messina LM, Gordon IL, Chute EP, et al. Immediate repair compared with surveillance of small abdominal aortic aneurysms. N Engl J Med. 2002;346(19):1437–44.
Mastracci TM, Cina CS. Screening for abdominal aortic aneurysm in Canada: review and position statement of the Canadian Society for Vascular Surgery. J Vasc Surg. 2007;45(6):1268–76.
Schermerhorn ML, O'Malley AJ, Jhaveri A, Cotterill P, Pomposelli F, Landon BE. Endovascular vs. open repair of abdominal aortic aneurysms in the Medicare population. N Engl J Med. 2008;358(5):464–74.
Moxon JV, Parr A, Emeto TI, Walker P, Norman PE, Golledge J. Diagnosis and monitoring of abdominal aortic aneurysm: current status and future prospects. Curr Probl Cardiol. 2010;35(10):512–48.
Mortality results for randomised controlled trial of early elective surgery or ultrasonographic surveillance for small abdominal aortic aneurysms. The UK Small Aneurysm Trial Participants. Lancet (London, England) 1998, 352(9141):1649–1655.
Reiner-Benaim A. FDR control by the BH procedure for two-sided correlated tests with implications to gene expression data analysis. Biom J. 2007;49(1):107–26.
Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57(1):289–300.
Shah US, Getzenberg RH. Fingerprinting the diseased prostate: associations between BPH and prostate cancer. J Cell Biochem. 2004;91(1):161–9.
Osman I, Bajorin DF, Sun TT, Zhong H, Douglas D, Scattergood J, Zheng R, Han M, Marshall KW, Liew CC. Novel blood biomarkers of human urinary bladder cancer. Clin Cancer Res. 2006;12(11 Pt 1):3374–80.
Damas JK, Smith C, Oie E, Fevang B, Halvorsen B, Waehre T, Boullier A, Breland U, Yndestad A, Ovchinnikova O, et al. Enhanced expression of the homeostatic chemokines CCL19 and CCL21 in clinical and experimental atherosclerosis: possible pathogenic role in plaque destabilization. Arterioscler Thromb Vasc Biol. 2007;27(3):614–20.
Nickel T, Pfeiler S, Summo C, Kopp R, Meimarakis G, Sicic Z, Lambert M, Lackermair K, David R, Beiras-Fernandez A, et al. oxLDL downregulates the dendritic cell homing factors CCR7 and CCL21. Mediat Inflamm. 2012;2012:320953.
Ivancic MM, Huttlin EL, Chen X, Pleiman JK, Irving AA, Hegeman AD, Dove WF, Sussman MR. Candidate serum biomarkers for early intestinal cancer using 15N metabolic labeling and quantitative proteomics in the ApcMin/+ mouse. J Proteome Res. 2013;12(9):4152–66.
Campbell JD, Alexandrov A. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. 2016;48(6):607–16.
Gamberi T, Puglia M, Guidi F, Magherini F, Bini L, Marzocchini R, Modesti A, Modesti PA. A proteomic approach to identify plasma proteins in patients with abdominal aortic aneurysm. Mol BioSyst. 2011;7(10):2855–62.
Wang F, Chang JT, Kao CJ, Huang RS. High expression of miR-532-5p, a tumor suppressor, leads to better prognosis in ovarian cancer both in vivo and in vitro. Mol Cancer Ther. 2016;15(5):1123–31.
Chen Z, Brand NJ, Chen A, Chen SJ, Tong JH, Wang ZY, Waxman S, Zelent A. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 1993;12(3):1161–7.
Hinterseher I, Erdman R, Elmore JR, Stahl E, Pahl MC, Derr K, Golden A, Lillvis JH, Cindric MC, Jackson K, et al. Novel pathways in the pathobiology of human abdominal aortic aneurysms. Pathobiology. 2013;80(1):1–10.
Alikhani M, Alikhani Z, Graves DT. FOXO1 functions as a master switch that regulates gene expression necessary for tumor necrosis factor-induced fibroblast apoptosis. J Biol Chem. 2005;280(13):12096–102.
Stohr R, Kappel BA, Carnevale D, Cavalera M, Mavilio M, Arisi I, Fardella V, Cifelli G, Casagrande V, Rizza S, et al. TIMP3 interplays with apelin to regulate cardiovascular metabolism in hypercholesterolemic mice. Molecular metabolism. 2015;4(10):741–52.
Cucina A, Pagliei S, Borrelli V, Corvino V, Stipa F, Cavallaro A, Sterpetti AV. Oxidised LDL (OxLDL) induces production of platelet derived growth factor AA (PDGF AA) from aortic smooth muscle cells. Eur J Vasc Endovasc Surg. 1998;16(3):197–202.
Li L, Blumenthal DK, Terry CM, He Y, Carlson ML, Cheung AK. PDGF-induced proliferation in human arterial and venous smooth muscle cells: molecular basis for differential effects of PDGF isoforms. J Cell Biochem. 2011;112(1):289–98.
Wilcox JN, Smith KM, Williams LT, Schwartz SM, Gordon D. Platelet-derived growth factor mRNA detection in human atherosclerotic plaques by in situ hybridization. J Clin Invest. 1988;82(3):1134–43.
Touyz RM, Deng LY, He G, Wu XH, Schiffrin EL. Angiotensin II stimulates DNA and protein synthesis in vascular smooth muscle cells from human arteries: role of extracellular signal-regulated kinases. J Hypertens. 1999;17(7):907–16.
Takahashi T, Kawahara Y, Okuda M, Ueno H, Takeshita A, Yokoyama M, Angiotensin II. Stimulates mitogen-activated protein kinases and protein synthesis by a Ras-independent pathway in vascular smooth muscle cells. J Biol Chem. 1997;272(25):16018–22.
Lopez-Candales A, Holmes DR, Liao S, Scott MJ, Wickline SA, Thompson RW. Decreased vascular smooth muscle cell density in medial degeneration of human abdominal aortic aneurysms. Am J Pathol. 1997;150(3):993–1007.
Randolph GJ. The fate of monocytes in atherosclerosis. J Thromb Haemost. 2009;7(Suppl 1):28–30.
Libby P, Ridker PM, Hansson GK. Inflammation in atherosclerosis: from pathophysiology to practice. J Am Coll Cardiol. 2009;54(23):2129–38.
Rateri DL, Howatt DA, Moorleghen JJ, Charnigo R, Cassis LA, Daugherty A. Prolonged infusion of angiotensin II in apoE(−/−) mice promotes macrophage recruitment with continued expansion of abdominal aortic aneurysm. Am J Pathol. 2011;179(3):1542–8.
Leeper NJ, Tedesco MM, Kojima Y, Schultz GM, Kundu RK, Ashley EA, Tsao PS, Dalman RL, Quertermous T. Apelin prevents aortic aneurysm formation by inhibiting macrophage inflammation. Am J Phys Heart Circ Phys. 2009;296(5):H1329–35.
Jones GT, Thompson AR, van Bockxmeer FM, Hafez H, Cooper JA, Golledge J, Humphries SE, Norman PE, van Rij AM. Angiotensin II type 1 receptor 1166C polymorphism is associated with abdominal aortic aneurysm in three independent cohorts. Arterioscler Thromb Vasc Biol. 2008;28(4):764–70.
Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts. J Biol Chem. 2006;281(27):18277–84.
Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, Feldman DS, Schmittgen TD, Elton TS. The human angiotensin II type 1 receptor +1166 a/C polymorphism attenuates microRNA-155 binding. J Biol Chem. 2007;282(33):24262–9.
Attenhofer Jost CH, Greutmann M, Connolly HM, Weber R, Rohrbach M, Oxenius A, Kretschmar O, Luscher TF, Matyas G. Medical treatment of aortic aneurysms in Marfan syndrome and other heritable conditions. Curr Cardiol Rev. 2014;10(2):161–71.
Low SK, Takahashi A, Cha PC, Zembutsu H, Kamatani N, Kubo M, Nakamura Y. Genome-wide association study for intracranial aneurysm in the Japanese population identifies three candidate susceptible loci and a functional genetic variant at EDNRA. Hum Mol Genet. 2012;21(9):2102–10.
Yasuno K, Bakircioglu M, Low SK, Bilguvar K, Gaal E, Ruigrok YM, Niemela M, Hata A, Bijlenga P, Kasuya H, et al. Common variant near the endothelin receptor type a (EDNRA) gene is associated with intracranial aneurysm risk. Proc Natl Acad Sci U S A. 2011;108(49):19707–12.
Ikeda Y. Aortic aneurysm: Etiopathogenesis and Clinicopathologic correlations. Ann Vasc Dis. 2016;9(2):73–9.
Pannu H, Tran-Fadulu V, Papke CL, Scherer S, Liu Y, Presley C, Guo D, Estrera AL, Safi HJ, Brasier AR, et al. MYH11 mutations result in a distinct vascular pathology driven by insulin-like growth factor 1 and angiotensin II. Hum Mol Genet. 2007;16(20):2453–62.
Zhu L, Vranckx R, Khau Van Kien P, Lalande A, Boisset N, Mathieu F, Wegman M, Glancy L, Gasc JM, Brunotte F, et al. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat Genet. 2006;38(3):343–9.
Tsujikawa K, Yayama K, Hayashi T, Matsushita H, Yamaguchi T, Shigeno T, Ogitani Y, Hirayama M, Kato T, Fukada S, et al. Hypertension and dysregulated proinflammatory cytokine production in receptor activity-modifying protein 1-deficient mice. Proc Natl Acad Sci U S A. 2007;104(42):16702–7.
Takahashi K, Matsumoto Y, Do e Z, Kanazawa M, Satoh K, Shimizu T, Sato A, Fukumoto Y, Shimokawa H. Combination therapy with atorvastatin and amlodipine suppresses angiotensin II-induced aortic aneurysm formation. PLoS One. 2013;8(8):e72558.
Acknowledgements
Not Applicable.
Funding
Not applicable
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
LW and JH drafted and revised the manuscript. LW, HN and GY performed the experiment and analyzed the data. JH designed the subject of the manuscript. All authors have read and agreed to the submission of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All participating individuals provided written informed consent with the approval of the ethics committee of the First Affiliated Hospital of Wenzhou Medical University (2017147).
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wan, L., Huang, J., Ni, H. et al. Screening key genes for abdominal aortic aneurysm based on gene expression omnibus dataset. BMC Cardiovasc Disord 18, 34 (2018). https://doi.org/10.1186/s12872-018-0766-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12872-018-0766-8