Abstract
Background
Fatty acid desaturases (FADs) are involved in regulating plant fatty acid composition by adding double bonds to growing hydrocarbon chain. Apart from regulating fatty acid composition FADs are of great importance, and are involved in stress responsiveness, plant development, and defense mechanisms. FADs have been extensively studied in crop plants, and are broadly classed into soluble and non-soluble fatty acids. However, FADs have not yet been characterized in Brassica carinata and its progenitors.
Results
Here we have performed comparative genome-wide identification of FADs and have identified 131 soluble and 28 non-soluble FADs in allotetraploid B. carinata and its diploid parents. Most soluble FAD proteins are predicted to be resided in endomembrane system, whereas FAB proteins were found to be localized in chloroplast. Phylogenetic analysis classed the soluble and non-soluble FAD proteins into seven and four clusters, respectively. Positive type of selection seemed to be dominant in both FADs suggesting the impact of evolution on these gene families. Upstream regions of both FADs were enriched in stress related cis-regulatory elements and among them ABRE type of elements were in abundance. Comparative transcriptomic data analysis output highlighted that FADs expression reduced gradually in mature seed and embryonic tissues. Moreover, under heat stress during seed and embryo development seven genes remained up-regulated regardless of external stress. Three FADs were only induced under elevated temperature whereas five genes were upregulated under Xanthomonas campestris stress suggesting their involvement in abiotic and biotic stress response.
Conclusions
The current study provides insights into the evolution of FADs and their role in B. carinata under stress conditions. Moreover, the functional characterization of stress-related genes would exploit their utilization in future breeding programs of B. carinata and its progenitors.
Similar content being viewed by others
Introduction
Brassicaceae family is comprised of 372 genera and 4006 species and members of this family are sources of fodder, oil, food, condiments, and biofuel. B. carinata (BBCC), an allotetraploid (2n = 4x = 34) came into birth after the natural hybridization between B. oleracea (CC) (2n = 2x = 18) and B. nigra (BB) (2n = 2x = 16) about 0.47 million years ago (MYA). The allotetraploid B. carinata is known for its climate resilience characteristics like heat, drought, and lodging resistance. It also has resistance against biotic stresses as well [1]. Brassicas are third source of oilseed after palm and Soybean. The B. carinata is popular for industrial use due to higher Erusic acid (22:1) [2]. However, its diploid progenitors are being used for condiments and vegetables. Vegetable oils are crucial part of the human diet carrying important fatty acids [3]. They also serve as an important energy source and are major constituent of most biological membranes [4]. The acetyl-CoA provides raw material to synthesize fatty acids in plastids [5]. Production of C16 to C18 carbon fatty acids require 30 enzymatic reactions in the stroma of plastids [6]. Fatty acids are categorized into polyunsaturated, mono-unsaturated, and saturated ones. Unsaturated fatty acids constitute 75% of total fatty acids and the concentration of unsaturated fatty acids is higher in higher plants compared to other living matter. Moreover, their proportion in oil also determines the quality of edible oil [7, 8].
FADs converts the saturated fatty acids to unsaturated fatty acids by modifying single bonds to double bonds at specific sites [9]. FADs are broadly categorized into soluble and non-soluble FADs [10]. Moreover, this gene family is further divided into five sub-gene families; ADS (Acyl-CoA-desaturase), DES (Sphingolipid Δ-4 desaturase), FAB (Δ-9 stearoyl-ACP desaturase), FAD, and SLD (Sphingolipid Δ-8 desaturase) [11]. Among soluble desaturases, only FAB2 have been identified in plants [12] which catalyzes the conversion of stearic acid (18:0) to oleic acid (18:1) by incorporating a double bond at Δ9 position [11]. Non soluble FADs are further classified into Omega-3 (ω3) desaturases (FAD3/FAD7/FAD8), and Omega-6 (ω6) desaturases (FAD2/FAD6, and FAD4). The ω3, ω6, and FAD4 are involved in regulation of linoleic acid (18:2), linolenic acid (18:3), and palmitoleic acid (16:1) biosynthesis respectively [13]. The FAB2, FAD4, FAD6, FAD7, and FAD8 are involved in lipid desaturation in plastids whereas FAD2 and FAD3 performed the same function in the endoplasmic reticulum [14, 15]. Previous reports suggested that FADs from the same family/subfamily possess conserved amino acid sequences. For instance, two (D/ EXXH) and three (H(X3 − 4 H/H(X)2−3HH/H/Q(X)2−3HH)) histidine motifs are conserved in FAB2s and membrane-bound FADs respectively [16].
Although FADs are mainly involved in determining edible oil quality but, their role in abiotic stress responses has also been reported. According to Iba [17] concentration of unsaturated fatty acids determines the plant tolerance level against heat and cold stresses. Moreover, unsaturated fatty acids are also required to maintain cell membrane fluidity during adverse environmental conditions [18]. For instance, expression of AtFAD8 gene abruptly increased under cold stress while expression of AtFAD6 and AtFAD8 genes induced at seedlings level against osmotic and salinity stresses [18,19,20,21]. Similarly, the GmFAD3 and GmFAD7 genes were found to be abundantly express under low temperature [22]. The LeFAD7 gene has been found to be negatively correlated with heat stress [23]. Moreover, heterologous expression of AtFAD3/AtFAD8 genes in tobbacco showed enhanced osmotic tolerance [24].
The availability of whole genome sequence of crop plants provides the opportunity to study any gene family. Consequently, the FAD gene family members have been studied in Arabidopsis [25], Cucumis sativus [14], Glycine max [26], Linum usitatissimum [27], Medicago trancula [28], Musa spp. [29], Cicer arietinum [13], Camelina sativa [30], Gossypium hirsutum [18], Medicago sativa [31], B. napus [11] and B. Juncea [16]. However, FADs have not been yet studied in B. carinata and its progenitors. Therefore, in this study, we have performed gene identification, phylogenetics, chromosomal localization, promoter and gene structure analyses in B. carinata and its progenitors. In addition, the role of FAD genes under normal and stressed conditions have also been explored using available transcriptomic data of different plant tissues. The findings of gene divergence analysis, and synteny analysis would certainly give insight into evolution of this gene family members.
Results
Gene identification and phylogenetics of FAD gene family
Genomes of B. carinata, B. nigra, and B. oleracea were searched against the query sequences of FAD proteins of A. thaliana to know the distribution of FAD gene family members in Brassica genomes. The conserved domain database (CDD) was used to check the conserve domains of each protein and genes having the conserve domain were used in further analysis. Finally, 67 ADS, 57 FAD, 28 FAB, 18 SLD and 04 DES genes were shortlisted after confirming their conserved domains (Fig. 1). The shortlisted genes were grouped into five sub-gene families (SLD, ADS, DES, FAD, and FAB) as suggested in previous literature of Brassicas [11, 16] (Table S1). The deleted incomplete domain length could be the result of evolution as genes might lost their part during evolution and considered as pseudo-genes [32]. Gene length varied between and within gene families and BolADS15 was the lengthiest gene followed by BcaFAD3.1 A, BcaFAB2.2.1 A, BcaDES02, and BolSLD1.2 genes (Table S1). Moreover, other gene related information like protein length, isoelectric point (pI) and coding sequence length are listed in Table S1. The identified genes were renamed according to their chromosomal position i.e., BniB01g012460.2 N.1 is the first gene located on B1 chromosome of B. nigra and renamed as BniADS01. Prediction of subcellular localization concluded that most membrane bound desaturases resided in endomembrane system, comparing to FABs that were predicted to be localized in chloroplast (Table S1). Since endomembrane system is comprised of complex membrane trafficking network crucial in material exchange and transport, and it might be possible that genes located within this system could be involved in transportation [33].
Phylogenetics and motif analysis of FAD proteins of Brassicas
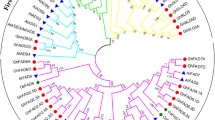
Reliable gene phylogeny could help to determine the function and structure of uncharacterized protein [34, 35]. Comparative phylogeny of membrane bound FADs (Figure S 1), and soluble FAB2s (Figure S 2) related proteins were carried out separately due to higher dissimilarities between them.
The phylogenetics of FAD proteins clustered into seven clusters. Cluster VII contained 76 proteins followed by cluster IV and cluster VII carrying 27 and 17 proteins from Brassicas, O. sativa, and A. thaliana, respectively. Cluster VII contained 25 orthologous gene pairs compared to cluster IV, cluster V and cluster III which had 06, 04, and 03 orthologous gene pairs, respectively. Cluster II was devoid of any orthologous gene pairs, and cluster I and cluster III each carried one orthologous gene pair. Moreover, it is interesting that each gene family from group I resided in separate clade that might be the indication of their independent evolution (Figure S 1). FAB proteins were grouped into four groups and group III had five orthologous gene pairs followed by group IV, and group II which carried four and three orthologous gene pairs respectively. Interestingly group II and III had not any O. sativa protein (Figure S 2).
Furthermore, MEME web server was used to identify the diversity of motifs in protein sequences. Finally, three types of motifs were identified in both FAD and FAB proteins. Motif 1 was common in all non-soluble FAD proteins except proteins of group IV. Protein of group I, II, III, and V had only motif 1 while group VI had all three motifs. The BolADS19 protein showed unique motif composition (Fig. 2). Like FADs, FAB2 proteins also had three motifs and proteins of group I had all three motifs except BcaFAB2.3.3 and BcaFAB2.3.2 proteins. The BcaFAB2.2.4 A, BniFAB2.2.4 A, and BolFAB2.2.2 A from clade III carried motif 2 and motif3 compared to BcaFAB2.2.1 A which had motif 1 and motif 3 respectively (Fig. 3).
Physical gene localization of FAD related genes
Genes were randomly and unevenly distributed on all chromosomes (chr). The ADS genes were localized in cluster form on both BB and CC chromosomes compared to FAD4 genes which were found in cluster form in B. carinata (Fig. 4a) chromosomes only that could be the indication of QTL formation. For ADS genes, B. nigra chromosomes contained 23 genes and among these chrB01, chrB06, and chrB07 each contained 04 genes (Fig. 4c). Four chromosomes viz., chrB01, chrB04, chrB08, and chrB02, each had only 01 gene. Twenty genes were resided on CC chromosomes of B. oleracea and chrC05 carried highest number of genes (06) followed by chrC03, chrC08, chrC04, and chrC09 which contained 05, 05, 02, and 01 genes respectively (Fig. 4b). Four DES genes were localized on different chromosomes and each gene resided on separate chromosomes. For FAB genes, 05 and 06 genes were localized on BB chromosomes and CC chromosomes of B. carinata respectively. The chrB08, chrC01, and chrC04 each had 02 genes.
Gene structure of FAD related genes of B. carinata and its parents
Gene structure analysis provides insights into evolutionary pattern of gene family [36]. Gene structure was conserved within sub-gene families and varied between sub-gene families. The DES and SLD genes were intron less and remained conserved during evolution. Interestingly gene structure were highly conserved within FAD3, FAD7, and FAD8 sub-gene family members. However, BcaFAD3.1 A showed variable intron distribution. The FAD6 related genes were enriched in introns and BcaFAD6.2 gene contained 15 introns. Moreover, genes carrying 9 introns constitute 60% of the gene family members (Fig. 5).
Out of total 67 ADS genes 53 had 04 introns and BolADS19 showed highest intron number (15). Each DES genes contained 01 intron and BcaDES02 gene showed variable intron/exon distribution compared to others. FAB genes have also showed conserved gene structure and genes with 02 and 03 introns shared similar intron and exon distribution (Fig. 6).
Prediction of Cis-regulatory elements in upstream regions of FAD and FAB genes
Presence of cis- regulatory elements in upstream regions of gene greatly influence its function [36]. Initially, identified cis elements were classed into developmental, stress, light, and hormonal responsive as suggested in earlier literature [11]. Developmental type of elements include MYC, MYB-recognition, MYB-like sequences, HD-zip3, Myb, GATA-motif, CAAT-box, CAT-box whereas, GT-1-motif, AE-box, and TCT-motif placed in light responsive type of elements. Moreover, ABRE, STRE, as-1, GA-motif, G-box, DRE core, W-box and MBS belong to stress responsive class. Whereas, GARE-motif and TGA-motif were classed as hormonal responsive elements. Primarily, TATA-box a developmental type of elements were in abundance in all genes suggesting their role in developmental processes. Further, genes having abundunt TATA box elements got upregulated against biotic and abiotic stress responses (Fig. 2) [37]. Similarly, five genes viz., BcaADS02, BcaFAD2.1 A, BcaFAD3.6 A, BniFAB2.3.2, and BolFAD3.1 A had stress type of elements indicating their potential in stress responsiveness (Figs. 2 and 3 and Table S2). However, compared to other elements, ABRE were abundantly found in most FADs might be the indication of their involvement in abiotic stress responsiveness especially cold, drought and salinity [38,39,40,41,42] (Figs. 2 and 3).
Gene divergence and syntenic analysis
Gene divergence is estimated through ka (Synonymous substitution)/ks (non-synonymous substitution) ratio, and this ratio is also used to predict the selection type [43]. If, Ka/ks remains > 1 it suggests positive selection [44], and equals to 1 or less than 1 indicates neutral [45] and negative type [46] of selection, respectively. The members of membrane bound FADs seemed to be go through extensive selection. Like, 12 and 19 orthologues gene pairs undergo positive and negative selection, respectively. Further, none of the genes showed neutral type of selection (Table S3). Compared to FAD genes, FAB seemed to have experienced lower selection pressure during evolution and only seven orthologous gene pairs depicted impact of evolution. Out of these seven pairs, three had experienced positive, three went through negative and only one pair had faced neutral selection (Table S3).
Moreover, Genomes evolution could be studied through doing multiple synteny along with model organism [47]. Chromosomes of B genome of B. carinata i.e., CB3, CB4, and CB5 shared collinear genes with B. nigra chromosomes viz., B3, B5, and B7 respectively. The CC5, CC6, CC7, and CC8 of B. carinata contributed maximum to C5, C6, C7, and C8 chromosomes of B. oleracea. Additionally, besides showing collinearity block multiple synteny also allows to explore the inversions and translocation within and between chromosomes. For instance, CB1, CB2, and CB6 showed inversions with B1, B2, and B8 chromosomes of B. nigra. Likewise, The CC chromosomes (CC2, CC4, and CC9) of B. carinata also have inversions with B. oleracea (C1, C3, and C4) chromosomes (Fig. 7).
Red lines showing ADS genes, whereas DES, FADs, SLD, and FAB2s genes are highlighted by yellow, blue, black, and green lines respectively.
RNA-seq data analysis of FAD genes in vegetative and reproductive tissues
RNA-seq data of both vegetative and reproductive tissues were analyzed to quantify the gene expression in different tissue. In vegetative tissues (Leaf, root, and stem) only genes from allotetraploid B. carinata showed abundant gene expression (BcaFAD4.1, BcaFAD7.1, BcaFAD2.1, BcaDES01, BcaDES02, BcaADS12, BcaADS20, and BcaADS27). likewise, during seed development, and embryo development 08 genes of B. carinata (BcaSLD1.3, BcaFAD2.2 A, BcaFAD2.3 A, BcaFAB2.2.1 A, BcaFAB2.3.1 A, BcaADS11, BcaADS14, and BcaADS24), and 06 genes of B. nigra (BniSLD2.1, BniFAD2.1 A, BniFAB2.3.1, BniFAB2.3.2, BniADS04, and BniADS19) were found to be abundantly expressed. Moreover, abundant expression of these genes in mature seed tissues could be an indication of their long transcription cycle and involvement in seed development (Figs. 2 and 3, and Table S2). Some genes expressed abundantly in all stages, but their expression markedly reduced in mature embryo and seed coat tissues. These gene include nine from B. carinata (BcaSLD1.1, BcaSLD1.4, BcaSLD2.5, BcaFAD2.1 A, BcaFAD8.1, BcaFAD8.2 A, BcaFAD6.2, BcaFAD7.3, BcaFAD6.4) and nine from B. nigra (BniSLD2.2, BniSLD1.1, BniSLD1.2, BniFAD4.1, BniFAD8.1 A, BniFAB2.2.3 A, BniADS08, BniADS12, and BniADS14). Interestingly, none of the genes from B. oleraceae showed higher expression in vegetative and reproductive tissues (Table S2).
FAD genes got upregulated under heat, and X. campestris stresses
RNA sequencing under stressed conditions gives opportunities to identify potential genes that might trigger plant defense systems against stresses [48]. Four genes viz., BolFAD6.4, BolADS13, BolFAD3.1 A, and BolFAD2.2 remained upregulated in all tissues regardless of temperature and day spans. Similarly, three genes (BolSLD2.1, BolADS1, and FAD2.1 A) were expressed at a higher rate in reproductive tissues (seed, endosperm, and embryo) only. These genes might have a role in fatty acid biosynthesis since majority of fatty acid biosynthesis occurs in seed tissues. Moreover, abundant expression of these genes under heat stress could also be an indication of their potential role in heat tolerance. In root tissues under phosphate and zinc stresses, six genes (BolSLD1.2, BolSLD1.1, BolADS9, BolADS14, BolADS16, BolFAB2.6) got upregulated. Among these six genes, BolSLD1.1 is the only gene that is expressed under both biotic and abiotic stresses in vegetative tissues highlighting its involvement in producing defense-related proteins to cope with heat and pathogen stress. Apart from abiotic stress, the expression of FADs was also quantified under X. campestris inoculation. It has been observed that five genes viz., BolFAD6.2, BolFAD4.3, BolFAD6.3, BolFAD4.1, and BolADS17 were upregulated under pathogen infection and could be further explored to confirm their resistance level against biotic stress (Fig. 8, and Table S4). Moreover, their functional characterization would further highlight their potential application in breeding climate-resilient Brassicas.
Discussion
Gene identification and their physio-chemical properties
Until now FADs have been identified in Musa spp. [29], Cicer arietinum [13], Camelina sativa [30], Gossypium hirsutum [18, 49], Medicago sativa [31], Brassica [16], and Medicago trauncatula [28]. Gene numbers varied from crop to crop, and we have identified 174 total FAD genes in B. carinata and its diploid progenitors [16]. The FAD genes are not following the genome sizes. For example, FAD genes are more in A. thaliana [25] compared to G. hirsutum [18], T. aestivum [50], and B. juncea [16] but equal to Musa spp. [29]. Moreover, FAD genes are less in number in Juglans regia [51], Cucumis sativus [14], and O. sativa [52] compared to A. thaliana. The same trend could also be observed in B. carinata and B. napus genomes where FAD genes are equal in both allotetraploids regardless of their genomes size. Further, identified genes could be classified as FAB, SLD, ADS, DES, and FAD2/ FAD3/ FAD4/ FAD6/ FAD7/ FAD8 as reported in other Brassica species [16] and T.aestivum [50] but C. arietinum genome contains FAD5 only [13]. Moreover, O. sativa and Musa spp. genomes lack ADS gene family implying that ADS came into birth after separation of monocots and dicots. During polyploidization Brassica genome might have lost ADS, FAB, and FAD genes as sum of genes of allotetraploid off-spring is less to the diploid progenitors genes (Fig. 1).
The FAD2 and FAD3 proteins were predicted to be localized in ER whereas FAD4, FAD6, and FAD7 proteins were predicted to be present in chloroplast thylakoid membrane, mitochondrial membrane, and chloroplast outer membrane accordingly as reported earlier in T. aestivum, Musa spp., H. annus, L. usitatissimum, and T. cacao genomes respectively [27, 29, 50, 53].
Gene structure is crucial in gene functioning and deviation in gene structure could provide insight into the evolution process [54]. The gene structure of FAD gene family is conserved across the species [11, 28, 29, 50, 52].
Phylogenetics of FAD proteins
Protein sequences of both soluble and non-soluble FAD proteins were subjected to phylogenetics since signal-to noise ratio of protein sequences are more suitable to analyse gene families [55]. Phylogenetics of soluble and non-soluble were separately performed due to higher dissimilarities at protein sequence level. Like, B. carinata FAB proteins are also dissimilar in C. arietinum [13] and B. napus [11] but are alike enough to construct a combine phylogenetic tree in Musa spp. [29], M. truncatula [28], M. sativa [31], and T. aestivum [50]. Deviation of protein sequences in sub-family protein might be due to the significant impact of evolution and polyploidization of genomes. Moreover, phylogenetic clade formation also helps to determine the homology between protein sequences. For example, non-soluble proteins of cluster I contain AtDES1 suggesting its homology with adjacent proteins whereas members of cluster II and cluster V had sequence similarities with AthFAD6, and AthFAD4 proteins, respectively. AthFAD4 involved in palmitic acid (16: 0) synthesis [56] and AthFAD6 [57] introduces double bond to 16:3 and proteins of these clusters might have relevant functions.
Cluster III contains 04 AtFAD proteins viz., AthFAD2A, AthFAD3A, AthFAD7, and AthFAD8A whereas, cluster VI contains 10 AtADS proteins viz., AthADS01, AthADS02, AthADS03, AthADS04, AthADS05, AthADS06, AthADS07, AthADS08, AthADS09, AthADS76. Some of these protein have role in carbon chain modification whereas some are involved in conversion of the fatty acids. For instance, the AthFAD2A changes oleic acid to linoleic acid and AthFAD3 coverts it to linolenic acid. Similarly, AthFAD8 encodes trienoic fatty acid (16:2 and 18:2) which serves as substrate for the activity of AthFAD7 and further desaturation of these products takes place in chloroplast by AthFAD7 [19, 58]. Although FAD proteins are mainly involved in FA elongation. However, they also have potential role in stress responsiveness.
Role of soluble and non-soluble FAD genes in biological processes
Exploring transcriptomic data under normal and stressed conditions helps to determine or to predict the gene’s function in biological processes [11]. Mainly, FAD genes are involved in pollen development [59], nodule and leaf development [60] and endosperm development [61]. The BolFAD2 got up-regulated during seed and endosperm development but in soybean this gene predominantly expressed in vegetative tissues only [62]. However, in B. napus this gene specifically expressed in developing seed tissues [63] confirming our current observations. Moreover, this gene is unevenly found in plants i.e., A. thaliana genome has one copy compared to B. napus, G.max, Z.mays, S. indicum, and G. hirsutum whose genomes has multiple copies of the FAD2 gene [64,65,66,67,68,69]. Variation in gene copies could cause differential expression patterns and one of the examples is highlighted in G. max where GmFAD2.1 strongly and exclusively expressed in developing embryos and GmFAD2.2, and GmFAD2.3 genes showed comparatively lower expression in all tissues [70].
The upstream regions of FAD and FAB genes are enriched in stress-related cis-elements [71]. Abiotic (heat) and diseases are major factors limiting oil seed production. In this regard, extensive studies have also been carried out to assess the expression pattern of FAD genes under multiple stresses including heat, drought, salinity, and cold stresses. For instance, the BolFAD2 and BolFAD7 gene induced their expression under elevated temperatures. These genes have already been reported as crucial genes in plant adaptation [14, 22, 25, 72]. Moreover, the FAD7 gene of M. truncatula also got up-regulated against higher temperatures validating our current observations [28]. Similarly, FADs were also found to be abundantly expressed against cold stress and production of polyunsaturated fatty acids is one of the most commons mechanism to combat cold stress/low temperature [73,74,75,76]. In addition to abiotic stress, we have also identified potential genes induced under X. campestris inoculum. Such investigation have also been carried out under different biotic stresses and in sunflower HaFAD3.1 and HaADS6 genes were found to be expressed at a higher rate under Orobanche cumana inoculum indicating that FADs have potential against multiple biotic stresses [53].
Conclusions
FADs add unsaturated bonds to the hydrocarbon chain of fatty acids. Broadly FADs are categorized into two classes: soluble and non-soluble. These two classes share no sequence similarities at the sequence level and evolved independently.
Upstream regions of most FAD genes are enriched with stress-responsive cis-regulatory elements highlighting their potential in stress response and their presence is also influenced gene expression as well.
Methods
Data base research, identification, and physio-chemical properties of FAD like genes in B. carinata, and its progenitors
Genomes, GFF (General feature format), CDS (coding sequence), and peptide sequence files of B. carinata, B. nigra, and B. oleracea were retrieved from Brassica data base BRAD (http://brassicadb.cn). The FAD genes of Arabidopsis were downloaded from TAIR data base (https://www.arabidopsis.org) and used as the query sequences. TB tools v 1.09832 was used to identify the all-FAD like genes in three Brassica genomes through BLASTp program against a threshold level of E value < 1e-5. The retrieved protein sequences were subjected to CDD to select the sequences with conserved domains only [77]. Moreover, the physio-chemical properties of identified proteins like protein length, molecular weight, and pI were calculated using the Expasy platform (https://web.expasy.org/compute_pi/) [78].
Multiple sequence alignment, phylogenetic and motif analysis
The retrieved protein of Brassicas along with A. thaliana and O. sativa were aligned in Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) [79] and MEGA X v 10.2.2 was used to construct an un-rooted neighbor-joining tree and adjusted the boost trap repeats to 1000- with JTT (Jones-Taylor-Thornton) + G (Gamma distributed) model [80]. Further, the retrieved protein sequences were submitted to the MEME suit server with default parameters to know the motif composition of the identified proteins.
Promotor, gene structure, and gene divergence analysis
The 1500 bp upstream nucleotide sequence from the start position of each gene (ATG) was retrieved using TB tool FASTA extract and submitted to online Plant care (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) data-base to identify the cis-regulatory elements in promotor regions [81].
Genes general information i.e., gene length, intron/exon distribution, and physical gene location were extracted from GFF files with the help of GFX select tool of TB tools [82]. Orthologues gene pairs were picked up from the adjacent nodes of the phylogenetic tree to estimate the gene divergence. The Ka and Ks values of orthologous gene pairs were calculated through TB tools. Moreover, the selection type was estimated using ka/ks ratio.
Transcriptomic data analysis of FAD like genes under normal and stressed conditions
The ENA EBI (https://www.ebi.ac.uk/ena/browser/home) database was used to retrieve the transcriptomic data of different tissues under normal and stressed conditions. The transcriptomic data of embryo development stages and seed coat development stages was acquired from bio project PRJNA641876 and expression data of these tissues under different temperature regimes was acquired from PRJNA524852. Expression data of PRJNA524852 was used to examine the gene expression of root tissues under phosphate and zinc stress. Further, transcriptomic data under X. campestris was obtained from PRJNA421190. The data was processed through the Galaxy platform using sailfish alignment tool and TPM (Transcript per Million) values were isolated by putting the putative gene IDs. The data was normalized (log2) and presented with the Morpheus heat map tool (https://software.broadinstitute.org/morpheus/).
Availability of data and materials
Genomes of Brassica species were acquired from (http://brassicadb.cn ) and query sequences of A. thaliana were retrieved from (https://www.arabidopsis.org ) whereas FAD related proteins of O. sativa were obtained from (http://rice.uga.edu/). Online transcriptomic data (PRJNA524852, PRJNA641876, PRJNA524852, and PRJNA421190) of different tissues under normal and stressed conditions were acquired from (https://www.ebi.ac.uk/ena/browser/home).
References
Huang J, Xue C, Wang H, Wang L, Schmidt W, Shen R, et al. Genes of ACYL CARRIER PROTEIN family show different expression profiles and overexpression of ACYL CARRIER PROTEIN 5 modulates fatty acid composition and enhances salt stress tolerance in Arabidopsis. Front Plant Sci. 2017;8:987.
Kumar S, Seepaul R, Small IM, George S, O’Brien GK, Marois JJ, et al. Interactive Effects of Nitrogen and Sulfur Nutrition on Growth, Development, and physiology of Brassica carinata A. Braun and Brassica napus L. Sustainability. 2021;13(13):7355.
Yang M, Xu Y. Oleate accumulation, induced by silencing of microsomal omega-6 desaturase, declines with leaf expansion in transgenic tobacco. J Plant Physiol. 2007;164(1):23–30.
Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(suppl2):W169–W75.
Browse J, Somerville C. Glycerolipid synthesis: biochemistry and regulation. Annu Rev Plant Biol. 1991;42(1):467–506.
Ohlrogge J, Browse J. Lipid biosynthesis. Plant Cell. 1995;7(7):957.
Pattee HE, Isleib TG, Moore KM, Gorbet DW, Giesbrecht FG. Effect of high-oleic trait and paste storage variables on sensory attribute stability of roasted peanuts. J Agric Food Chem. 2002;50(25):7366–70.
Simopoulos AP. Evolutionary aspects of the dietary omega–6: omega–3 fatty acid ratio: medical implications. A balanced omega-6/omega-3 fatty acid ratio, cholesterol and coronary heart disease. 2009;100:1–21.
Zhu S, Zhu Z, Wang H, Wang L, Cheng L, Yuan Y, et al. Characterization and functional analysis of a plastidial FAD6 gene and its promoter in the mesocarp of oil palm (Elaeis guineensis). Sci Hort. 2018;239:163–70.
Shanklin J, Cahoon EB. Desaturation and related modifications of fatty acids. Annu Rev Plant Biol. 1998;49:611.
Xue Y, Chen B, Wang R, Win AN, Li J, Chai Y. Genome-wide survey and characterization of fatty acid desaturase gene family in Brassica napus and its parental species. Appl Biochem Biotechnol. 2018;184(2):582–98.
Singh SC, Sinha RP, Hader D-P. Role of lipids and fatty acids in stress tolerance in cyanobacteria. Acta Protozool. 2002;41(4):297–308.
Saini R, Kumar S. Genome-wide identification, characterization and in-silico profiling of genes encoding FAD (fatty acid desaturase) proteins in chickpea (Cicer arietinum L). Plant Gene. 2019;18:100180.
Dong C-J, Cao N, Zhang Z-G, Shang Q-M. Characterization of the fatty acid desaturase genes in cucumber: structure, phylogeny, and expression patterns. PLoS ONE. 2016;11(3):e0149917.
Yin D-D, Xu W-Z, Shu Q-Y, Li S-S, Wu Q, Feng C-Y, et al. Fatty acid desaturase 3 (PsFAD3) from Paeonia suffruticosa reveals high α-linolenic acid accumulation. Plant Sci. 2018;274:212–22.
Xue Y, Chai C, Chen B, Shi X, Wang B, Mei F, et al. Whole-genome mining and in silico analysis of FAD gene family in Brassica juncea. J Plant Biochem Biotechnol. 2020;29(1):149–54.
Iba K, ACCLIMATIVE RESPONSE TO TEMPERATURE STRESS. IN HIGHER PLANTS: approaches of. Annu Rev Plant Biol. 2002;53:225–45.
Feng J, Dong Y, Liu W, He Q, Daud M, Chen J, et al. Genome-wide identification of membrane-bound fatty acid desaturase genes in Gossypium hirsutum and their expressions during abiotic stress. Sci Rep. 2017;7(1):1–12.
Zhang J-T, Zhu J-Q, Zhu Q, Liu H, Gao X-S, Zhang H-X. Fatty acid desaturase-6 (Fad6) is required for salt tolerance in Arabidopsis thaliana. Biochem Biophys Res Commun. 2009;390(3):469–74.
Zhang J, Liu H, Sun J, Li B, Zhu Q, Chen S, et al. Arabidopsis fatty acid desaturase FAD2 is required for salt tolerance during seed germination and early seedling growth. PLoS ONE. 2012;7(1):e30355.
Lou Y, Schwender J, Shanklin J. FAD2 and FAD3 desaturases form heterodimers that facilitate metabolic channeling in vivo. J Biol Chem. 2014;289(26):17996–8007.
Román Á, Andreu V, Hernández ML, Lagunas B, Picorel R, Martínez-Rivas JM, et al. Contribution of the different omega-3 fatty acid desaturase genes to the cold response in soybean. J Exp Bot. 2012;63(13):4973–82.
Liu XY, Yang JH, Li B, Yang XM, Meng QW. Antisense-mediated depletion of tomato chloroplast Omega‐3 fatty acid desaturase enhances thermal tolerance. J Integr Plant Biol. 2006;48(9):1096–107.
Zhang M, Barg R, Yin M, Gueta-Dahan Y, Leikin‐Frenkel A, Salts Y, et al. Modulated fatty acid desaturation via overexpression of two distinct ω‐3 desaturases differentially alters tolerance to various abiotic stresses in transgenic tobacco cells and plants. Plant J. 2005;44(3):361–71.
Liu W, Li W, He Q, Daud MK, Chen J, Zhu S. Characterization of 19 genes encoding membrane-bound fatty acid desaturases and their expression profiles in Gossypium raimondii under low temperature. PLoS ONE. 2015;10(4):e0123281.
Xiang D, Venglat P, Tibiche C, Yang H, Risseeuw E, Cao Y, et al. Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Physiol. 2011;156(1):346–56.
You FM, Li P, Kumar S, Ragupathy R, Li Z, Fu Y-B, et al. Genome-wide identification and characterization of the gene families controlling fatty acid biosynthesis in flax (Linum usitatissimum L). J Proteom Bioinform. 2014;7(10):310–26.
Zhang Z, Wei X, Liu W, Min X, Jin X, Ndayambaza B, et al. Genome-wide identification and expression analysis of the fatty acid desaturase genes in Medicago truncatula. Biochem Biophys Res Commun. 2018;499(2):361–7.
Cheng C, Liu F, Sun X, Wang B, Liu J, Ni X, et al. Genome-wide identification of FAD gene family and their contributions to the temperature stresses and mutualistic and parasitic fungi colonization responses in banana. Int J Biol Macromol. 2022;204:661–76.
Ahmadizadeh M, Rezaee S, Heidari P. Genome-wide characterization and expression analysis of fatty acid desaturase gene family in Camelina sativa. Gene Rep. 2020;21:100894.
Zhang Z, Jin X, Liu Z, Zhang J, Liu W. Genome-wide identification of FAD gene family and functional analysis of MsFAD3. 1 involved in the accumulation of α‐linolenic acid in alfalfa. Crop Sci. 2021;61(1):566–79.
Panchy N, Lehti-Shiu M, Shiu S-H. Evolution of gene duplication in plants. Plant Physiol. 2016;171(4):2294–316.
Morita MT, Shimada T. The plant endomembrane system—a complex network supporting plant development and physiology. Plant Cell Physiol. 2014;55(4):667–71.
Koonin EV, Tatusov RL, Galperin MY. Beyond complete genomes: from sequence to structure and function. Curr Opin Struct Biol. 1998;8(3):355–63.
Koonin EV, Tatusov RL, Rudd KE. [18] Protein sequence comparison at genome scale. In Methods in enzymology. Academic Press; 1996;266:295–322.
Li P, Jiang Z, Wang Y, Deng Y, Van Nostrand JD, Yuan T, et al. Analysis of the functional gene structure and metabolic potential of microbial community in high arsenic groundwater. Water Res. 2017;123:268–76.
Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389(1):52–65.
Leung J, Giraudat J. Abscisic acid signal transduction. Annu Rev Plant Biol. 1998;49(1):199–222.
Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14(suppl1):15–S45.
Himmelbach A, Yang Y, Grill E. Relay and control of abscisic acid signaling. Curr Opin Plant Biol. 2003;6(5):470–9.
Chak RK, Thomas TL, Quatrano RS, Rock CD. The genes ABI1 and ABI2 are involved in abscisic acid-and drought-inducible expression of the Daucus carota L. Dc3 promoter in guard cells of transgenic Arabidopsis thaliana (L.) Heynh. Planta. 2000;210(6):875–83.
Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 2005;10(2):88–94.
Messier W, Stewart C-B. Episodic adaptive evolution of primate lysozymes. Nature. 1997;385(6612):151–4.
Trabesinger-Ruef N, Jermann T, Zankel T, Durrant B, Frank G, Benner SA. Pseudogenes in ribonuclease evolution: a source of new biomacromolecular function? FEBS Lett. 1996;382(3):319–22.
Duret L. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 2000;16(7):287–9.
Anisimova M, Bielawski JP, Yang Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 2001;18(8):1585–92.
Baek JH, Kim J, Kim CK, Sohn SH, Choi D, Ratnaparkhe MB, et al. MultiSyn: A webtool for multiple synteny detection and visualization of user's sequence of interest compared to public plant species. Evolutionary Bioinformatics. 2016;12:EBO-S40009.
Hu L, Li H, Chen L, Lou Y, Amombo E, Fu J. RNA-seq for gene identification and transcript profiling in relation to root growth of bermudagrass (Cynodon dactylon) under salinity stress. BMC Genomics. 2015;16(1):1–12.
Yurchenko OP, Park S, Ilut DC, Inmon JJ, Millhollon JC, Liechty Z, et al. Genome-wide analysis of the omega-3 fatty acid desaturase gene family in Gossypium. BMC Plant Biol. 2014;14(1):1–15.
Hajiahmadi Z, Abedi A, Wei H, Sun W, Ruan H, Zhuge Q, et al. Identification, evolution, expression, and docking studies of fatty acid desaturase genes in wheat (Triticum aestivum L). BMC Genomics. 2020;21(1):1–20.
Liu K, Zhao S, Wang S, Wang H, Zhang Z. Identification and analysis of the FAD gene family in walnuts (Juglans regia L.) based on transcriptome data. BMC Genomics. 2020;21(1):1–12.
Chen C, Yang J, Tong H, Li T, Wang L, Chen H. Genome-wide analysis of fatty acid desaturase genes in rice (Oryza sativa L). Sci Rep. 2019;9(1):1–11.
Li J, Liu A, Najeeb U, Zhou W, Liu H, Yan G, et al. Genome-wide investigation and expression analysis of membrane-bound fatty acid desaturase genes under different biotic and abiotic stresses in sunflower (Helianthus annuus L). Int J Biol Macromol. 2021;175:188–98.
Cao J, Shi F. Evolution of the RALF gene family in plants: gene duplication and selection patterns. Evolutionary Bioinf. 2012;8:EBO.
Agosti D, Jacobs D, DeSalle R. On combining protein sequences and nucleic acid sequences in phylogenetic analysis: the homeobox protein case. Cladistics. 1996;12(1):65–82.
Gao J, Ajjawi I, Manoli A, Sawin A, Xu C, Froehlich JE, et al. FATTY ACID DESATURASE4 of Arabidopsis encodes a protein distinct from characterized fatty acid desaturases. Plant J. 2009;60(5):832–9.
Lusk HJ, Neumann N, Colter M, Roth MR, Tamura P, Yao L, et al. Lipidomic analysis of Arabidopsis T-DNA insertion lines leads to identification and characterization of C-terminal alterations in FATTY ACID DESATURASE 6. Plant and Cell Physiology. 2022;63(9):1193–204.
McConn M, Hugly S, Browse J, Somerville C. A mutation at the fad8 locus of Arabidopsis identifies a second chloroplast [omega]-3 desaturase. Plant Physiol. 1994;106(4):1609–14.
McConn M, Browse J. The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell. 1996;8(3):403–16.
Lakhssassi N, Colantonio V, Flowers ND, Zhou Z, Henry J, Liu S, et al. Stearoyl-acyl carrier protein desaturase mutations uncover an impact of stearic acid in leaf and nodule structure. Plant Physiol. 2017;174(3):1531–43.
Troncoso-Ponce MA, Barthole G, Tremblais G, To A, Miquel M, Lepiniec L, et al. Transcriptional activation of two delta-9 palmitoyl-ACP desaturase genes by MYB115 and MYB118 is critical for biosynthesis of omega-7 monounsaturated fatty acids in the endosperm of Arabidopsis seeds. Plant Cell. 2016;28(10):2666–82.
Schlueter JA, Vasylenko-Sanders IF, Deshpande S, Yi J, Siegfried M, Roe BA, et al. The FAD2 gene family of soybean: insights into the structural and functional divergence of a paleopolyploid genome. Crop Sci. 2007;47:S–14.
Lee K-R, Sohn SI, Jung JH, Kim SH, Roh KH, Kim J-B, et al. Functional analysis and tissue-differential expression of four FAD2 genes in amphidiploid Brassica napus derived from Brassica rapa and Brassica oleracea. Gene. 2013;531(2):253–62.
Heppard EP, Kinney AJ, Stecca KL, Miao G-H. Developmental and growth temperature regulation of two different microsomal [omega]-6 desaturase genes in soybeans. Plant Physiol. 1996;110(1):311–9.
Jin U-H, Lee J-W, Chung Y-S, Lee J-H, Yi Y-B, Kim Y-K, et al. Characterization and temporal expression of a ω-6 fatty acid desaturase cDNA from sesame (Sesamum indicum L.) seeds. Plant Sci. 2001;161(5):935–41.
Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell. 1994;6(1):147–58.
Pirtle IL, Kongcharoensuntorn W, Nampaisansuk M, Knesek JE, Chapman KD, Pirtle RM. Molecular cloning and functional expression of the gene for a cotton ∆-12 fatty acid desaturase (FAD2). Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression. 2001;1522(2):122–9.
Xiao G, Zhang H, Peng Q, Guan C. Screening and analysis of multiple copy of oleate desaturase gene (fad2) in Brassica napus. Acta Agron Sinica. 2008;34(9):1563–8.
Yang Q, Fan C, Guo Z, Qin J, Wu J, Li Q, et al. Identification of FAD2 and FAD3 genes in Brassica napus genome and development of allele-specific markers for high oleic and low linolenic acid contents. Theor Appl Genet. 2012;125(4):715–29.
Martínez-Rivas JM, Sperling P, Lühs W, Heinz E. Spatial and temporal regulation of three different microsomal oleate desaturase genes (FAD2) from normal-type and high-oleic varieties of sunflower (Helianthus annuus L). Mol Breeding. 2001;8(2):159–68.
Chen L, Wang L, Wang H, Sun R, You L, Zheng Y, et al. Identification and characterization of a plastidial ω-3 fatty acid desaturase EgFAD8 from oil palm (Elaeis guineensis Jacq.) And its promoter response to light and low temperature. PLoS ONE. 2018;13(4):e0196693.
Chen M, Markham JE, Cahoon EB. Sphingolipid ∆8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J. 2012;69(5):769–81.
Guschina IA, Harwood JL. Mechanisms of temperature adaptation in poikilotherms. FEBS Lett. 2006;580(23):5477–83.
Martz F, Kiviniemi S, Palva TE, Sutinen M-L. Contribution of omega-3 fatty acid desaturase and 3-ketoacyl-ACP synthase II (KASII) genes in the modulation of glycerolipid fatty acid composition during cold acclimation in birch leaves. J Exp Bot. 2006;57(4):897–909.
Nishida I, Murata N. Chilling sensitivity in plants and cyanobacteria: the crucial contribution of membrane lipids. Annu Rev Plant Biol. 1996;47(1):541–68.
Williams JP, Khan MU, Mitchell K, Johnson G. The effect of temperature on the level and biosynthesis of unsaturated fatty acids in diacylglycerols of Brassica napus leaves. Plant Physiol. 1988;87(4):904–10.
Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32(suppl2):W327–W31.
Bjellqvist B, Basse B, Olsen E, Celis JE. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis. 1994;15(1):529–39.
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7(1):539.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9.
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–7.
Chen C, Chen H, He Y, Xia R. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv. 2018;289660(101101):289660.
Acknowledgements
Not applicable.
Funding
The study did not receive any external fundings.
Author information
Authors and Affiliations
Contributions
I.A.R., N.S., and U.M.K conceived the idea. I.A.R., U.M.K., and N.S designed the experiments. U.B.Z and A.F. retrieved and curated the data. U.B.Z. and A.F. prepared the figures and arranged the tables. U.M.K. and N.S. performed analyses and wrote the manuscript. S.H.K., S.A., M.T.A., and R.M.A reviewed and edited the manuscript. I.A.R. and S.H. supervised the experiments, manuscript writeup and acquired funding for research. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
This study was conducted in the absence of any competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Phylogenetic distribution of FAD genes along with A. thaliana and O. sativa.
Additional file 2: Figure S2.
Phylogenetic distribution of FAB genes along with A. thaliana and O. sativa.
Additional file 3: Table S1.
Molecular and physio-chemical parameters of both FAD and FAB like genes of Brassicas
Additional file 4: Table S2.
Cis-regulatory elements of FAD and FAB like genes and their expression in different tissues.
Additional file 5: Table S3.
Gene divergence analysis of membrane bound FAD and FAB orthologous gene pairs.
Additional file 6: Table S4.
Transcriptomic data of FAD and FAB like genes in diferent tissues under biotic and abiotic stresses.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Shaheen, N., Khan, U.M., Farooq, A. et al. Comparative transcriptomic and evolutionary analysis of FAD-like genes of Brassica species revealed their role in fatty acid biosynthesis and stress tolerance. BMC Plant Biol 23, 250 (2023). https://doi.org/10.1186/s12870-023-04232-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-023-04232-9