Abstract
Background
Proanthocyanidins (PAs) have always been considered as important medicinal value component. In order to gain insights into the PA biosynthesis regulatory network in I. stachyodes roots, we analyzed the transcriptome of the I. stachyodes in Leaf, Stem, RootI (one-year-old root), and RootII (two-year-old root).
Results
In this study, a total of 110,779 non-redundant unigenes were obtained, of which 63,863 could be functionally annotated. Simultaneously, 75 structural genes that regulate PA biosynthesis were identified, of these 6 structural genes (IsF3′H1, IsANR2, IsLAR2, IsUGT72L1-3, IsMATE2, IsMATE3) may play an important role in the synthesis of PAs in I. stachyodes roots. Furthermore, co-expression network analysis revealed that 34 IsMYBs, 18 IsbHLHs, 15 IsWRKYs, 9 IsMADSs, and 3 IsWIPs hub TFs are potential regulators for PA accumulation. Among them, IsMYB24 and IsMYB79 may be closely involved in the PA biosynthesis in I. stachyodes roots.
Conclusions
The biosynthesis of PAs in I. stachyodes roots is mainly produced by the subsequent pathway of cyanidin. Our work provides new insights into the molecular pathways underlying PA accumulation and enhances our global understanding of transcriptome dynamics throughout different tissues.
Similar content being viewed by others
Introduction
Proanthocyanidins (PAs) are the polymers or oligomers of flavan-3-ol units, usually epicatechin (EC) and catechin (C), and are widely distributed in grape seed and tea plant. In the current research progress, PA extract has a variety of medical values, and can be used for anti-aging, prevention of cardiovascular and tumor, etc [1]. According to the types of flavan bonds, PAs are mainly divided into A-type and B-type. Of which B-type PA is the most frequent found in plant kingdom, their constitutive units are singly linked by C4–C8 or C4–C6 bonds, such as procyanidins B1, B2, B3, and B4 [1,2,3].
The biosynthesis of PAs is a part of the flavonoid pathway that has been well-characterized over the past two decades with the identification of numerous structural, regulatory, and transport-related genes [1, 4, 5]. Genes involved in each biosynthetic step from phenylalanine to flavan-3-ols (( +)-catechin and ( −)-epicatechin) have been well characterized, including phenylalanine ammonia lyase (PAL), cinnamate-4-hydroxylase (C4H), 4-coumarate ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), dihydroflavonol reductase (DFR), flavonoid 3′ hydroxylase (F3′H), anthocyanidin synthase/leucoanthocyanidin dioxygenase (ANS/LDOX), anthocyanidin reductase (ANR) and leucoanthocyanidin reductase (LAR). The synthesis of PAs and anthocyanins share common steps leading to flavan3,4-diols (such as leucoanthocyanidin), which can be converted to catechin (2,3-trans-flavan-3-ol) by LAR [6] or to anthocyanidin by ANS [7, 8]. Anthocyanidin then either serves as the substrate for the synthesis of epicatechin (2,3-cis-flavan-3-ol) by ANR [9]. Flavan-3-ol precursor will be glycosylated and transferred to vacuoles for polymerization. It is clear that epicatechin is glycosylated to form epicatechin 3′-O-glucoside with the participation of UDP-glycosyltransferase (UGT72L1), and then epicatechin-3′-O-glucoside is translocated into the vacuole via specific transporters of the multidrug detoxification and extrusion (MATE) factor family [10, 11]. However, the details of the polymerization process controlled by TT10(LAC15) in vacuole are still unclear [12]. Moreover, these pathway structural genes are regulated by a variety of transcription factors (TFs). To date, TFs of R2R3-MYB [13], bHLH [14, 15], WD40 [16], WIP [17], MADs [18], and WRKY [19, 20] families have been found to regulate PA biosynthesis. Among them, MYB TFs play a key role in the regulation of PA biosynthesis.
Indigofera stachyodes Lindl (Papilionoideae family) is distributed mainly in Guizhou, Yunnan, and Guangxi provinces. Its roots were known as Xuerenshen in Chinese and commonly used as the Miao traditional medicine for the treatment of cold fever, cough, etc. The distinguishing feature of I. stachyodes is its "blood" (i.e., it is reddish-brown after root bark is scraped off). This phenomenon is affected by many internal and external factors, but flavonoids content and type are among the most important factors that determine root color [21]. Flavonoids will gradually accumulate as the plant grows, and roots over three years old are regularly regarded as the harvesting standard of medicinal materials [22, 23]. Previous phytochemical studies indicated the presence of over 30 compounds in I. stachyodes, including epicatechin, stigmasterol, stigmast-4-en-3-one, l-maackiain, etc. [21, 24,25,26]. In the current research, we studied the flavonoid composition in I. stachyodes, and further found that procyanidin B2, catechin (C), epicatechin (EC), and epicatechin gallate (ECG) were the main flavonoids in I. stachyoides roots [27]. The flavonoids extracted from its root have important roles in anti-inflammatory [28], anti-oxidation [29], liver protection [30], anti-tumor [31], etc. There are increasing evidences that clinically valuable traits of I. stachyodes roots benefit from flavonoids, but the PA accumulation and biosynthesis in I. stachyoides roots is still unknown.
Currently, the regulation mechanism of PA synthesis at the gene level by transcriptome analysis has been deeply studied in other plants, such as persimmon [32], Malus Crabapple [33], Brassica napus [34], pinto bean [35], cranberry beans [36]. However, there is still a lack of genomic data on the regulatory mechanism of PA biosynthesis in I. stachyodes roots, which has affected the breeding process of red root varieties. In this study, we performed RNA-seq analysis on I. stachyodes in different tissues (Fig. 1a) to identify candidate regulators of PA accumulation. Furthermore, we conducted a TGMI network analysis to investigate PA biosynthesis pathway‑specific regulators involved in I. stachyodes roots. The results of our research can provide help for the study of the PA biosynthesis mechanism of I. stachyodes roots.
Results
Quantitation of PA content in four different tissues
Figure 1a shows the morphological features of the four tissues Leaf, Stem, RootI, and RootII at the same period. From RootI to RootII, the color of root bark kept rising. RoootI and RootII showed distinct PA accumulation patterns compared to Stem and Leaf (Fig. 1b). Levels of procyanidin B2 differed significantly between ground parts (leaves, stems) and underground parts (roots). Procyanidin B2 levels for RootII (208.4 µg/g FW) and RootI (233.7 µg/g FW) were high and there is no significant difference between the two (Additional file 1: Table S1). The content of another polymeric procyanidin B1 in roots is not higher than that in stems. But due to the proportion of the bark in the sample taken in RootI was significantly more than that in RootII, thus the content of proanthocyanidins in RootII was slightly lower than that in RootI.
Transcriptome sequencing and de novo assembly of I. stachyodes
In order to fully construct the transcriptome of I. stachyodes, four major tissues, including Leaf, Stem, RootI, and RootII, were sampled for RNA isolation. Distinct cDNA libraries of those tissues were constructed and sequenced, resulting in a total of 102 G raw reads. After the removal of adapters, poly-N-containing reads, and low-quality sequences from the raw data, approximately 98 G clean reads were obtained with 98.36% Q20 and 94.89% Q30 bases, and the clean data of each sample is above 7.18 G. The GC percentage in ground parts (leaves, stems) and underground parts (roots) were an average of 44.6% and 44.65%, respectively (Additional file 2: Table S2). Trinity assembly program was used for de novo assembly of all sample clean data, after optimizing the assembly results, a set of 110,779 non-redundant unigenes was obtained. The total length of the unigenes was 92,992,355 bp, with an average length of 839.44 bp and the N50 and E90N50 value of 1,540 and 3,117 bp, respectively. In the 110,779 unigenes, 24,442 unigenes (22.1%) were greater than 1 kb in length (Table 1). The size distribution of unigenes is shown in Additional file 3: Figure S1.
Further, the clean reads of each sample are compared with the Trinity-assembled transcriptome. The average alignment rate was 83.66%, indicating that a high-quality de novo assembled transcriptome was obtained.
Functional annotation and classification
After assembly, the 110,779 transcriptome sequences were annotated by six databases (NR, Swiss-Prot, Pfam, COG, GO, and KEGG) to obtain similarity sequence and the corresponding annotation information. Gene annotation showed that 63,863 unigenes were successfully annotated in Pfam, Swiss-Prot, NR, COG, KEGG, and GO databases. The number and mapping rates of unigenes against the Pfam, Swissprot, GO, COG, and KEGG databases were 42.05%(46,581)ˎ 42.03%(46,559)ˎ 40.05%(45,036)ˎ 48.11%(53,295)ˎ 29.74%(32,950), respectively (Fig. 2a) (Table 1). 34,092 unigenes had high similarity (greater than 80%) in mapped sequences with Nr database and 36,632 unigenes (63.39%) had significant homology (e-value < 1e-30) (Fig. 2b and 2c). Species distribution analysis showed that only 20,060 unigenes (34.71%) had high homology with the genes from Quercus suber, followed by Abrus precatorius (7,272, 12.58%), Spatholobus suberectus (4,505, 7.8%), while 9,478 unigenes had high homology with sequences from other organisms (Fig. 2d).
Blast results of the assembled unigenes and the I. stachyodes transcriptome homology searches against the NR database. a Functional annotation of unigene. b Similarity distribution of top BLAST hits for each unigene. (c) E-value distribution of BLAST hits with a cut off E-value of 1e-5. d Species distribution for top BLAST hits in the Nr database
The functions of all unigenes were classified by using the Nr annotation and Gene Ontology (GO) classification, and a total of 45,036 unigene gene functions were described under three main divisions (biological process, cellular component, and molecular function) (Additional file 4: Table S3). The predominant group in each of the biological processes, cellular components, and molecular functions was “cellular process” (21,820, 50.13%), “cell part” (22,578, 50.13%), and “binding” (26,213, 26.21%), respectively (Additional file 3: Figure S2). To further understand the biological functions and interactions of transcripts, the unigenes of assembled sequences were assigned by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. A total of 31,215 unigenes were assigned to 148 KEGG pathways using BLASTx, with an e-value < 1e-5, and were assigned to six main categories. “Translation” had the largest number of unigenes (6,455 unigenes) followed by “Carbohydrate metabolism” (4,069 unigenes), “Energy metabolism” (2,764 unigenes), “Amino acid metabolism” (2,457 unigenes), “Folding, sorting and degradation” (2,036 unigenes), and “Transport and catabolism” (1,853 unigenes) (Additional file 3: Figure S3) (Additional file 5: Table S4).
Analysis of differentially expressed genes
The unigenes from different tissues of I. stachyodes were compared using assembled data as a reference (Fig. 3a). Under the criteria of p-adjust < 0.05 and |log2FC|≥ 2, a total of 11,648 differentially expressed genes (DEGs) between Leaf and Stem were identified. Among them, 9,058 genes were up-regulated, and 26,711 genes were down-regulated. In addition, 35,490 (7,589 up-regulated and 27,901 down-regulated), 37,234 (10,250 up-regulated and 26,984 down-regulated), 12,467 (5,143 regulated and 7,324 down-regulated), 14,989 (7,866 up-regulated and 7,123 down-regulated), 11,648 (7,430 up-regulated and 4,218 down-regulated) were identified in the comparison of Leaf vs RootI, Leaf vs RootII, Stem vs RootI, Stem vs RootII, RootI vs RootII, respectively (Fig. 3b). To obtain a comprehensive understanding of DEGs, gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG)-based functional enrichment was conducted. According to GO assignments, a total of 27,999 up-regulated DEGs (Additional file 6: Table S5) and 56,166 down-regulated DEGs (Additional file 6: Table S6) were divided into three main categories: biological process, cellular component, and molecular function. Overall, the up-regulated and down-regulated DEGs in different groups were significantly enriched in the same or different GO terms (Additional file 3: Figure S4). Among the KEGG pathway analysis, biosynthesis of secondary metabolites such as “Phenylpropanoid biosynthesis” and “Flavonoid biosynthesis” represented the top twenty enriched KEGG pathways, especially in the up-regulated DEGs of group Leaf vs Stem, Leaf vs RootI, Stem vs RootI, RootI vs RootII, and the down-regulated DEGs of group Stem vs Root and RootI vs RootII (Additional file 6: Table S7, Table S8). Notably, the down-regulated DEGs in group Leaf vs RootI and Leaf vs RootII were significantly enriched in Ribosome, and only group Stem vs RootI had up-regulated DEGs that significantly enriched in the “Phenylalanine metabolism pathway”, containing 70 upregulated DEGs (Fig. 3c). DEGs between root tissue and other tissues, such as PAL, 4CL, CHS, CHI, C4H, DFR, ANR, ANS, and LAR, were in the significantly enriched pathway terms “Phenylpropanoid biosynthesis”(map00940) and “Flavonoid biosynthesis”(map00941), seemed relevant to the goal of our study (Table 2).
The number and KEGG enrichment of DEGs. a Distribution of the unigenes of the four libraries. b The red columns indicate the up-regulated DEGs and the green columns represent the down-regulated DEGs in six pair-wise comparisons (FDR ≤ 0.05 and an absolute value of log 2 Ratio ≥ 2 was used as the significant threshold for DEGs). c The top 20 enriched KEGG pathways of DEGs. The y axis shows the metabolic pathway terms, and the x axis shows the different comparison groups. The size of the plotted circle indicates the Sample number in this pathway terms. The fill color is scaled to the -log10(FDR). (FDR < 0.05)
Expression patterns of PA biosynthesis potential pathway structural genes in different tissues
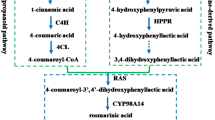
A large number of PAs were detected in I. stachyodes roots, and the two-year-old I. stachyodes root with red color is usually used as the medicinal harvesting standard, which means the content of flavonoids in the root of two-year-old I. stachyodes is higher [27]. To further investigate these important findings, the transcriptome of different tissues were compared to dig out the key genes in the metabolism of red root related to the growth years of I. stachyodes. In total, 75 unigenes that encoded 14 enzymes in the flavonoid and PA biosynthesis pathways were identified using BLASTp with previously identified Arabidopsis thaliana genes annotated in KEGG and additional literatures [37,38,39] (Additional file 7: Table S9). The normalized expression profiles of all the putative PA biosynthesis unigenes found in the I. stachyodes transcriptome were shown in Fig. 4. The biosynthesis pathway structural genes of PAs have been mainly divided into three parts [34]. The general biosynthetic genes (GBGs) including PAL, C4H, and 4CL are marked in blue, the EBGs including CHS, CHI, F3H, and F3′H are marked in green, while the LBGs including DFR, ANS, ANR, LAR, UGT72L1, MATE and LAC15 are marked in red.
An overview of PA biosynthesis pathway and PA-related structural genes expression across I. stachyodes organ type. Abbreviations are as follows: PAL (Phenylalanine ammonia lyase), C4H (cinnamate 4-hydroxylase), 4CL (4-coumaroyl CoA ligase), CHS (chalcone synthase), CHI (chalcone isomerase), F3H (flavanone 3-hydroxylase), F3′H (flavonoid 3′-monooxygenase), DFR (dihydroflavonol-4-reductase), LAR (leucoanthocyanidin reductase), ANS (anthocyanidin synthase), ANR (anthocyanidin reductase), MATE (multidrug detoxification and extrusion), UGT72L1 (UDP-glycosyltransferase), and LAC15 (laccase 15). yellow is high expression, blue is low expression
In general, most of the pathway structural genes had significantly tissue-specific expression, which must be related to the accumulation level of PAs in various tissues. IsUGT72L1-2, IsUGT72L1-3, IsUGT72L1-4, IsUGT72L1-7, IsUGT72L1-8, IsANR2 had the same expression pattern with IsLAR2 (TRINITY_DN26566_c0_g1), showed specific high expression in RootII tissue. In the result of correlation between the expression of pathway structural genes and the measured PA content, the correlation with procyanidin B2 is IsLAR2(0.479), IsANR2(0.654), IsUGT72L1-3(0.84), sequentially show a strong positive correlation (Additional file 3: Figure S5). Similarly, IsF3′H1ˎ IsMATE2ˎ IsMATE3 showed highly up-regulated expression in RootII tissue, and had a strong positive correlation with the accumulation level of procyanidin B2 and B1. Interestingly, IsCHS6, IsANS, IsUGT72L1-4, IsDFR2 showed specific low expression in RootI tissue, the correlation with epicatechin is IsANS(0.371), IsDFR2(0.509), IsCHS6(0.739) sequentially show a strong positive correlation. IsCHS1, IsCHS4, IsCHS7, IsCHS8, IsCHI5, IsC4H1, IsC4H2, IsUGT72L1-12, Is4CL11, IsPAL7 showed specific high expression in RootI tissue, and showed highly strong positive correlation with catechin and procyanidin B2 (Additional file 3: Figure S5). From the results of our association analysis, it can be seen that the synthesis of PA synthesis precursors is mainly regulated in RootI, while the synthesis of procyanidin B2 is critically regulated in RootII, and genes involved in this process may play a key regulatory role.
Identification of regulators of PA biosynthesis genes in I. stachyodes roots
PA biosynthesis is controlled by regulatory networks that consist of TFs or regulatory complexes in different species [13, 40]. In order to comprehensively reveal the regulatory network of PA biosynthesis, the expression data of the PA pathway genes and all the TFs were extracted from I. stachyodes transcriptome dataset (Additional file 7: Table S9; Additional file 8: Table S10), and were applied to co-expression analysis using the TGMI algorithm. The triple gene blocks were identified by the TGMI algorithm with a cut-off significance level of 0.05 (Additional file 9: Table S11). The interference frequencies of TFs on pathway genes were displayed in descending order (Additional file 10: Table S12). Among the top 185 TFs regulators, which interfere with the pathway genes with the highest frequencies, in the lists identified by TGMI, 34 IsMYBs, 18 IsbHLHs, 15 IsWRKYs, 9 IsMADSs, 3 IsWIPs are known PA pathway regulators supported by literature. These TFs were further combined to generate a circular network, as shown in Fig. 5. It is perceivable that the core pathway regulator MYB highlighted in a light coral color, is considered to be a top candidate that plays a central role in the expression regulation of pathway structural genes [13, 14].
Network analysis of the most positive known PA biosynthesis pathway regulators MYB, bHLH, WRKY, MADS, and WIP. Regulatory network of GBGs (top) and regulatory network of EBGs and LBGs (bottom) generated by TGMI algorithm for the I. stachyodes PA biosynthesis pathway using the high-throughput data yielded from treatment versus control. Blue nodes represent GBGs. Green nodes represent EBGs. Orange-red nodes represent LBGs. All other nodes are TFs regardless of what colors they are. Light coral nodes represent the most positive known pathway regulators MYB
In brief, co-expression analysis identified numerous potential interactive regulators of PA biosynthesis, involving 34 IsMYBs, 18 IsbHLHs, 15 IsWRKYs, 9 IsMADSs, and 3 IsWIPs (Additional file 11: Table S13) were chosen for heatmap analysis (Additional file 3: Figure S6). It is worth noting that IsWRKY45, IsMYB24, IsbHLH33, IsMYB80, IsMYB9, IsMYB52, IsMADS12, IsMYB68, IsbHLH14, IsMYB79, IsMYB69, IsMYB23 exhibited a higher expression level in RootII. Genes with same or similar expression patterns are often under the regulation of the same molecular mechanism [41]. Thus, we should focus on the network in RootII in order to further dig out key regulators affecting the synthesis of PAs in roots. Two mainly different subnetworks were detected. In sub-network one, four MYBs (IsMYB23, IsMYB79, IsMYB9, IsMYB80), one IsWRKY45, one IsMADS12, and 2 LBGs (IsMATE2, IsMATE3) were co-expressed. Meanwhile, in sub-network two, four MYBs (IsMYB24, IsMYB52, IsMYB68, IsMYB69), two bHLHs (IsbHLH14, IsbHLH33), and only with 1 LBGs IsANR2 co-expressed. These TFs also have a strong correlation with the level of PAs in I. stachyodes (Additional file 3: Figure S7). R2R3-MYB generally plays a central role in regulating target genes in PA pathways [13, 14]. To further screen out the PA-related R2R3-MYB proteins and predict their functions, we constructed a phylogenetic tree comprising the 34 IsMYBs proteins along with 126 Arabidopsis R2R3-MYB proteins and 16 proteins related to this process in other plant species (Fig. 6). IsMYB79 with higher expression level in RootII than other tissues, was clustered in subgroup 6 of the MYB gene family, such as AtMYB90, AtMYB75, AtMYB114, and AtMYB113 in Arabidopsis thaliana [42]. The overexpression of AtMYB75 or AtMYB90 in purple transgenic tobacco plants strongly enhances anthocyanin contents via upregulating all of the anthocyanin biosynthetic genes [43]. IsMYB24, another higher expressed in RootII, which was clustered in subgroup 5 of the MYB gene family, AtMYB123, and TT2-type genes were involved in anthocyanin and PA biosynthesis regulation [13, 44]. IsMYB75 clustered in subgroup 7 and IsMYB22 clustered in subgroup 5, but they specifically expressed in leaf tissues not in roots (Fig. 6). To determine the characterization of IsMYB24 and IsMYB79, homologous sequence alignment was carried out using deduced amino acid sequences and other published flavonoid-related genes amino acid sequences (Fig. 7). The results show that IsMYB79 and IsMYB24 have the general characteristics of R2R3-MYB gene family, and contained R2 and R3 domains. IsMYB79 was closely related to other published anthocyanin-related MYBs, which were promoting pigmentation, such as CmMYB6 [45], MaAN2 [46], LrMYB15 [47], StMYB113 [48], PpMYB10 [49], MrMYB1 [50] had been studied in model plant tobacco, the molecular mechanism of regulating anthocyanin accumulation has been basically clarified. EsMYBA1 influences pigmentation in the leaves, flowers, and flower buds [51]. LhMYB12 and LhSorMYB12 in the Lilium species control anthocyanin pigmentation in whole tepals [52]. The transcriptional activation of RsMYB1 [53] resulted the anthocyanin pigmentation. The highly homology indicated that the function of these MYBs was similar. In addition, IsMYB24 was closely related to VvMYBPA2, which plays crucial roles in regulating PA biosynthesis [54]. Therefore, IsMYB79 and IsMYB24 were similar to other flavonoid-related genes, which may play an important role in promoting root pigmentation.
Phylogenetic analyses of the combinatorial TFs MYB. Phylogenetic tree constructed with MYBs of Arabidopsis thaliana and proteins related to flavonoids biosynthesis in other species, including Vitis vinifera VvMYBPA1 (NP_001268160.1), Raphanus sativus RsMYB1 (AKM95888.1), Camellia sinensis CsMYB2 (AEI83426.1), Brassica napus BnTT2 (ABI13035.1), Lilium hybrid division VII LhSorMYB12(BAJ22983.1), Lilium hybrid division I LhMYB12(BAO04194.1), Lilium regale LrMYB15(BAU29930.1), Prunus persica PpMYB10 (ADK73605.1), Litchi chinensis LcMYB1 (APP94121.1), Chrysanthemum x morifolium CmMYB6 (QUP79395.1), Fragaria x ananassa FaMYB10 (QIZ03070.1), Morella rubra MrMYB1 (ADG21957.1), Epimedium sagittatum EsMYBA1 (AGT39060.1), Euproctus montanus EsAN2 (AFY04089.1), Solanum tuberosum StMYB113 (AND01219.1), Muscari armeniacum MaAN2 (ASF20090.1), Vitis vinifera VvMYBPA2 (NP_001267953.1), Malus domestica MdMYBPA1(NP_001315766.1). IsMYBs protein sequences screened from the TGMI algorithm are labelled with triangles. Proteins labelled with red triangles belong to the clades of proanthocyanidin synthesis. The tree was constructed with the NJ method (1000 replications of bootstrap test) using the MEGAX program
Multiple alignments analyses of key MYB TFs. a Multiple alignments of IsMYB79 and IsMYB24 b amino acid sequences and other published flavonoid-related MYBs. Black lines indicate R2 and R3 domain in MYB family. Jnetpred means secondary structure prediction results of IsMYB79 and IsMYB24 proteins, red indicates tubes, and green arrows indicate sheets
Discussion
PA compounds in I. stachyodes roots
The result of our study showed that procyanidin B2 is the most important flavonoid in I. stachyodes roots, composed of two molecules of epicatechin [55]. Therefore, in I. stachyodes roots, PAs maybe are primarily epicatechin-based, similar to the situation in seed coats of the model plants Arabidopsis thaliana and Medicago truncatula [56]. Moreover, Cyanidin (Cy) was the main coloration anthocyanin component in I. stachyodes roots. Similarly, in apple, one of the most common anthocyanin pigments is cyanidin, which, in the form of cyanidin 3-O-galactoside, is the pigment chiefly responsible for red skin coloration [57]. In this regard, Cy appears to be the main anthocyanins determining the red color of I. stachyodesin roots. Once formed, the unstable Cy would be converted to the colorless epicatechin, which would eventually form procyanidin B2 via later glycosylation and other reactions.
The PA biosynthesis pathway in I. stachyodes roots
The KEGG database revealed that upregulated DEGs were significantly enriched in “Phenylpropanoid biosynthesis”, which provided a precursor for the biosynthesis of flavonoids including flavonol, anthocyanidin, and PA [58]. In addition, 75 DEGs correlated with PA biosynthesis were identified and found to encode PAL, C4H, 4CL, CHS, CHI, F3H, F3′H, DFR, ANS, ANR, LAR, UGT72L1, MATE, and LAC15. Of these, IsF3′H1, as a key rate-limiting enzyme in the process of flavonoids biosynthesis [59], showed a higher expression level at RootII compared with other tissues. The high expression of genes encoding F3′H would catalyze and synthesize a large amount of dihydroquercetin. ANR enzyme first catalyzes anthocyanins to generate flav-enol intermediates, then ANR enzyme catalyzes flav-en-ol intermediates to generate flavan-3-ol or flavan-3-ol carbocation, which participates in the subsequent transport and polymerization of PA [60]. In this study, we found that IsANR2 was up-regulated in RootII, leading to the accumulation of (-)-Epicatechin. IsLAR2 is also found up-regulated in RootII, which can not only convert anthocyanins into ( +)-catechins, but also convert 4β-(S-cysteinyl)-epicatechin back to epicatechin, the starter unit in PAs, thereby regulating the relative proportions of starter and extension units and consequently the degree of PA oligomerization [4].
The synthesis of dimeric flavan-3-ols (procyanidin B2) is the key metabolic pathway of PAs synthesis in I. stachyodes roots. Studies have reported that procyanidin B2 is produced by the polymerization of (–)-epicatechin carbocation and (–)-epicatechin [4]. Glycosylation was the precondition for flavonoids to be transported from endoplasmic reticulum to vacuoles, and epicatechin glycoside was the potential precursor of PA polymerization [61]. UGT72L1 can catalyze the glycosylation of epicatechin to produce epicatechin glycoside [10]. In this study, we found IsUGT72L1-3 up-regulated in RootII, which has a great connection with the transport process of PA synthesis in I. stachyodes roots. Two genes (IsMATE2, IsMATE3) code MATE also found up-regulated in RootII in our study, can preferentially transport epicatechin-3′-O-glucoside across membranes in yeast assay systems [11]. So far, the only known enzyme involved in PA oxidation and polymerization in Arabidopsis thaliana is TT10 (LAC15), and other enzymes involved in polymerization and oxidation still need to be identified. The result of this study found LAC15 not expressed in the root tissue, thus we deduced that the polymerization of PAs maybe not the key step in I. stachyodes roots.
Identification of PA biosynthesis key genes and TFs in I. stachyodes root
TGMI has been used to study lignin biosynthesis pathway in Arabidopsis thaliana [62], Populus [63], and Populus trichocarpa [64], for identifying which regulatory genes potentially control wood formation. In this study, we also applied the TGMI algorithm to true pathway regulators of PA biosynthesis in I. stachyodes roots based on the tissue-specific I. stachyodes gene expression datasets. As anticipated, our study identified 34 IsMYBs, 18 IsbHLHs, 15 IsWRKYs, 9 IsMADSs, and 3 IsWIPs regulators that potentially regulate PA biosynthesis in I. stachyodes and ranked them to the top of candidate regulatory gene lists (Additional file 3: Figure S6). IsMYB24, a homologous gene of PA-related MYB genes in subgroup 5, showed strong correlation with PA biosynthetic genes IsANR2. In many plant species, TT2 (AtMYB123) and its homologs are direct activators of genes encoding ANR, LAR, and other enzymes in the PA biosynthesis pathway [13]. Furthermore, TT2 forms a ternary complex with TT8 (bHLH) and TTG1 (WD40) to activate genes related to PA biosynthesis [65]. Similarly, in our study, IsMYB24 may form transcriptional complexes with IsbHLH (IsbHLH14, IsbHLH33), co-expressed with IsANR2, to regulate PA biological processes. In addition, IsMYB79 clustered in subgroup 6 could regulate the expression of LBGs (IsMATE2, IsMATE3) and biosynthesis of late anthocyanins [14, 66]. As our result, IsMATEs (IsMATE2, IsMATE3) was also co-expressed with the RootII-specific expression TFs (IsMYB79, IsWRKY45, IsMADS12). MdWRKY11 can increase the expression of F3H, FLS, DFR, ANS, and UFGT to promote anthocyanin accumulation in apples [67], binds to W-box cis elements in MdMYB10, MdMYB11 and MdUFGT promoters [68]. Thus, we speculate IsWRKY45 could bind to IsMYB79 to affect the synthesis of PAs. But for IsMADS12, there is still no research showing interactive relationship between MYB and MADS.
In summary, MYB TF is the core member of transcriptional complex, and overexpression of transgenic MYB alone will obviously promote PA biosynthesis [69, 70]. Our TGMI algorithm analysis, hierarchical clustering, and PA-related MYB evolutionary trees together determined two important TFs IsMYB24 and IsMYB79. However, the mechanism of action for them is not yet clear and needs further research.
Conclusions
In this study, the complete transcriptome of I. stachyodes was de novo-assembled and annotated for the first time, generating a total of 110,779 non-redundant unigenes, of which 63,863 could be functionally annotated. The high content of procyanidin B2 in I. stachyodes roots was associated with up-regulated genes involved in the early and late steps of PA biosynthesis (F3′H, ANR, LAR, UGT72L1, and MATE), which produce the dihydroquercetin, (-)-Epicatechin, ( +)-catechins, and epicatechin-3′-O-glucoside, ultimately yield procyanidin B2 during these steps of PA synthesis. Simultaneously, IsANR2 might be regulated by IsMYB24, while IsMATE (IsMATE2, IsMATE3) could be regulated by IsMYB79. These results may enable further metabolomic and gene functional study in I. stachyodes.
Methods
Sample preparation and RNA extraction
I. stachyodes was grown in Dechangxiang I. stachyodes Planting Base in Xiuwen County, Guizhou. The collection date of four tissues (Leaf, Stem, RootI, and RootII) in the year 2019 is 21th, June. And each sample was composed of three biological replicates. The total RNA was extracted from tissue samples using TRIzol® Reagent (Plant RNA Purification Reagent for plant tissue) according the manufacturer’s instructions (Invitrogen, Carlsbard, CA, USA) and genomic DNA was removed using DNaseI (TaKara). Then the integrity and purity of the total RNA quality were determined by 2100 Bioanalyser (Agilent Technologies, Inc., SantaClaraCA, USA) and quantified using the ND-2000 (NanoDrop Thermo Scientific, Wilmington, DE, USA). Only high-quality RNA sample (OD260/280 = 1.8 ~ 2.2, OD260/230 ≥ 2.0, RIN ≥ 8.0,28S:18S ≥ 1.0, > 1 μg) was used to construct sequencing library.
Measurement of PAs
A 1-g tissue of each sample was finely grinded into powder for PAs extraction. The PAs were extracted in 40% Ethanol solution at 50℃, then the Continued filtrate was drained under 60 °C water bath. The residue was reconstituted, filtered through a 0.22 microporous membrane, and loaded for analysis.
cDNA library preparation and transcriptome sequencing
The construction of cDNA library and RNAseq was performed by Shanghai Majorbio Bio-Isarm Technology Co., Ltd. (Shanghai, China). Firstly, mRNA was purified from 12 µg of total RNA from four tissues (Leaf, Stem, RootI, and RootII) by using Oligo(dT) magnetic beads, respectively. Then, the mRNA samples were randomly broken into 300 bp fragments and added with fragmentation buffer. The first-strand cDNA was formed via reverse transcription using reverse transcriptase and random hexamer primer using mRNA as a template. Then, second-strand cDNA was synthesized, forming a stable double-stranded structure. These cDNA fragments were ligated with the Illumina paired-end sequencing adaptors. Finally, these libraries were sequenced on a paired-end flow cell using Illumina Novaseq 6000 platform. We obtained 7.18 G of reads from each sample for de novo assembly.
De novo assembly and Gene annotation
The raw paired-end reads were trimmed and quality controlled by SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) with default parameters. Then clean data from the samples (RootI-1, RootI-2, RootI-3, RootII-1, RootII-2, RootII-3, Stem-1, Stem-2, Stem-3, Leaf-1, Leaf-2, Leaf-3) were used to do de novo assembly with Trinity ([71]. Then, the assembly results were filtered by using TransRate software (http://hibberdlab.com/transrate/) and CD-HIT software (http://weizhongli-lab.org/cd-hit/). Finally, the results of optimized assembly were evaluated by using BUSCO (Benchmarking Universal Single-Copy Orthologs, http://busco.ezlab.org) [72]. Annotation of the assembled unigenes was conducted using BLASTX [73] searches against the KEGG, Pfam, Swissprot, and non-redundant (NR) databases, with the public database (E < 1e-5). The gene ontology (GO) annotation information of these unigenes was obtained from the NCBI Nr database by using the program Blast2GO and contains molecular functions, biological processes, and cellular components [74]. Furthermore, the program WEGO [75] classified all unigenes based on the GO annotation information.
Analysis of DEGs
To identify DEGs between two different samples, the expression level of each transcript was calculated according to the transcripts per million reads (TPM) method. RSEM (http://deweylab.biostat.wisc.edu/rsem/) was used to quantify gene abundances. Essentially, differential expression analysis was performed using the DESeq2/DEGseq/EdgeR with Q value ≤ 0.05, DEGs with |log2FC|> 1 and Q value ≤ 0.05(DESeq2 or EdgeR) /Q value ≤ 0.001(DEGseq) were considered to be significantly different expressed genes. In addition, functional-enrichment analyses including GO and KEGG were performed to identify which DEGs were significantly enriched in GO terms and metabolic pathways at Bonferroni-corrected P-value ≤ 0.05 compared with the whole-transcriptome background. GO functional enrichment and KEGG pathway analysis were carried out by Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do).
Co-expression network analysis and Network visualization
Co-expression networks were generated using the R package Triple Gene Mutual Interaction (TGMI) [62]. Pathway genes were first evaluated by conditional mutual information plus a novel mutual interaction measure (MIM) we discovered. This MIM reflects the regulatory strength exerted by the TF on two pathway genes in the triple gene block. The larger the MIM, the more significant the TF controls two pathway genes. In order to meet the criteria for TGMI, a cut-off significance level of 0.05 was used in the calculation. This resulted in a final network of 183 nodes (genes and TFs) connected by 1,357 edges (str values). Cytoscape (v 3.8.2) [76] was used to visualize the resulting network using the Allegro Layout plugin with an edge-weighted Allegro Fruchterman-Reingold layout algorithm.
Homolog search, gene identification, and distance analysis
The coding sequence of AtR2R3-MYB was acquired from the TAIR (http://www.arabidopsis.org/) databases. The amino acids of the MYB proteins were used to perform Phylogenetic analysis using MEGAX software with the neighbour-joining statistical method and 1000 bootstrap replicates.
Availability of data and materials
The RNA sequencing reads are available in the Sequence Read Archive database of NCBI (BioProject ID: PRJNA817883).
Abbreviations
- PAs:
-
Proanthocyanidins
- TFs:
-
Transcription factors
- F3′H:
-
Flavonoid 3′ hydroxylase
- ANS/LDOX:
-
Anthocyanidin synthase/Leucoanthocyanidin dioxygenase
- ANR:
-
Anthocyanidin reductase
- LAR:
-
Leucoanthocyanidin reductase
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- MATE:
-
Multidrug detoxification and extrusion
- DEG:
-
Differentially Expressed Gene
- GBGs:
-
General biosynthetic genes
- LBGs:
-
Late biosynthetic genes
- EBGs:
-
Early biosynthetic genes
- Cy:
-
Cyanidin
References
Dixon RA, Xie DY, Sharma SB. Proanthocyanidins – a final frontier in flavonoid research? New Phytol. 2005;165:9–28.
Ou K, Gu L. Absorption and metabolism of proanthocyanidins. J Funct Foods. 2014;7:43–53.
Luca SV, Bujor A, Miron A, et al. Preparative separation and bioactivity of oligomeric proanthocyanidins. Phytochem Rev. 2019;19:1093–140.
Liu C, Wang X, Shulaev V, Dixon RA. A role for leucoanthocyanidin reductase in the extension of proanthocyanidins. Nat Plants. 2016;2:16182.
Lepiniec L, Debeaujon I, Routaboul JM, et al. Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol. 2006;57:405–30.
Tanner GJ, Francki KT, Abrahams S, Watson JM, Larkin PJ, Ashton AR. Proanthocyanidin biosynthesis in plants. Purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J Biol Chem. 2003;278:31647–56.
Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR. The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J. 2003;35:624–36.
Saito K, Kobayashi M, Gong Z, Tanaka Y, Yamazaki M. Direct evidence for anthocyanidin synthase as a 2-oxoglutarate-dependent oxygenase: molecular cloning and functional expression of cDNA from a red forma of Perilla frutescens. Plant J. 1999;17:181–9.
Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science. 2003;299:396–9.
Pang Y, Peel GJ, Sharma SB, Tang Y, Dixon RA. A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc Natl Acad Sci U S A. 2008;105:14210–5.
Zhao J, Dixon RA. MATE transporters facilitate vacuolar uptake of epicatechin 3’-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell. 2009;21:2323–40.
Zhao J, Pang Y, Dixon RA. The mysteries of proanthocyanidin transport and polymerization. Plant Physiol. 2010;153:437–43.
Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2099–114.
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–80.
Li P, Chen B, Zhang G, Chen L, Dong Q, Wen J, Mysore KS, Zhao J. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 2016;210:905–21.
Pang Y, Wenger JP, Saathoff K, Peel GJ, Wen J, Huhman D, Allen SN, Tang Y, Cheng X, Tadege M, Ratet P, Mysore KS, Sumner LW, Marks MD, Dixon RA. A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development. Plant Physiol. 2009;151:1114–29.
Sagasser M, Lu GH, Hahlbrock K, Weisshaar B. A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev. 2002;16:138–49.
Nesi N, Debeaujon I, Jond C, Stewart AJ, Jenkins GI, Caboche M, Lepiniec L. The The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell. 2002;14:2463–79.
Lloyd A, Brockman A, Aguirre L, Campbell A, Bean A, Cantero A, Gonzalez A. Advances in the MYB-bHLH-WD Repeat (MBW) Pigment Regulatory Model: Addition of a WRKY Factor and Co-option of an Anthocyanin MYB for Betalain Regulation. Plant Cell Physiol. 2017;58:1431–41.
Amato A, Cavallini E, Zenoni S, Finezzo L, Begheldo M, Ruperti B, Tornielli GB. A Grapevine TTG2-Like WRKY Transcription Factor Is Involved in Regulating Vacuolar Transport and Flavonoid Biosynthesis. Front Plant Sci. 2016;7:1979.
Yang YX, Liao SG, Wang Z, et al. Analysis of Water-soluble Chemical constituents of Indigoferae Stachyoidis Radix by UHPLC-DAD-Q-TOF-MS / MS. Chin J Exp Tradit Med Formulae. 2014;20:63–7.
Zhang SY, Piao HS, Song CY. Study on the relation between duration of cultivation of plant and content of chemical components in Astragalus. J Med Sci Yanbian Univ. 2005;28:87–9.
Feng W, Wang WQ, Zhao PR. Content variation of saponins and flavonoids from growing and harvesting time of Glycyrrhiza uralensis. J Chin Med Mater. 2008;31:184–6.
Jian FU, Liang GY, Zhang JX, et al. Chemical constituents in Indigoferae stachyoidis. Drugs & Clinic. 2013;28:265–8.
Qiu L, Liang Y, Tang GH, et al. Chemical constituents from the roots of Indigoferae stachyoidis. Chin Tradit Pat Med. 2013;35:320–3.
Zhong L, Zhu XY, Yang YS, et al. Hepatoprotective chemical constituents from thylacetate extract of Indigofera stachyoides Radix. Chin J Exp Tradit Med Formulae. 2018;24:56–63.
Li J, Wang CM, Zhang YP, et al. Analysis of Flavonoids from the Roots of Indigofera stachyoides. Mol Plant Breeding. 2021; 1–22.
Dan CL, Zhang YY, Zhang YP, et al. Study on anti-inflammatory activity of radix Indigofera extract based on transgenic zebrafish mode-screening. Lishizhen Med and Mater Med res. 2016;27:2617–20.
Wu XF, Wang XG, Zhang RG. Comparative study on antioxidant activity and alpha glucosidase inhibitory activity of different parts of Indigofera Stachyodes. J of Qiannan Med Coll for Natl. 2017;30:161–4.
Zhu X, Luo H, Maoqiu HE, et al. Study on the Quality Standard of Miao Medicine Indigofera stachyoides. China Pharm. 2016;27:3829–31.
Duan L, Zhang YP, Miao YY, et al. In vivo and in vitro effects of Miao medicine Indigofera stachyoides extracts on breast cancer 4T1 cells. Chin Tradit Herb Drugs. 2018;49:2902–7.
Zheng Q, Chen W, Luo M, Xu L, Zhang Q, Luo Z. Comparative transcriptome analysis reveals regulatory network and regulators associated with proanthocyanidin accumulation in persimmon. BMC Plant Biol. 2021;21:356.
Li H, Han M, Yu L, Wang S, Zhang J, Tian J, Yao Y. Transcriptome Analysis Identifies Two Ethylene Response Factors That Regulate Proanthocyanidin Biosynthesis During Malus Crabapple Fruit Development. Front Plant Sci. 2020;11:76.
Hong M, Hu K, Tian T, Li X, Chen L, Zhang Y, Yi B, Wen J, Ma C, Shen J, Fu T, Tu J. Transcriptomic Analysis of Seed Coats in Yellow-Seeded Brassica napus Reveals Novel Genes That Influence Proanthocyanidin Biosynthesis. Front Plant Sci. 2017;8:1674.
Duwadi K, Austin RS, Mainali HR, Bett K, Marsolais F, Dhaubhadel S. Slow darkening of pinto bean seed coat is associated with significant metabolite and transcript differences related to proanthocyanidin biosynthesis. BMC Genomics. 2018;19:260.
Freixas Coutin JA, Munholland S, Silva A, Subedi S, Lukens L, Crosby WL, Pauls KP, Bozzo GG. Proanthocyanidin accumulation and transcriptional responses in the seed coat of cranberry beans (Phaseolus vulgaris L.) with different susceptibility to postharvest darkening. BMC Plant Biol. 2017;17:89.
Sharma SB, Dixon RA. Metabolic engineering of proanthocyanidins by ectopic expression of transcription factors in Arabidopsis thaliana. The Plant J. 2005;44:62–75.
Jin JQ, Ma JQ, Yao MZ, Ma CL, Chen L. Functional natural allelic variants of flavonoid 3′,5′-hydroxylase gene governing catechin traits in tea plant and its relatives. Planta. 2016;245:523–38.
Bogs J, Downey MO, Harvey JS, Ashton AR, Tanner GJ, Robinson SP. Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol. 2005;139:652–63.
Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot. 2011;62:2465–83.
Clements M, van Someren EP, Knijnenburg TA, Reinders MJ. Integration of Known Transcription Factor Binding Site Information and Gene Expression Data to Advance from Co-Expression to Co-Regulation. Genomics Proteomics Bioinf. 2007;5:86–101.
Chen S, Kong Y, Zhang X, Liao Z, He Y, Li L, Liang Z, Sheng Q, Hong G. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20:735–47.
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–94.
An XH, Tian Y, Chen KQ, Liu XJ, Liu DD, Xie XB, Cheng CG, Cong PH, Hao YJ. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015;56:650–62.
Hong Y, Li M, Dai S. Ectopic Expression of Multiple Chrysanthemum (Chrysanthemum x morifolium) R2R3-MYB Transcription Factor Genes Regulates Anthocyanin Accumulation in Tobacco. Genes (Basel). 2019;10:777.
Chen K, Liu H, Lou Q, Liu Y. Ectopic Expression of the Grape Hyacinth (Muscari armeniacum) R2R3-MYB Transcription Factor Gene, MaAN2, Induces Anthocyanin Accumulation in Tobacco. Front Recent Dev Plant Sci. 2017;8:965.
Yamagishi M. A novel R2R3-MYB transcription factor regulates light-mediated floral and vegetative anthocyanin pigmentation patterns in Lilium regale. Mol Breeding. 2015;36:3.
Liu Y, Lin-Wang K, Espley RV, Wang L, Yang H, Yu B, Dare A, Varkonyi-Gasic E, Wang J, Zhang J, Wang D, Allan AC. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J Exp Bot. 2016;67:2159–76.
Zhou H, Lin-Wang K, Wang H, Gu C, Dare AP, Espley RV, He H, Allan AC, Han Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. The Plant J. 2015;82:105–21.
Huang YJ, Song S, Allan AC, et al. Differential activation of anthocyanin biosynthesis in Arabidopsis and tobacco over-expressing an R2R3 MYB from Chinese bayberry. Plant Cell Tiss ORG. 2013;113:491–9.
Huang W, Khaldun AB, Lv H, Du L, Zhang C, Wang Y. Isolation and functional characterization of a R2R3-MYB regulator of the anthocyanin biosynthetic pathway from Epimedium sagittatum. Plant Cell Rep. 2016;35:883–94.
Yamagishi M, Shimoyamada Y, Nakatsuka T, Masuda K. Two R2R3-MYB genes, homologs of Petunia AN2, regulate anthocyanin biosyntheses in flower Tepals, tepal spots and leaves of asiatic hybrid lily. Plant Cell Physiol. 2010;51:463–74.
Lim SH, Song JH, Kim DH, Kim JK, Lee JY, Kim YM, Ha SH. Activation of anthocyanin biosynthesis by expression of the radish R2R3-MYB transcription factor gene RsMYB1. Plant Cell Rep. 2015;35:641–53.
Terrier N, Torregrosa L, Ageorges A, Vialet S, Verriès C, Cheynier V, Romieu C. Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 2009;149:1028–41.
Esatbeyoglu T, Wray V, Winterhalter P. Dimeric procyanidins: screening for B1 to B8 and semisynthetic preparation of B3, B4, B6, And B8 from a polymeric procyanidin fraction of white willow bark (Salix alba). J Agric Food Chem. 2010;58:7820–30.
Ito C, Oki T, Yoshida T, Nanba F, Yamada K, Toda T. Characterisation of proanthocyanidins from black soybeans: isolation and characterisation of proanthocyanidin oligomers from black soybean seed coats. Food Chem. 2013;141:2507–12.
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. The Plant JThe Plant Journal. 2007;49:414–27.
Wang Y, Zhou LJ, Wang Y, Liu S, Geng Z, Song A, Jiang J, Chen S, Chen F. Functional identification of a flavone synthase and a flavonol synthase genes affecting flower color formation in Chrysanthemum morifolium. Plant Physiol Biochem. 2021;166:1109–20.
Wu Y, Wang T, Xin Y, Wang G, Xu LA. Overexpression of the GbF3′H1 Gene Enhanced the Epigallocatechin, Gallocatechin, and Catechin Contents in Transgenic Populus. J Agr Food Chem. 2020;68:998–1006.
Wang P, Liu Y, Zhang L, Wang W, Hou H, Zhao Y, Jiang X, Yu J, Tan H, Wang Y, Xie DY, Gao L, Xia T. Functional demonstration of plant flavonoid carbocations proposed to be involved in the biosynthesis of proanthocyanidins. Plant J. 2020;101:18–36.
Zerbib M. Etude de la glycosylation de flavanols dans le raisin et incidence dans les vins. Université Montpellier. 2018.
Gunasekara C, Zhang K, Deng W, Brown L, Wei H. TGMI: an efficient algorithm for identifying pathway regulators through evaluation of triple-gene mutual interaction. Nucleic Acids Res. 2018;46:e67-e.
Zhang J, Tuskan GA, Tschaplinski TJ, Muchero W, Chen JG. Transcriptional and Post-transcriptional Regulation of Lignin Biosynthesis Pathway Genes in Populus. Front Plant Sci. 2020;11:652.
Hong J, Gunasekara C, He C, Liu S, Huang J, Wei H. Identification of biological pathway and process regulators using sparse partial least squares and triple-gene mutual interaction. Sci Rep. 2021;11:13174.
Xu W, Grain D, Bobet S, Le Gourrierec J, Thévenin J, Kelemen Z, Lepiniec L, Dubos C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014;20:132–44.
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant J. 2008;53:814–27.
Wang N, Liu W, Zhang T, Jiang S, Xu H, Wang Y, Zhang Z, Wang C, Chen X. Transcriptomic Analysis of Red-Fleshed Apples Reveals the Novel Role of MdWRKY11 in Flavonoid and Anthocyanin Biosynthesis. J Agric Food Chem. 2018;66:7076–86.
Liu W, Wang Y, Yu L, Jiang H, Guo Z, Xu H, Jiang S, Fang H, Zhang J, Su M, Zhang Z, Chen X, Chen X, Wang N. MdWRKY11 Participates in Anthocyanin Accumulation in Red-Fleshed Apples by Affecting MYB Transcription Factors and the Photoresponse Factor MdHY5. J Agric Food Chem. 2019;67:8783–93.
Constabel CP. Molecular Controls of Proanthocyanidin Synthesis and Structure: Prospects for Genetic Engineering in Crop Plants. J Agr Food Chem. 2018;66:9882–8.
Hassani D, Fu X, Shen Q, Khalid M, Rose JKC, Tang K. Parallel Transcriptional Regulation of Artemisinin and Flavonoid Biosynthesis. Trends Plant Sci. 2020;25:466–76.
Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52.
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–2.
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421.
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6.
Ye J, Zhang Y, Cui H, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–7.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Acknowledgements
Not applicable.
Materials statement
All authors comply with the IUCN Policy Statement on Research Involving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora.
General guideline statement
The materials involved in this research are artificially planted, and the collection of materials conforms to Good Agricultural Practice for Chinese Crude Drugs.
Permission statement
All authors declare that the collection application complies with relevant national or local regulations, and the collection method, time, location, and quantity of the collection application are appropriate.
Funding
Joint Fund Project of National Natural Science Foundation of China and Guizhou Provincial People's Government(U1812403), National Key R&D Program of China (NO. 2019YFC1712500), National Natural Science Foundation of China (NO. 81860667), Science and Technology Department of Guizhou Province (QKHHBZ[2020]3003).
Author information
Authors and Affiliations
Contributions
Jun Li collected the samples, performed the experiments and data analyses. Chongmin Wang made the figures and tables, and wrote the manuscript. Haijun Jin and Xiaoqing Liu joined the samples collection and edited the manuscript. Yongping Zhang conceived and designed the study. Tao Zhou supervised the experiments and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All the plant materials used in this study were provided by Dechangxiang I. stachyodes Planting Base in Xiuwen County, Guizhou. The field experiments were conducted under local legislation and permissions.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, C., Li, J., Zhou, T. et al. Transcriptional regulation of proanthocyanidin biosynthesis pathway genes and transcription factors in Indigofera stachyodes Lindl. roots. BMC Plant Biol 22, 438 (2022). https://doi.org/10.1186/s12870-022-03794-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-022-03794-4