Abstract
Background
Metal homeostasis is critical for plant growth, development and adaptation to environmental stresses and largely governed by a variety of metal transporters. The plant ZIP (Zn-regulated transporter, Iron-regulated transporter-like Protein) family proteins belong to the integral membrane transporters responsible for uptake and allocation of essential and non-essential metals. However, whether the ZIP family members mediate metal efflux and its regulatory mechanism remains unknown.
Results
In this report, we provided evidence that OsZIP1 is a metal-detoxified transporter through preventing excess Zn, Cu and Cd accumulation in rice. OsZIP1 is abundantly expressed in roots throughout the life span and sufficiently induced by excess Zn, Cu and Cd but not by Mn and Fe at transcriptional and translational levels. Expression of OsZIP-GFP fusion in rice protoplasts and tobacco leaves shows that OsZIP1 resides in the endoplasmic reticulum (ER) and plasma membrane (PM). The yeast (Saccharomyces cerevisiae) complementation test shows that expression of OsZIP1 reduced Zn accumulation. Transgenic rice overexpressing OsZIP1 grew better under excess metal stress but accumulated less of the metals in plants. In contrast, both oszip1 mutant and RNA interference (RNAi) lines accumulated more metal in roots and contributed to metal sensitive phenotypes. These results suggest OsZIP1 is able to function as a metal exporter in rice when Zn, Cu and Cd are excess in environment. We further identified the DNA methylation of histone H3K9me2 of OsZIP1 and found that OsZIP1 locus, whose transcribed regions imbed a 242 bp sequence, is demethylated, suggesting that epigenetic modification is likely associated with OsZIP1 function under Cd stress.
Conclusion
OsZIP1 is a transporter that is required for detoxification of excess Zn, Cu and Cd in rice.
Similar content being viewed by others
Background
Metal homeostasis is a mechanism essential for plant growth, development and adaptation to diverse environmental stresses [1,2,3]. It is built up by metal uptake and transport across cells or within the cells, which largely relies on a variety of metal transporters [4, 5]. The plant ZIP (Zn-regulated transporter, Iron-regulated transporter-like Protein) proteins belong to the metal transporter family involved in uptake and allocation of Zn, Cu, Cd, iron (Fe) or manganese (Mn) [1, 2, 6,7,8]. In Arabidopsis the ZIP family genes encode 14 protein members. AtIRT1 is a high affinity Fe uptake transporter [9,10,11,12]. AtIRT2 and AtIRT3 are responsible for Fe and Zn uptake in roots [13, 14]. AtZIP1 serves as a vacuolar transporter remobilizing Mn and Zn from vacuole to cytoplasm in root cells [12, 15]. AtZIP2 is located in the PM and may mediate Mn (and possibly Zn) uptake into root stele cells or Mn/Zn mobility in the stele to the xylem parenchyma [12]. While AtZIP3 is supposed to transport Zn and Fe from soil to plant [16], AtZIP4 is likely involved in Zn transport across tissues [2]. The rest of AtZIPs transport Mn, Zn and Fe [12]. The rice genome houses 18 ZIP members [17]. OsIRT1 and OsIRT2 are homologs primarily for Fe and Zn transport in roots [18, 19]. OsZIP1 is thought of a Zn uptake transporter whose expression is induced under Zn deficiency [20,21,22], however, its biological functions in rice under metal stress are not fully understood. OsZIP2–5 and OsZIP8 are expressed in roots for Zn uptake or distribution [20, 21, 23,24,25,26,27]. OsZIP6 is induced in rice when Fe, Zn and Mn are deprived [28]. OsZIP7 is expressed in shoots under Zn deficiency [18] and in roots when Fe is limiting [26]. The functions of OsZIP9-OsZIP16 are less understood.
Cadmium is a non-essential and toxic metal to plants. Uptake of excess Cd jeopardizes crop production and food security [29]. Since no specific transporters for Cd are available, uptake of Cd into cells is shared by Fe, Zn and Mn transport systems [29]. To date, only a few studies are available on the transport of Cd out of plants. AtPDR8 is a PM transporter and can be induced by Cd exposure; mutantion of AtPDR8 leads to hypersensitivity to Cd, whereas AtPDR8 overexpression plants show Cd tolerance and accumulate less of Cd than wild-type [30]. Another example is rice Cal1 which can chelate Cd in cytosol and get it into the outer space [31]. Recently, the epigenetic modifications such as DNA methylation, histone modification and small interference RNAs have emerged as additional modulators for plant adaptation to biotic and abiotic stresses [32,33,34,35,36,37,38]. DNA methylation is a kind of post-modification predominantly found in cytosine residues (5-methyl cytosine, 5mC) of the dinucleotides of CG and to less extent, of CHG and CHH (where H is A, C or T). In plants, addition of a methyl group to the cytosine residue in the three contexts is mediated by a group of DNA methyltransferases such as MET1 (METHYLTRANSFERASE 1) mainly for CG, CMT3 (CHROMOMETHYLASES) for CHG, and DRM2 (DOMAINS REARRANGED METHYLTRANSFERASE 2) for CHH [39, 40]. Since DNA methylation/demethylation is a dynamic process, some demethylation enzymes such as ROS1 (REPRESSOR OF SILENCING), DME (DEMETER), DML2 (DEMETER-LIKE) and DML3 [41] are also involved in the process. While DNA methylation is the major epigenetic mechanism for regulating gene expression, the histone modifications at lysine or arginine are also engaged in the epigenetic process [42]. For example, the histone methylation at H3K9me2 is essential for DNA methylation particularly at the CHG catalyzed by CMT3 [43]. Studies demonstrate that Cd exposure can alter the genomic DNA methylation pattern in plants [37, 44, 45]. However, whether Cd-induced DNA methylation marks are able to mediate transcription of target genes and functional consequences is largely unknown.
Rice (Oryza sativa L.) is a model plant species for environmental research owning to its abundant germplasm resources. It is also one of the major crops. Many rice cultivars genotypes are gifted with special traits contributing to accumulating essential and nonessential metals or gain defense and tolerant traits [37, 46,47,48]. Understanding the regulatory mechanism for uptake, transport and accumulation of toxic metals like Cd is critical for developing strategies of hyper-accumulating or minimizing the toxic metals in crops growing on the metal-contaminated soils [29, 37, 49]. We previously identified several metal transporters including OsZIP1 from transcriptome and methylome of Cd-exposed rice plants [37]. The underlying regulatory mechanisms for OsZIP1 mediating Cd or other metals remain to be investigated. In this report, we provided the new evidence that OsZIP1 is localized in both plasma membrane and endoplasmic reticulum. OsZIP1 overexpression detoxified rice plants exposed to excess Zn, Cu and Cd by limiting accumulation of the metals in rice tissues. Furthermore, we precisely demonstrate that Cd exposure induced DNA and histone H3K9me2 demethylation in the transcribed region of OsZIP1 which is likely associated with OsZIP1 function under Cd stress.
Results
Expression of OsZIP1 is upregulated under excess Zn and Cu and Cd stress
The genomic sequence of OsZIP1 (LOC Os01g74110) is 1909 bp in length, with a 1059 bp coding DNA sequence (CDS). The CDS was predicted to encode a 352-amino acid protein with eight transmembrane domains (TMDs) and a variable region between the 3th and 4th TMDs relevant to the metal binding and transport (Additional file 1: Figure S1).
A qRT-PCR analysis showed that OsZIP1 was transcriptionally expressed in rice throughout the life span (Additional file 2: Figure S2). In roots OsZIP1 was abundantly expressed, but its expression was weak in shoots. We sought to explore the challenge of metal excess to OsZIP1 expression. Two week-old rice plants were exposed to the excess metals Zn, Cu, Fe, Mn and Cd, and the transcripts of OsZIP1 were measured by qRT-PCR. A high level (> 6 fold) of OsZIP1 transcripts was detected in roots exposed to 1–10 mM Zn over the control (Fig. 1a). Treatment with 30 μM Cu moderately affected expression of OsZIP1, with 4 fold over the control (Fig. 1a). There was no effect of excess Mn and Fe on OsZIP1 transcripts. Transcripts of OsZIP1 in roots were increased under 40–160 μM Cd exposure (Fig. 1b), which is similar to the previous study [37]. The pOsZIP1::GUS vector was constructed and transformed into tobacco leaves by Agrobacterium tumefaciens transformation. The GUS transcripts under Cd stress were increased more 2.5 fold than those under the control (Additional file 3: Figure S3). Expression of OsZIP1 in roots induced by excess Zn, Cu and Cd was confirmed by Western blot analysis (Fig. 1c-e).
Analyses of OsZIP1 expression under metal stress conditions using qRT-PCR and Western blot. Two week-old rice plants grew in the nutrient solutions with 1000 and 10,000 μM Zn, 500 and 5000 μM Mn, 10 and 30 μM Cu, 100 and 1000 μM Fe and 0–160 μM Cd for 3 h. For Western blot analysis, seedlings were exposed to the metals at the indicated concentrations for 4 d. a Response of OsZIP1 transcription to excess Zn, Mn, Cu and Fe. b Response of OsZIP1 transcription to Cd. c-e Western blotting analysis of OsZIP1 expression in roots under excess Zn and Cu and Cd stress. Vertical bars represent standard deviation. Asterisks indicate that the mean values of three replicates are significantly different between the treatments and control (p < 0.05)
OsZIP1 resides in the PM and ER
OsZIP1 is expressed in the vascular bundles and epidermal cells in rice roots, and is proposed to be a Zn uptake transporter [20]. We therefore constructed a recombinant plasmid connecting the green fluorescent protein (GFP) to OsZIP1, which was expressed in the rice mesophyll protoplasts. Confocal microscopy analysis revealed overlapped PM biomarker fluorescence (PIP2A-RFP, red) and OsZIP1-GFP fluorescence (green) signals in the rice mesophyll protoplasts (Fig. 2 a-d). We then tested several other specific biomarkers and found that only ER marker (RFP-KDEL, red) was probed, with strong signals overlapped with the fusion proteins (Fig. 2 e-h). To confirm the observation, the OsZIP1-GFP fusion was transformed and expressed in tobacco leaf epidermal cells [50]. Using the same ER marker, clear signals of co-localization of the fusion proteins and marker around the ER area were visualized (Fig. 2 i-k). Moreover, strong signals of GFP only and the merged images of GFP and PM-marker were detected (Fig. 2 l-n), suggesting that OsZIP1 is also localized to plasma membrane.
Subcellular localization of OsZIP1-GFP fusion proteins with plasma membrane (PM) and endoplasmic reticulum (ER) markers in rice mesophyll protoplast (a-h) and tobacco leaf epidermal cells (i-k) detected by Confocal images under normal metal supply. a OsZIP1-GFP. b PM marker PIP2A-RFP. c Bright field. d Merged images of a and b. e OsZIP1-GFP. f ER marker RFP-KDEL. g Bright field. h Merged images of e and f. i OsZIP1-GFP. j ER marker RFP-KDEL. k Merged images of i and j. l OsZIP1-GFP. m PM marker PIP2A-RFP. n Merged images of l and m. Arrows indicate the part of the ER or PM areas where OsZIP1-GFP fusion images were overlapped with the corresponding markers
Expressing OsZIP1 in yeast reduced Zn accumulation in cells
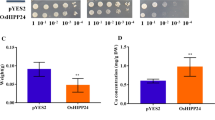
The wild-type and mutants of yeast (Saccharomyces cerevisiae) were used to identify the role of OsZIP1 in mediating metal resistance and accumulation in the cells. The empty vector pYES2 and the vector carrying OsZIP1 were transformed into the zrc1 mutants. OsZIP1-transformed zrc1 cells exposed to 3 and 6 mM Zn showed more resistance to excess Zn than the empty vector cells (Additional file 4: Figure S4). Analyses by ICP-AES revealed that the OsZIP1-expressing yeast cells accumulated significantly lower levels of Zn than its control cell (Additional file 4: Figure S4).
OsZIP1 knockdown repressed and OX lines promoted the rice growth under excess Zn, Cu and Cd stress
A T-DNA insertion mutant oszip1 was ordered from the rice database [51]. PCR analysis showed a single insertion in the intron (Additional file 5: Figure S5a). The OsZIP1 transcripts in the oszip1 line were reduced to 10.0% of its wild-type and the protein level was also drastically lowered (Additional file 5: Figure S5 b-d). Additionally, a set of independent knockdown lines of OsZIP1 were generated using RNA interference approach. Transcript analysis showed that the RNAi lines contained only 20.1–30.2% transcripts of the wild-type (Additional file 5: Figure S5 e). We experimented with the knockdown lines exposed to the high level of metals for a short time and the low level of metals for a long time. Under control condition, no differences in growth response between oszip1 or RNAi lines and wild-type were observed; however, when under 1000 μM Zn, 20 μM Cu or 80 μM Cd, both oszip1 and RNAi lines displayed the reduced shoot growth and dry weight compared to the wild-type (Fig. 3 a-l; Fig. 4 a,b, d, e, g, h). A similar response was found with the long-term study (Fig. 4 j).
Growth response to excessive Zn, Cu and Cd stress in rice wild-type (WT), oszip1 mutant, RNAi lines (i) and OsZIP1-transgenic lines (OX). Two weeks-old young rice plants were grown in the nutrient solution supplemented with 0.4 (normal) and 1000 (high) μM Zn or 0.2 (normal) and 20 (high) μM Cu for 10 days or with 0 and 80 μM Cd for 4 d. a-d, m, n Phenotypes and shoot length of WT, oszip1, RNAi and OX plants exposed to Zn excess. e-h, o, p Phenotypes and shoot length of WT, oszip1, RNAi and OX plants exposed to Cu excess. i-l, q, r Phenotypes and shoot length of WT, oszip1, RNAi and OX plants exposed to Cd. Vertical bars represent standard deviation. Asterisks indicate that the mean values are significantly different between the oszip1 mutant/RNAi lines/OX lines and wild-type (p < 0.05)
Growth response to excess Zn, Cu and Cd stress in wild-type (WT), oszip1 mutant, RNAi and OX lines. Two weeks-old young rice plants were grown in the nutrient solution supplemented with 0.4 (normal) and 1000 (high) μM Zn or 0.2 (normal) and 20 (high) μM Cu for 10 days or with 0 and 80 μM Cd for 4 d or 1 μM Cd for 30 d. a-c Dry weight of WT, oszip1, RNAi and OX plants exposed to Zn excess. d-f Dry weight of WT, oszip1, RNAi and OX plants exposed to Cu excess. g-k Dry weight of WT, oszip1, RNAi and OX plants exposed to Cd. Vertical bars represent standard deviation. Asterisks indicate that the mean values are significantly different between the transgenic plants/mutants and wild-type (p < 0.05)
We further generated OsZIP1-overexpressing lines (OXs) under the control of cauliflower mosaic virus 35S promoter. The OX lines used in this study showed 25.1 to 41.5-fold higher transcripts of OsZIP1 and higher protein levels than the wild-type (Additional file 5: Figure S5f, g). There were no growth differences between the OX lines and wild-type plants grown in the non-Cd medium (Fig. 3 m-r). When exposed to 1000 μM Zn, 20 μM Cu or 80 μM Cd, the shoot of OX lines grew longer, with 1.3–1.5 fold for Zn, 1.2–1.3 fold for Cu, and 1.2–1.3 fold for Cd over the wild-type (Fig. 6 m-r). The enhanced dry biomass was obtained in the OX lines (Fig. 4 c, f, i, k).
OsZIP1 knockdown and OX lines depicted a contrasting metal accumulation under excess metal stress
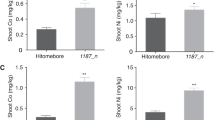
Under normal conditions (0.4 μM Zn and 0.2 μM Cu), the oszip1/RNAi, OX and wild-type plants had the similar Zn and Cu concentrations in their shoots. In roots, the oszip1/RNAi lines had a lower concentration of Zn and Cu, while the OX lines had a higher level of the metals compared to the wild-type (Fig. 5 a-f). Examination of metal concentrations in plants exposed to the excess levels (1000 μM Zn, 20 μM Cu and 80 μM Cd) shows that both oszip1 and RNAi lines accumulated more Zn, Cu and Cd in their roots than the wild-type, whereas no differences in metal concentrations were observed in the shoots (Fig. 5 g, h, j, k, m, n). Under the same conditions, the OX lines accumulated less Zn, Cu and Cd in roots and shoots than the wild-type (Fig. 5 i, l, o). The OsZIP1-mediated metal accumulation is similar to the previous report on a metal efflux transporter OsHMA9, as mutation of OsHMA9 led to increased accumulation of more Zn, Cu and Cd in roots [52].
Zn, Cu and Cd concentrations in shoots and roots of WT, oszip1 mutant, RNAi lines and OX lines under excess metal stress. Two week-old rice seedlings were grown in nutrient solutions supplemented with 1000 μM Zn or 20 μM Cu for 10 d or supplemented with 80 μM Cd for 4 d. 0.4 μM Zn and 0.2 μM Cu were used as the controls. Zn (a-c) and Cu (d-f) concentrations in WT and oszip1 mutants, RNAi lines and OXs lines under normal conditions. Zn (g-i) and Cu (j-l) concentrations in WT, oszip1 mutant, RNAi lines and OX lines under excess Zn and Cu stress. m-o Cd concentrations in WT, oszip1 mutant, RNAi lines and OX lines exposed to 80 μM. Vertical bars represent standard deviation. Asterisks indicate that the mean values are significantly different between the oszip1 mutant/ RNAi lines/OX lines and wild-type (p < 0.05)
We identified the role of OsZIP1 in mediating accumulation of metals under low supply conditions. Treatments with the lower levels of Zn (0.1 μM) and Cu (0.05 μM) led to lower accumulation of Zn and Cu in the roots of oszip1 and RNAi lines but higher accumulation in the OX lines, compared to the normal condition (Fig. 6 a-f). Analyzing Cd concentration in rice exposed to the lower level of Cd showed a higher concentration of Cd in the oszip1 and RNA1 roots but no difference in shoots (Fig. 6 g-i). These results suggested that knockdown of OsZIP1 led to lower accumulation of Zn and Cu but higher accumulation Cd in roots, whereas OsZIP1 overexpression led to a contrasting metal accumulation.
Zn, Cu and Cd concentrations in shoots and roots of WT, oszip1 mutant, RNAi lines and OX lines exposed to limiting Zn and Cu or lower concentration of Cd. Two day-old young plants were grown in the nutrient solutions with 0.1 µM Zn (a-c), 0.2 µM Cu (d-f) or supplemented with 1 µM Cd (g-i) for two weeks. Vertical bars represent standard deviation. Asterisks indicate that the mean values are significantly different between the oszip1 mutant/ RNAi lines/OX lines and wild-type (p<0.05)
Cd stress reduced DNA methylation in the transcribed region of OsZIP1
Our previous studies show that Cd exposure is able to induce DNA demethylation of OsZIP1, however, the detailed epigenetic processes remain elusive. To clarify it, three regions including the promoter (R1), intragenic (gene body) (R2) and intergenic (downstream) (R3) sequences of OsZIP1 were profiled (Fig. 7a). Using the BS-seq datasets [37], we found a significant demethylation region (+ 222~ + 464) within R2 under Cd stress (Fig. 7a), where the total cytosine methylation was reduced by 24.4% compared to the control (−Cd treatment); among the three contexts, CHG, CG and CHH methylation was reduced by 28.6, 22.5 and 20.0%, respectively. The Cd-induced body demethylation was confirmed by PCR-based DNA methylation assay (Fig. 7b). A long-term experiment with the rice plants exposed to 1 μM Cd for 30 days was performed, but the body demethylation at R2 of OsZIP1 was not detected under the condition (Additional file 6: Figure S6). Similarly, the transcripts of OsZIP1 did not increase in rice exposed to the low dose of Cd (Additional file 7: Figure S7).
Identification of DNA and histone demethylation of OsZIP1 in rice exposed to excess Zn, Cu, Mn, Fe and Cd. a Diagram of the OsZIP1 genomic region with CG, CHG and CHH demethylation pattern in R2 with Cd (+Cd) and without Cd (−Cd) exposure. b DNA methylation status At R1-R3 of OsZIP1 with -Cd and + Cd was determined by McrBC-qPCR assays (Note: a lower value of DNA amplification of (+McrBC/−McrBC) indicates the higher methylation). c: Histone methylation state was detected using an anti-H3K9me2 ChIP-qPCR assay at R1-R3 of the genomic regions of OsZIP1 under –Cd and + Cd conditions. d DNA methylation status at R1-R3 of OsZIP1 with the normal and excessive Zn, Cu, Mn and Fe supply. Two weeks-old young rice plants were grown in the nutrient solution supplemented with 0.4 (normal) and 1000 (high) μM Zn, 0.2 (normal) and 20 (high) μM Cu, 0.5 (normal) and 500 (high) μM Mn, and 10 (normal) and 1000 (high) μM Fe, or with 0 and 80 μM Cd for 4 d. The rice Ubi2 and Actin1 were tested in parallel as negative controls. Anti-H3 was used as an internal reference in the ChIP-qPCR assay. Vertical bars represent standard deviation. Asterisks indicate that the mean values of three replicates are significantly different between the metal treatments and control (p < 0.05)
H3K9me2 occurs around the repressed euchromatic regions and plays an important role in DNA methylation-mediated gene silencing [43, 53]. The rice SDG714 encodes an H3K9me2-specific histone methyltransferase and coordinates CMT3 for DNA methylation mainly at the CHG sequence context [53,54,55]. By analyzing chromatin immunoprecipitation (ChIP) using a specific H3K9me2 antibody, we found the reduced H3K9me2 marks in the R2 transcribed regions of OsZIP1 under Cd stress (Fig. 7c). The DNA methylation and H3K9me2 pattern was also investigated under the Zn and Cu stress condition, however, no change at the specific site was found (Fig. 7d). Likewise, both Fe and Mn excess failed to change the DNA methylation of OsZIP1 (Fig. 7d).
Since OsZIP1 methylation pattern was affected by Cd, it is essential to investigate whether Cd stress could affect the expression of DNA methylation-related protein genes. To address the question, qRT-PCR was used to test the transcriptional response of several genes to Cd stress. The OsCMT3a expression was repressed in rice under Cd stress (Fig. 8). The reduced transcripts of OsSDG714 and OsMET1 were also found in rice under Cd stress (Fig. 8). In contrast, expression of OsROS1 was slightly induced by Cd stress. Expression of OsDRM2 had no response to Cd stress. We further examined the response of these genes in expose to the low Cd and found no significant changes (Additional file 8: Figure S8). Finally, transcripts of the genes under excess Zn, Cu, Fe and Mn were examined and none of them were significantly changed (Additional file 9: Figure S9).
Effect of Cd on the transcripts of DNA methylation modifier genes. Two week-old young rice plants were grown in the nutrient solution supplemented with 0, 80 and 160 μM Cd for 4 d. Vertical bars represent standard deviation. Asterisks indicate that the mean values of three replicates are significantly different between the treatment and control (p < 0.05)
Mutation of OsSDG714 led to increase in OsZIP1 transcription
We assessed the DNA methylation status and OsZIP1 transcripts using a set of mutants defective in activities of DNA methylation/demethylation, histone modification and small RNA generation. Identification of T-DNA insertion of the mutants was previously described [37]. Since oscmt3a was unavailable and OsSDG714 is necessary for CHG methylation, the ossdg714 mutant was used in the study. Mutation of SDG714 caused massive loss of total H3K9me2 in OsZIP1 under -Cd and + Cd conditions (Fig. 9a) and consequently, the CHG methylation of OsZIP1 in ossdg714 was significantly lost (Fig. 9b). Meanwhile, the OsZIP1 transcripts were drastically increased (Fig. 9c), suggesting that loss of H3K9me2 and CHG methylation in ossdg714 may associate the increased OsZIP1 transcripts. We checked on the DNA methylation and H3K9me2 levels of OsZIP1 in the rest of the mutants. The DNA methylation of OsZIP1 in osmet1 was increased (Fig. 9d). In the absence of Cd, there was no change in H3K9me2 marks between osmet1 and wild-type plants; however, the H3K9me2 level in osmet1 was significantly lower than those of wild-type under Cd stress (Fig. 9e). According to DNA methylation status, the transcript levels of OsZIP1 in osmet1 were significantly lower than those in wild-type (Fig. 9f). There was no change in DNA methylation and H3K9me2 marks as well as transcripts of OsZIP1 in osdrm2–2, osros1, and osrdr2i-6 (Fig. 9g-l).
Identification of DNA and histone demethylation and expression of OsZIP1 in the wild-type (WT) and epigenetic modified mutant plants exposed to Cd. DNA methylation (a, d, g and j), H3K9me2 mark (b, e, h and K) and transcript levels (c, f, i and l) of OsZIP1 in ossdg714, osmet1, osrdr2i-6, osdrum2 and osros1 mutants and their wild-types with or without Cd exposure. Two weeks-old young rice plants were grown in the nutrient solution supplemented with 0 and 80 μM Cd for 4 d. The rice Actin1 was used as a negative control. Anti-H3 was used as an internal reference in the ChIP-qPCR assay. Vertical bars represent standard deviation. Asterisks indicate that the mean values of three replicates are significantly different between the wild-type and mutants (p < 0.05)
DNA methylation inhibitor-mediated detoxification of Cd-exposed rice depends on OsZIP1
To verify that the OsZIP1 demethylation was able to alter the phenotype of rice growth under Cd stress, we performed an additional experiment with a global DNA methylation inhibitor, azacitidine (Aza) [56]. Young rice plants were exposed to 80 μM Cd and 20 μM Aza for 4 d, and the growth response was assessed by measuring the shoot and root elongation. As shown in Fig. 10, Aza provision elongated the shoots and roots of wild-type under Cd stress, whereas no change in shoot or root growth of oszip1 mutants was observed between the Cd and Cd + Aza treatments, indicating that Aza failed to relieve Cd-induced inhibition of shoot and root growth due to the mutation of OsZIP1 and suggesting that Aza-mediated detoxification in Cd-exposed plants depends on OsZIP1.
Effects of DNA methyltransferase inhibitor 5-aza-2-deoxycytidine (Aza) on the growth of oszip1 mutants and wild-type rice plants exposed to Cd. Seven day-old young rice plants were treated with Cd (80 μM) and/or Aza (20 μM) for 4 d. Thereafter, the phenotype of rice seedlings were photographed (Scale bar = 2 cm). a Phenotype of rice plants. b Shoot length of rice plants. c Root length. Vertical bars represent standard deviation of the mean of three biological replicates. Asterisks indicate that the mean values are significantly different between the treatments (p < 0.05)
Discussion
The toxic heavy metal Cd is a non-essential element for plant growth and development, therefore, it must be under the tight control; likewise, even for essential metals such as Zn, Mn or Cu, the concentrations must be also restricted to a narrow range [29]. To prevent toxicity of excess metals, plants develop elaborate mechanisms for governing metal uptake, translocation and homeostasis. One of them is concerning the efflux transport systems. To date, several efflux transporters have been functionally characterized in rice [31, 52]. However, compared to those from Arabidopsis, the rice metal exporters are less reported.
OsZIP1 has long been considered as a metal uptake transporter for Zn and Cd in rice [15, 17, 21, 22]. Evidence to support the view relies on the increased transcripts of OsZIP1 under Zn deficiency and the transport activity for Zn in a OsZIP1-transformed yeast (ZHY3) [15, 17, 21, 22]. This study presented several lines of new evidence that both OsZIP1 transcripts and proteins can be substaintially incrased under excess Zn, Cu and Cd stress. Yeast complementation test supported the activity of OsZIP1 for the metal export and detoxification. In an accordace with it, OsZIP1 overexpression reduced concentrations of Zn, Cu and Cd in rice. As a consequence, the growth status was improved under metal stress. These results support the notion that OsZIP1 would be a transporter responsible for metal exfllux in rice. The phenotype of OsZIP1 resembles many metal efflux transporters such as OsHMA9 [52]; TcHMA4 [57]; AtHMA4 [58]; AtPDR8 [30]; CAL1 [31] and AtPCR2 [59].
Since most of the metal exporters localize to the PM, we addressed the question by examining OsZIP1 fusion proteins in both rice protoplasts and tobacco leaves. The fluorescence signals of OsZIP1-GFP occurred in both PM and ER. The dual localization suggests that OsZIP1 may play roles under changing environmental condition. Our data corroborate a model that has been recently illustrated from in Caryophyllaceae (Silene vulgaris) [60]. SvHMA5II is a Cu-tolerant transporter isolated from S. vulgaris and resides in the ER and upon Cu exposure, it re-localizes to the PM where the excess Cu is supposed to be fluxed out of cells [60]. Meanwhile, a study with yeast (Saccharomyces cerevisiae) provided a similar a model for extrusion of Pca1 (a P-type ATPase metal transporter) from the ER to PM, where it functions in the efflux of Cd across the PM [61]. Pca1 is a short-lived and primarily ER-localized protein that is targeted by a degron for ubiquitination and proteasomal degradation before approaching cell surface. Cd can upregulate Pca1 rapidly in the ER and lead to the degron bound to Pca1, which prevents Pca1 from being degraded in the so called endoplasmic reticulum-related degradation system [61]. Whether there is a similar metal exclusion mechanism in higher plants will be an interesting topic of research.
DNA methylation is an important epigenetic mechanism for regulating many plant biological processes [32, 35, 36, 38, 62]. Whether the epigenetic mechanism functionally participates in plant response to heavy metal stress is largely unknown. This study precisely characterized the OsZIP1 methylation regions and found that the gene body is the region most demethylated under Cd stress. In euchromatin, genes with methylated regions in their promoters are usually those inducible, tissue-specific developmental-regulator or environmental stress response genes [63,64,65], whereas genes densely methylated in their transcribed regions are those actively expressed [66,67,68,69,70,71]. The gene body methylation has been thought to be conserved and to slowly evolve [66, 67, 71, 72]. However, compared to the promoter-methylated genes, the biological significance of the body-methylated genes relevant to the evolution, functionality and cytosine methylation pattern is poorly understood [63, 71]. Given that the body methylation preferentially occurs in the constitutively expressed genes [66, 67, 73] and is associated with active transcription [63, 70, 71], most of them are proposed to play pivotal roles in biological functions [71]. On the other hand, the body methylation genes tend to be weakly expressed and even be transcriptionally repressed [72, 74]. Our study is partially consistent with the view that the densely methylated context is associated with the low level of OsZIP1 expression and, upon Cd exposure the methylation was reduced. The detailed mechanism for the dynamic methylation and demethylation change under Cd stress remains elusive. By identifying a suit of DNA methylation-defective mutants, only in ossdg714 was found the methylation in the transcribed region of OsZIP1 to be lower and the OsZIP1 transcripts to be higher (Figs. 8, 9), suggesting that the reduced H3K9me2 marks would be involved in the process.
The non-CG methylation was found to be involved in the epigenetic modification of OsZIP1 under Cd stress. This is different from the previous observation from Arabidopsis, where non-CG methylation in gene bodies could be hardly detected [38]. It may be the fact that crop plants with larger genomes have much more transposons close to genes and thereby DNA methylation has more responsibilities for gene regulation in crops than in Arabidopsis [38]. DNA methylation at the CG context is found dominantly in the transcribed regions of many constitutively expressed genes [73]. In plants, maintenance of the CG methylation requires MET1 activity [68]. Mutation of OsMET1 led to a global loss of 75% CmG in rice, causing developmental abnormities [64]. The level of OsZIP1 methylation in osmet1 was increased, whereas the OsZIP1 transcripts were reduced. The mechanism is currently unknown. A recent study has shown that Cd-induced CHG hypomethylation is transgenerationally inherited without entailing an alteration of CG methylation in rice [45]. Gene body methylation is likely established by de novo methylation activity via the RNA-dependent DNA methylation (RdDM) pathway, followed by the maintenance of MET1 [73]. In this case, the occasional antisense transcripts may form the double-stranded RNAs by base-pairing with sense transcripts, leading to generation of small interfering RNAs (siRNAs) which provide a scaffold for DRM2 to methylate the target gene [73]. We examined the OsZIP1 transcripts in osdrm2–2 knockout and osrdr2i-6 knockdown mutant lines but it turned out to have no change in DNA methylation, H3K9me2 marks and transcripts of OsZIP1, indicating that the siRNAs-guided RdDM pathway was not involved in the process.
In animals and human cells, Cd as a non-essential metal is the major cause of many chronic diseases through epigenetic modification such as DNA methylation [75,76,77]. Maintaining DNA methylation depends on the persistent activity of epigenetic modifiers and disturbing the activity of DNA methyltransferases alters the existing DNA methylation patterns [68]. Cd is a selective inhibitor or stimulator of DNA methyltransferases [75, 76]. The Cd-induced genomic demethylation by inhibiting DNMT1 and DNMT3 led to dysregulation of many functional genes [77]. The present study shows that OsMET1, OsCMT3 and OsSDG714 were transcriptionally downregulated under Cd stress. Although growing evidence shows that Cd is able to change DNA methylation in eukaryotes [37, 78, 79], the direction and specificity of Cd influence on the epigenetic modifications remain to be investigated [77, 79]. The Cd-induced OsZIP1 expression can be also attributed to the upregulation of some putative transcription factors in the promoter region, as our results show that the pOsZIP1-GUS fusion transcripts increased under Cd stress. This is consistent with the study using a DNA methyltransferase inhibitor (5-aza-2-deoxycytidine, Aza) [37], in which OsZIP1 methylation was reduced with Cd or Aza or Cd + Aza treatment, and its relative expression was significantly increased with +Cd or Aza + Cd samples. The Cd-induced transcripts of OsZIP1 were much more than those of the Aza-treated sample. In this case, Cd not only triggers the promoters but induces demethylation of OsZIP1 as well.
Conclusions
Based on the results of this study and others, we propose a likely model illustrating the role of OsZIP1 in transport of its substrates in different ways. Under the normal condition, the low and constant expression of OsZIP1 in the ER would be required to keep OsZIP1 at a minimum level that ensures proper allocation of Zn and Cu inside cells (or tissues) and prevents the loss of the essential metals. When excess metals are present in the environment, the expression of OsZIP1 can be upregulated. This work will broaden our understanding of the regulatory mechanism underlying cadmium-induced epigenetic modification and metal resistance in rice when challenged to the metal-contaminated soils.
Methods
Plant culture and treatment
Rice (Oryza sativa L. japonica, c.v. Nipponbare, Kitaake (KT) and Dong Jing (DJ) were used in this study. The T-DNA insertion mutants Osmet1, Oszip1, Osdrm2–2, Ossdg714 and Osros1 with their corresponding wild-types were ordered from Kyung Hee University, Korea [51]. Osrdr2–6 RNAi (RNA interference) was kindly provided by Dr. Yi Jun Qi from Tsinhua University, China. Seeds were surface-sterilized by 5% NaClO, rinsed thoroughly with distilled water and germinated under the conditions of 28 °C and darkness for 2 d. After germination, seedlings were transferred to a polyetheylene container, floating on a 0.5 mM CaCl2 solution, and grown under the condition of a 14/10 light/dark cycle at 28/25 ± 1 °C (day/night) and 200 μmol m− 2 s− 1 light intensity for 5 d. The young plants were then transferred to the half-strength Kimura B solution [27]. Plants were then exposed to Cd for a short-term at different concentrations (0–160 μM) based on the previous report [52]. For long-term experiments, rice plants were exposed to 0 and 1 μM Cd for 30 d based on the previous report [27]. Treatment solutions were renewed other day.
DNA methylation analysis
The sequences of twenty cloned PCR products derived from bisulfite-treated genomic DNA samples were obtained from Feng et al. 2016 [37]. Using these data, the percentages of CG, CHG and CHH methylation were re-analyzed. We then used the McrBC-based DNA methylation assay [80] to validate the results of the bisulfite PCR analysis.
RNA isolation and transcript analyses by PCR
Total RNA was isolated by TRIzol reagent (Invitrogen) and one μg of total RNA (treated with DNase) was used for quantitative reverse transcription polymerase chain reaction (qRT-PCR). One-step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech) and qPCR were performed by special primers (Additional file 10: Table S1). OsACTIN was used as an internal control with the primers of OsACTIN-F (10 μM) and OsACTIN-R (10 μM). The samples were pre-incubated at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 10 s and annealing at 60 °C for 1 min. The reaction was performed in the 7500 real-time PCR System (Applied Biosystems, Foster City, CA, USA) using iTaqTM Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, USA).
OsZIP1 transformation in rice
The coding sequence (CDS) of OsZIP1 (LOC_Os01g74110) was PCR-amplified using the specific primers (Additional file 10: Table S1) [81]. The synthesized OsZIP1 sequence was digested and inserted into the corresponding sites of pCAMBIA1300 driven by CaMV35S as a promoter. The embryonic callus of rice induced by mature embryo was infected by Agrobacterium tumefaciens carrying OsZIP1. For RNAi transformation, a 355 bp cDNA fragment of OsZIP1 was used and PCR-amplified using the primers listed in Table S1. The PCR products were inserted into the corresponding sites of LH-FAD1390RNAi. The constructed vectors were introduced into A. tumefaciens EHA105 by thermal activation. At least twenty 35S::OsZIP1 and twenty RNAi lines (T3 homozygotes) were generated.
Immunoblot detection of OsZIP1 proteins in plants
Preparing antibody and detecting of OsZIP1 protein were performed by the method described previously with some modifications [82]. Briefly, polyclonal antibodies were prepared by immunizing Rabbit with purified OsZIP1 protein. Detection of heat shock protein was used as a control for equal loading. Plant tissues were homogenized in a medium containing 20 mM HEPES (Hepes free acid)-KOH (pH 7.5), 200 mM sorbitol, 1 mM DL-Dithiothreitol, 1 mM Phenylmethanesulfonyl fluoride and a protease inhibitor cocktail. OsZIP1-specific antibody was used to detect OsZIP1 protein.
Metal quantification
Samples were dried at 70 °C in an air-forced oven for 72 h and weighted. The dried samples were digested with nitric acid. The metal concentrations in the samples were quantified using inductively coupled plasma-atomic emission spectrometry (ICP-AES) (Optimal 2100DV, Perkin Elmer Instruments, Waltham MA, USA) [81].
Subcellular localization of OsZIP1
The coding sequence of OsZIP1 was amplified by RT-PCR and inserted into pCAMBIA 1305-GFP vector driven by 35S promoter. The OsZIP1-GFP fusion vector was co-transformed with the ER-marker and PM-marker into rice leaf mesophyll protoplasts and tobacco leaves [50]. The fluorescence signal was visualized using Confocal laser scanning microscopy (Confocal System-UitraView VOX, Perkin Elmer).
Transient expression of pZIP1::GUS in tobacco leaves
The 2.1 kb sequence in the OsZIP1 promoter region was retrieved and constructed into a vector containing GUS (β-glucuronidase) reporter gene. The constructed pZIP1::GUS vector was transformed into tobacco (Nicotiana benthamiana) leaves by Agrobacterium tumefaciens transformation [83]. The transformed cells were exposed to 80 μM Cd for 4 h. qRT-PCR analysis was performed to assess the GUS mRNAs based on the methods described previously [84, 85].
Chromatin immunoprecipitation assay
The ChIP assay was performed by the method described previously [80]. Anti-H3K9me2 (Abcam; ab1220) antibodies (7 mL) were used. The amount of immunoprecipitated OsZIP1 chromatin was determined by qPCR on different regions of OsZIP1. The rice Ubi-10 and Actin1 were used as internal controls. The relative abundance was normalized to the DNA immunoprecipitated by the histone 3-specific antibody.
Yeast complementation assay
The cDNA fragments containing an entire open reading frame of OsZIP1 were amplified. The fragments were cloned into pEGM-T Easy vector (with Kpn1 and EcoR1), correctly introduced into vector pYES2 and incubated at certain condition [49]. The resulting plasmids were transformed into the yeast strain zrc1 (wild-type BY4741). zrc1 complementation by drop-spotting assays was performed on the synthetic defined (SD)-Ura medium, which contained 2% galactose, 0.67% yeast nitrogen base (sigma), 2% agar, and supplemented with 0.025, 3 and 6 mM Zn at pH 5.8.
Statistical analysis
The result was shown as the mean of at least three replicated treatments and each treatment contained at least 9–18 plants. The significant differences between treatments were statistically evaluated by standard deviation (SD) and analysis of variance (ANOVA). The data between differently treated groups were compared statistically by ANOVA followed by the least significant difference (LSD) test if the ANOVA result is significant at p < 0.05. The statistic one-way analysis was performed using Statistical Package for the Social Science (SPSS) 22.0 (https://www.ibm.com/analytics/spss-statistics-software).
Availability of data and materials
All the data supporting our findings is contained within the manuscript. Constructs and seeds are available upon request from ZMY.
Abbreviations
- Aza:
-
azacitidine
- CDS:
-
Coding sequence
- ChIP:
-
Chromatin immunoprecipitation
- CMT3:
-
CHROMOMETHYLASE3
- DJ:
-
Dong Jing
- DME:
-
DEMETER
- DMGs:
-
Differentially methylated genes
- DML2:
-
DEMETER-LIKE2
- DRM2:
-
DOMAINS REARRANGED METHYLTRANSFERASE 2)
- ER:
-
Endoplasmic reticulum
- GFP:
-
Green fluorescent protein
- KT:
-
Kitaake
- LSD:
-
Least significant difference
- MET1:
-
METHYLTRANSFERASE 1
- OXs:
-
Overexpressing transgenic lines
- PM:
-
Plasma membrane
- RNAi:
-
RNA interference
- ROS1:
-
REPRESSOR OF SILENCING
- TMDs:
-
Transmembrane domains
- ZIP:
-
Zn-Regulated transporter, Iron-regulated transporter-like Protein
References
Grotz N, Fox T, Connolly E, Park W, Guerinot ML, Eide D. Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc Natl Acad Sci U S A. 1998;95:7220–4.
Guerinot ML. The ZIP family of metal transporters. BBA-Gene Regul Mech. 2000;1465:190–8.
Krämer U, Clemens S. Functions and homeostasis of zinc, copper, and nickel in plants. Plant Physiol. 2006:214–72.
Stein RJ, Höreth S, de Melo JR, Syllwasschy L, Lee G, Garbin ML, Clemens S, Krämer U. Relationships between soil and leaf mineral composition are element-specific, environment-dependent and geographically structured in the emerging model Arabidopsis halleri. New Phytol. 2017;213:1274–86.
Zhang XD, Meng JG, Zhao KX, Chen X, Yang ZM. Annotation and characterization of cd-responsive metal transporter genes in rapeseed Brassica napus. Biometals. 2018b;31:107–21.
Eide D, Broderius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci U S A. 1996;93:5624–8.
Krämer U, Talke IN, Hanikenne M. Transition metal transport. FEBS Lett. 2007;581:2263–72.
Vatansever R, Ozyigit II, Filiz E. Essential and beneficial trace elements in plants, and their transport in roots: a review. Appl Biochem Biote. 2017;181:464–82.
Korshunova Y.O., Eide D., Clark W.G., Guerinot M.L. & Pakrasi H.B. 1999; The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol. 40:37-44.
Connolly EL, Fett JP, Guerinot ML. Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell. 2002;14:1347–57.
Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. Irt1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002;14:1223–33.
Milner MJ, Seamon J, Craft E, Kochian LV. Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J Exp Bot. 2013;63:695–709.
Lin YF, Liang HM, Yang SY, Boch A, Clemens S, Chen CC, Wu JF, Huang JL, Yeh KC. Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter. New Phytol. 2009;182:392–404.
Vert G, Barberon M, Zelazny E, Séguéla M, Briat JF, Curie C. Arabidopsis IRT2 cooperates with the high-affinity iron uptake system to maintain iron homeostasis in root epidermal cells. Planta. 2009;229:1171–9.
Ramesh SA, Choimes S, Schachtman DP. Over-expression of an Arabidopsis zinc transporter in Hordeum vulgare increases short-term zinc uptake after zinc deprivation and seed zinc content. Plant Mol Biol. 2004;54:373–85.
Schmidt W, Buckhout TJ. A hitchhiker's guide to the Arabidopsis ferrome. Plant Physiol and Bioch. 2011;49:462–70.
Chen WR, Feng Y, Chao YE. Genomic analysis and expression pattern of OsZIP1, OsZIP3, and OsZIP4 in two rice Oryza sativa L.; genotypes with different zinc efficiency. Russ J Plant Physiol. 2008;55:400–9.
Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Nakanishi H, Mori S, Nishizawa NK. Rice plants take up iron as an Fe3+−phytosiderophore and as Fe2+. Plant J. 2006;45:335–46.
Lee S, An G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 2009;32:408–16.
Ramesh SA, Shin R, Eide DJ, Schachtman DP. Differential metal selectivity and gene expression of two zinc transporters from rice. Plant Physiol. 2003;133:126–34.
Bashir K, Ishimaru Y, Nishizawa NK. Molecular mechanisms of zinc uptake and translocation in rice. Plant Soil. 2012;361:189–201.
Ramegowda Y, Venkategowda R, Jagadish P, Govind G, Hanumanthareddy RR, Makarla U, Guligowda SA. Expression of a rice Zn transporter, OsZIP1, increases Zn concentration in tobacco and finger millet transgenic plants. Plant Biotechnol Rep. 2013;7:309–19.
Ishimaru Y, Suzuki M, Kobayashi T, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. OsZIP4, a novel zincregulated zinc transporter in rice. J Exp Bot. 2005;56:3207–14.
Lee S, Jeong H, Kim S, Lee J, Guerinot M, An G. OsZIP5 is a plasma membrane zinc transporter in rice. Plant Mol Biol. 2010a;73:507–17.
Lee S, Kim S, Lee J, Guerinot M, An G. Zinc deficiency inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol Cells. 2010b;29:551–8.
Yang X, Huang J, Jiang Y, Zhang HS. Cloning and functional identification of two members of the ZIP Zrt, Irt-like protein; gene family in rice Oryza sativa L. Mol Biol Rep. 2009;36:281–7.
Sasaki A, Yamaji N, Mitani-Ueno N, Kashino M, Ma JF. A node-localized transporter OsZIP3 is responsible for the preferential distribution of Zn to developing tissues in rice. Plant J. 2015;84:374–84.
Kavitha PG, Kruvilla S, Mathew MK. Functional characterization of a transition metal iron transporter OsZIP6 from rice Oryza sativa L. Plant Physiol and Bioch. 2015;97:165–74.
Clemens S, Aarts MGM, Thomine S, Verbruggen N. Plant science: the key to preventing slow cadmium poisoning. Trends Plant Sci. 2013;18:92–9.
Kim DY, Bovet L, Maeshima M, et al. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007;50:207–18.
Luo JS, Huang J, Zeng DL, et al. A defensin-like protein drives cadmium efflux and allocation in rice. Nat Commun. 2018;9:645.
Grafi G. Epigenetics in plant development and response to stress. BBA Gene Regul Mech. 1809;2011:351–2.
Zhou ZS, Zeng HQ, Liu ZP, Yang ZM. Genome-wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant Cell Environ. 2012b;35:8699.
Chen X, Zhou DX. Rice epigenomics and epigenetics: challenges and opportunities. Curr Opin Plant Biol. 2013;16:164–9.
Mirouze M, Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol. 2011;14:1–8.
Yaish MW. DNA methylation-associated epigenetic changes in stress tolerance of plants. In: Molecular stress physiology of plants Rout GR and Das AB, eds. Springer India. 2013;427–440.
Feng SJ., Liu XS., Tao H, Tan SK., Chu SS., Oono Y, Zhang XD, Chen J and Yang Z.M. Variation of DNA methylation patterns associated with gene expression in rice Oryza sativa; exposed to cadmium. Plant Cell Environ 2016;39:2629–2649.
Zhang H, Lang Z, Zhu JK. Dynaimics and function of DNA methylation in plants. Nat Rev Mol Cell Bio. 2018a;19:489–506.
Chan SW-L, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–60.
Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–20.
Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci U S A. 2007;104:6752–7.
Chandler VL, Stam M. Chromatin conversations: mechanisms and implications of paramutation. Nat Rev Genet. 2004;5:532–44.
Du JM, Xuehua Z, Yana VB, Hume S, Suhua F, Elena C, Ajay AV, Jolyon T, Hang GC, Andy T, Jonathan H, James AW, Sriharsa P, Dinshaw JP, Steven EJ. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151:167–80.
Greco M, Chiappetta A, Bruno L, Bitonti MB. In Posidonia oceanic cadmium induces changes in DNA methylation and chromatin patterning. J Exp Bot. 2012;63:695–709.
Ou X, Zhang Y, Xu C, Lin X, Zang Q, Zhuang T, Jiang L, Wettstein D, Bao L. Transgenerational inheritance of modified DNA methlylation patterns and enhanced tolerance induced by heavy metal stress in rice Oryza sativa L. PLoS One. 2012;7:e41143.
Ogo Y, Kakei Y, Itai RN, Kobayashi T, Nakanishi H, Takahashi H, Nakazono M, Nishizawa NK. Spatial transcriptomes of iron-deficient and cadmium-stressed rice. New Phytol. 2014;201:781–94.
Zhang JJ, Gao S, Xu JY, Lu YC, Lu FF, Ma LY, Su XN, Yang H. Degrading and Phytoextracting atrazine residues in Rice (Oryza sativa) and growth media intensified by a phase II mechanism modulator. Environ Sci Technol. 2017;51:11258–68.
Mitra S, Pramanik K, Sarkar A, Kumar P, Ghosh P, Maiti TK. Bioaccumulation of cadmium by Enterobacter sp. and enhancement of rice seedling growth undercadmium stress. Ecotox Environ Safe. 2018;156:183–96.
Meng JG, Zhang XD, Tan SK, Zhao KX, Yang ZM. Genome-wide identification of cd-responsive NRAMP transporter genes and analyzing expression of NRAMP1 mediated by miR167 in Brassica napus. Biometals. 2017;30:917–31.
Ma G, Li J, Li J, Li Y, Gu D, Chen C, Cui J, Chen X, Zhang W. OsMTP11, a trans-Golgi network localized transporter, is involved in manganese tolerance in rice. Plant Sci. 2018;274:59–69.
Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, Kim J, Shin M, Jung M, An G. Generation of flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–32.
Lee S, Kim YY, Lee Y, An G. Rice P1B-type heavy metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol. 2007;145:831–42.
Ding Y, Wang X, Su L, Zhai JX, Cao SY, Zhang DF, Liu CY, Bi YP, Qian Q, Cheng ZK, Chu CC, Cao XF. SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell. 2007b;19:9–22.
Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, HenikoV S, Jacobsen SE. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–80.
Ebbs ML, Bartee L, Bender J. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol Biol Cell. 2005;25:10507–15.
Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, Shao Y, Giovannoni JJ. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol. 2013;31:154–9.
Papoyan A, Kochian LV. Identification of Thlaspi caerulescens genes that may be involved in heavy metal hyperaccumulation and tolerance. Characterization of a novel heavy metal transporting ATPase. Plant Physiol. 2004;136:3814–23.
Mills RF, Francini A, Ferreira da Rocha PS, Baccarini PJ, Aylett M, Krijger GC and Williams LE. The plant P1Btype ATPase AtHMA4 transports Zn and Cd and plays a role in detoxification of transition metals supplied at elevated levels. Febs Letters. 2005;579:783–791.
Song WY, Choi KS, Kim DY, Geisler M, Park J, Vincenzetti V, Schellenberg M, Kim SH, Lim YP, Noh EW, Lee Y, Martinoia E. Arabidopsis PCR2 is a zinc exporter involved in both zinc extursion and long-distance zinc transport. Plant Cell. 2010;22:2237–52.
Li Y, Iqbal M, Zhang Q, Spelt C, Bliek M, Hakwoort HWJ, Quattrocchio FM, Koew R, Schat H. Two Silene vulgaris copper transporters residing in different cellular compartments confer copper hypertolerance by distinct mechanisms when expressed in Arabidopsis thaliana. New Phytol. 2017;215:1102–14.
Adle DJ, Wei W, Smith N, Bies JJ, Lee J. Cadmium-mediated rescue from ER-associated degradation induces expression of its exporter. Proceedings of the National Academy of Sciences, USA. 2009;106:10189–94.
Boyko A, Kovalchuk I. Epigenetic control of plant stress response. Environ Mol Mutagen. 2008;49:61–72.
To TK, Saze H, Kakutani T. DNA methylation within transcribed regions. Plant Physiol. 2015;168:1219–25.
Wang X, Zhang Z, Fu T, Hu L, Xu C, Gong L, Wendel JF, Lui B. Gene-body CG methylation and divergent expression of duplicate genes in rice. Sci Rep. 2017;7:2675.
Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22.
Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecke JR. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–201.
Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–9.
Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–36.
Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 2010;107:8689–94.
Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, Zilberman D. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci U S A. 2010;107:18729–34.
Takuno S, Gaut BS. Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol Biol and Evol. 2012;29:219–27.
Li X, Wang X, He K, Ma Y, Su N, He H, Stolc V, Tongprasit W, Jin W, Jiang J, Terzaghi W, Li S, Deng XW. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell. 2008;20:259–76.
Bewick AJ, Jib L, Niederhuth CE, Willing EM, Hofmeister BT, Shi X, Wang L, Lu Z, Rohr NA, Hartwig B, Kiefer C, Deal RB, Schmutz J, Grimwood J, Stroud H, Jacobseng SE, Schneeberger K, Zhang X, Schmitz RJ. On the origin and evolutionary consequences of gene body DNA methylation. Proc Natl Acad Sci U S A. 2016;113:9111–6.
He G, Zhu X, Elling AA, Chen L, Wang X, Guo L, Liang M, He H, Zhang H, Chen F, Qi YJ, Chen RS, Deng XW. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell. 2010;22:17–33.
Jiang G, Xu L, Song S, Zhu CC, Wu Q, Zhang L, Wu L. Effects of long-term low-dose cadmium exposure on genomic DNA methylation in human embryo lung fibroblast cells. Toxicology. 2008;244:49–55.
Doi T, Puri P, McCann A, Bannigan J, Thompson J. Epigenetic effect of cadmium on global De novo DNA hypomethylation in the cadmium-induced ventral body wall defect VBWD; in the chick model. Toxicol Sci. 2011;120:475–80.
Sanders AP, Smeester L, Rojas D, DeBussycher T, Wu MC, Wright FA, Zhou YH, Laine JE, Rager JE, Swamy GK, Ashley-Koch A, Miranda ML, Fry RC. Cadmium exposure and the epigenome: Exposureassociated patterns of DNA methylation in leukocytes from mother-baby pairs. Epigenetics. 2014;9:212–21.
Cheng C, Yoshiaki T, Akio M, Tasuku I, Muneo Y, Hiroaki S, Eigo F, Hirohiko H. Loss of function mutations in the rice chromomethylase OsCMT3a cause a burst of transposition. Plant J. 2015;83:1069–81.
Wang B, Li Y, Shao C, Tan Y, Cai L. Cadmium and its epigenetic effects. Curr Med Chem. 2012;19:2611–20.
Mao H, Wang H, Liu S, Li Z, Yang X, Yan J, Li J, Tran LS, Qin F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat Commun. 2015;6:8326.
Shen Q, Jiang M, Li H, Che LL, Yang ZM. Expression of a Brassica napus heme oxygenase confers plant tolerance to mercury toxicity. Plant Cell Environ. 2011;34:752–63.
Li H, Song JB, Zhao WT, Yang ZM. AtHO1 is involved in iron homeostasis in a NO-dependent manner. Plant Cell Physiol. 2013;54:1105–17.
Song JB, Huang SQ, Dalmay T, Yang ZM. Regulation of LEAF morphology by microRNA394 and its target LEAF CURLING RESPONSIVENESS. Plant Cell Physiol. 2012;53:1283–94.
Xu R, Wang Y, Zheng H, Lu W, Wu C, Huang J, Yan K, Yang G, Zheng C. Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J Exp Bot. 2015;66:5997–6008.
Guo C, Xu Y, Shi M, Lai Y, Wu X, Wang H, Zhu Z, Poethig RS, Wu G. Repression of miR156 by miR159 regulates the timing of the juvenile-to-adult transition in Arabidopsis. Plant Cell. 2017;29:1293–304.
Acknowledgments
Not applicable.
Funding
This study was funded by the National Natural Science Foundation of China (21777072). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Z.M.Y. conceived, designed and drafted the manuscript; X.S.L, S.J.F, B.Q.Z, M.Q.W, and H.W.C. carried out the molecular and physiological experiments; J.K.R. modified the manuscript; and CX provided technical support on yeast assay. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1. Basic information of OsZIP1. (DOC 4190 kb)
Additional file 2:
Figure S2. Analysis of OsZIP1 transcripts under normal growth condition. (DOC 794 kb)
Additional file 3:
Figure S3. Transient expression of GUS reporter genes fused to OsZIP1 promoter under –Cd and + Cd exposure. (DOC 61 kb)
Additional file 4:
Figure S4. Identification of oszip1 mutant, RNAi and OX lines. (DOC 1477 kb)
Additional file 5:
Figure S5. Zn transport activity and detoxification response assay of of OsZIP1-transgenic yeast (Saccharomyces cerevisiae). (DOC 1815 kb)
Additional file 6:
Figure S6. Identification of DNA demethylation of OsZIP1 in rice exposed to the low level of Cd stress. (DOC 84 kb)
Additional file 7:
Figure S7. Transcriptional expression of OsZIP1 under the low level of Cd stress. (DOC 118 kb)
Additional file 8:
Figure S8. Effects of Cd on the transcripts of DNA methylation modifier genes. (DOC 652 kb)
Additional file 9:
Figure S9. Effects of Zn, Cu, Mn and Fe excess on the transcription of DNA methylation and demethylation modifier genes. (DOC 3871 kb)
Additional file 10:
Table S1. Primer sequences used for qRT-PCR in this study. (DOC 42 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, X.S., Feng, S.J., Zhang, B.Q. et al. OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol 19, 283 (2019). https://doi.org/10.1186/s12870-019-1899-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-019-1899-3