Abstract
Endophytic fungi, particularly from higher plants have proven to be a rich source of antimicrobial secondary metabolites. The purpose of this study is to examine the antimicrobial potential of three endophytic fungi Aspergillus sp. SA1, Aspergillus sp. SA2, and Aspergillus sp. SA3, cultivated from Nigella sativa seeds against Staphylococcus aureus (ATCC 9144), Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853), Klebsiella pneumoniae (ATCC 13883), MRSA (ATCC 33591), and human pathogen Candida albicans (ATCC 10231). Furthermore, the most active cultivated endophytic fungi were molecularly identified via internal transcribed spacer (ITS) sequencing. HR-ESIMS guided approach has been used successfully in chemical profiling of 26 known bioactive secondary metabolites (1–26), which belongs to different classes of natural compounds such as polyketides, benzenoids, quinones, alcohols, phenols or alkaloids. Finally, in-silico interactions within active site of fungal Cyp51 and bacterial DNA gyrase revealed possibility of being a hit-target for such metabolites as antimicrobials.
Similar content being viewed by others
Introduction
Endophytes are fungi, bacteria, and yeasts that live in healthy plant tissues for at least a stage of their life cycles without showing any symptoms of disease or producing a harmful impact on their host plants [1]. Endophytes can survive in plant tissues for a long time and protect the plant from biotic and abiotic stress [2]. Endophytic fungal bioactive metabolites are useful as new drugs due to their diversity of biological activities. A variety of bioactive metabolites in endophytes, including antiviral, anticancer, anti-diabetic, and antibacterial compounds, have been reported [3]. Infectious diseases are a long-lasting and a serious public health problem that affects people all over the world [4]. Today, numerous bacteria, including Staphylococci, Enterococci, Gonococci, Streptococci, and others, demonstrate multidrug resistance [5]. There is an urgent need to look for novel compounds for drug development, industry, and agricultural applications [6,7,8,9]. Therefore, finding new antibiotics is crucial to combat these resistant bacteria [10]. Natural compounds from endophytic fungi are potential candidates for new antibiotics against pathogenic bacteria [11]. Therefore, in recent years, more attention has been given to the endophytic fungi of medicinal plants, which represent a rich source of new and useful natural compounds of interest to the pharmaceutical and agricultural industries and a potential source for the discovery of novel microorganisms [12,13,14]. Nigella Sativa seeds (Ranunculaceae), commonly known as black seed or black cumin, traditionally used for treatments of various illness affected lungs, kidney, GIT, circulatory and immune systems [15, 16]. The powder of N. sativa seeds has antibacterial activity comparable with other antibiotics such as, tetracycline, ampicillin, levofloxacin, gentamycin and streptomycin [17, 18]. In this work, three endophytic fungi have been isolated and identified from Nigella sativa (F. Ranunculaceae) seeds growing in Egypt. The isolated fungi were identified morphologically and microscopically up to species to be Penicillium sp. SA1, Aspergillus sp. SA2, and Aspergillus sp. SA3. The isolated fungal strains were cultivated using the OSMAC approach (One Strain many Compounds) method “is a simple and effective approach for activating metabolic pathways and has been successfully applied”, this terms including the alteration in culture conditions resulting in diversity in the microorganisms production and subsequently variation in secondary metabolites. Moreover, the chemical profiles were explored by LC‒MS-based metabolomics. Furthermore, we have investigated these three strains against five bacterial strains (Staphylococcus aureus, Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae, MRSA) and Candida albicans to evaluate their antimicrobial activity. In-silico studies were performed within crystal structure of two protein, fungal sterol 14α-demethylase and bacterial DNA gyrase.
Experimental
Plant material
Fresh plant materials (seeds) were collected from the Agricultural research center in Malawi, EL- Minia, Egypt. Prof. Nasser Barakat (Department of Botany and Microbiology, Faculty of Science, Minia University) identified the investigated plant.
Isolation and identification of endophytic fungi
Isolation of endophytic fungi from seeds of Nigella sativa was carried out using the protocol by Strobel et al. with slight modifications [19].
Nigella sativa seeds were collected, and subjected to surface sterilization first by distilled water followed 70% EtOH for 1 min, then distilled sterilized water, 1.0% sodium hypochlorite (NaOCl) (v/v) for 1 min, and finally with sterilized water. Nigella sativa seeds were crushed into small particles using sterilized pestle and mortar and homogenized with sterilized water. Each 100 µL of the suspension was spread on the surface of potato dextrose agar medium plate (PDA, 200 g potato, 20 g glucose, and 15 g agar in 1 L distilled water, PH 6.0) supplemented with gentamycin (100 mg/ L) and amoxicillin (100 mg/ L) to suppress bacterial contamination. The plates were then incubated for 21 days in the incubator, and the plates were examined daily during the incubation period and the pure strains of fungi were detected and isolated [20]. The isolated fungal colonies with distinct morphological appearance were isolated, purified and stored in glycerol stock at – 70 ºC for long storage and on a new agar slants at 4 °C for further studies [13]. Endophytes were deposited in the Microbial Repository of Botany and Microbiology (MRBM) Department, Faculty of Science, Minia University, Minia, Egypt, where they were stored at 4 °C. Based on morphological characteristics, out of 20 isolated colonies, three isolates with distinct morphology were selected for further work and named SA1, SA2 and SA3 respectively.
Molecular identification and phylogenetic analysis
Molecular identification of the most active fungal isolated strains recovered from Nigella sativa seeds was achieved by sequence analysis of the fungal internal transcribed spacer (ITS) region including ITS1, the 5.8 S rRNA gene, and ITS2 sequences according to [21]. In brief, DNA was extracted from fungal biomass using the MasterPure Yeast DNA extraction kit (epientre, Madison, Wisconsin). DNA amplification was performed with universal fungal primers NS1 [22] (x) and ITS-4 [23]. Sanger sequencing with primer ITS-4 was performed by LGC Genomics (Berlin, Germany). Manual sequence corrections and phylogenetic analyses were performed with MEGA11 version 11.0.1 [24]. Next related taxa were determined by BLASTn analysis against the ITS RefSeq Targeted Loci project database (BioProject PRJNA177353; update 2023/03/22) provided in the BLASTn tool of the NCBI (https://www.ncbi.nlm.nih.gov/). ITS sequences of next related reference strains were imported into MEGA11 and aligned with ClustalW [25]. All nucleotide positions were considered with uniform rates. The phylogenetic tree was constructed with the maximum likelihood method based on the Kimura 2-parameter model [26]. The phylogeny was tested by the bootstrap method (100 replications). A total of 39 sequences and 620 nucleotide positions were in the final dataset. Two Penicillium spp. were included as outgroup sequences. The ITS sequences of Aspergillus sp. SA1, Aspergillus sp. SA2 and Aspergillus sp. SA3 weredeposit in Genbank/EMBL/DDBJ with Accession numbers OQ652078 to OQ652080.
The three fungal strains SA1, SA2 and SA3 were compared by genomic fingerprinting using rep- and RAPD-PCR techniques to determine if the strains are clonal or genetically identical. Analysis was performed according to Glaeser et al. [27] using primer (GTG)5 and two fungal specific primers, GACA (5´- GACAGACAGACAGACA-3´) and NS3 (5´- GCAAGTCTGGTGCCAGCAGCC-3´) [28]. PCRs were performed in a total volume of 15 µl with 1 x DreamTaq buffer, 0.2 mM of each dNTP, 1 µM primer (one per reaction), 0.2 mg/ml bovine serum albumin, 0.02 U/µL DreamDNA polymerase (all chemicals expect primers, Thermo Fisher Scientific, ) and 3 µl cell lysate of fungal biomass. Following cycling conditions were used for GTG5 PCR: 95 °C, 3 min, followed by 30 cycles 94 °C for 30 s, 53 °C for 60 s, 70 °C for 8 min, and finally 70 °C for 16 min and fungal fingerprintings: 95 °C for 3 min, followed by 40 cycles 95 °C for 30 s, 36.5 °C (GACA) and 48.5 °C (NS3) for 30 s, 72 °C for 1 min, and finally 72 °C for 6 min. PCR products were resolved by agarose gel electrophoresis (1.5% agarose, 3.33 V/cm, 150 min) and stained with ethidium bromide.
Fermentation and extraction
The three fungal endophytes; Aspergillus sp. SA1, Aspergillus sp. SA2, and Aspergillus sp. SA3 (depending on molecular identification) were fermented using the solid approach [29]. In the solid treatment, 150 µL of each strain were inoculated and streaked on ten solid plates (petri dishes : 15 cm )of the media: PDA (200 g potato, 20 g glucose, and 15 g agar in 1 L distilled water). The agar plates were cut into pieces and extracted with 300 mL of ethyl acetate (3 times). Ethyl acetate was then evaporated using rotary evaporator (Heidolph ® 125, 35 °C, 154 rpm) and the yielded dry extract was kept in refrigerator for further analysis.
LC-MS metabolomic analysis
Metabolomic profiling was performed on the crude fungal extracts on an Acquity Ultra Performance Liquid Chromatography system coupled to a Synapt G2 HDMS quadrupole time-of-flight hybrid mass spectrometer (Waters, Milford, CT, USA). Chromatographic separation was carried out on a BEH C18 column (2.1 × 100 mm, 1.7 μm particle size; Waters, Milford, CT, USA) with a guard column (2.1 × 5 mm, 1.7 μm particle size) and a linear binary solvent gradient of 0–100% eluent B over 6 min at a flow rate of 0.3 mL·min−1, using 0.1% formic acid in water (v/v) as solvent A and acetonitrile as solvent B. The injection volume was 2.0 µL and the column temperature was 40 ◦C. Ms Converter software was used to convert the raw data into divided positive and negative ionization files. Obtained files were then subjected to the data mining software MZmine 3 (Okinawa Institute of Science and Technology Graduate University, Japan) for deconvolution, peak picking, alignment, deisotoping, and formula prediction. The databases used for the identification of compounds were MarinLit: http://pubs.rsc.org/marinlit/, and Dictionary of Natural Products(DNP)2018: http://dnp.chemnetbase.com/faces/chemical/ChemicalSearch.xhtml [30, 31].
Antimicrobial activity
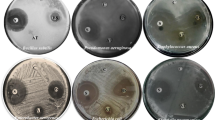
The antimicrobial activity of different extracts of Aspergillus sp. SA1, Aspergillus sp. SA2, and Aspergillus sp. SA3 were tested against five pathogenic bacteria Staphylococcus aureus (ATCC 9144), Escherichia coli (ATCC 25,922), Pseudomonas aeruginosa (ATCC 27,853), Klebsiella pneumonia (ATCC 13,883), MRSA (ATCC 33,591), and human pathogen Candida albicans (ATCC 10,231) using Microtiter bioassay [32]. The dried extracts from each fungus were dissolved in dimethyl sulfoxide (DMSO) to a concentration of 100 mg /mL. Overnight culture in Mueller Hinton broth (Sigma Aldrich, SA) at 37 °C in shaker incubator from each bacterial and the fungal strain was adjusted to 1 McFarland standard which is equivalent to 3.0 × 108 CFU/ mL. First, 100 µL of plain and sterile Mueller Hinton broth was dispensed to all wells of a microtiter plate. 100 µL of the tested extract was added to the first well and serial dilution by 100 µL (1:1) was done from the first well to the 8th well. 100 µL from the last well was discarded. Thus, the concentration of each tested extract was as follow; 50, 25, 12.5, 6.25, 3.125, 1.5625, 0.78125 and finally 0.3906 mg / mL. 5 µL of the bacterial (or fungal) suspension were added to all wells of a microtiter plate except for raw for sterility control. Ciprofloxacin antibiotic was used as positive control for all bacterial strains [33] with the same concentrations as the tested extracts. Fluconazole antifungal agent was used as positive control for the fungal strain [34, 35] with the same concentrations as the tested extracts. Raw contain only the plain sterile medium and the microorganism was used as negative control. The microtiter plates were incubated at 37 ºC for 24 h. After incubation, the plate was measured by ELISA reader at a wavelength of 570 nm using an ELISA plate reader.
In-Silico molecular docking studies
Molecular Orbital Environment (MOE®) software package was used for running molecular docking simulations. The dereplicated identified compounds 1–26 (SA1:7), (SA2:10), (SA3:9) were drawn using ChemDraw®Ultra (ver. 8, 2013) and their energy were minimized using MMFF94x Forcefield energy minimization capability of MOE software with a gradient RMS of 0.0001 kcal/mol and prepared ligands were saved as Microsoft Access Data Base (mdb) file. Crystal structures of two target proteins: Fungal sterol 14a-demthyalse (CYP51; PDB ID: 1EA1; Cytochrome P450 14 alpha-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with fluconazole) and bacterial DNA gyrase (topoisomerase II; PDB ID: 5BTC; Crystal structure of a topoisomerase II complex with ciprofloxacin) were downloaded from RCSB Protein Data Bank website (https://www.rcsb.org/ ). Both proteins were prepared in MOE program using protein Quick prep capability to protonate their structures and removal of water molecules. Validation of prepared proteins was performed by docking their co-crystallized ligands (Fluconazole for 1EA1 and Ciprofloxacin for 5BTC) and their docking score (S; Kcal/mol) and RMSD (Å) were in-agreement to reported ones (Informatics in Medicine Unlocked 26 (2021) 100,748 and www.pnas.org/cgi/doi/10.1073/pnas.1525047113. ). The prepared ligands were docked into active site using MOE Alpha triangle placement method and refinements were done by Forcefield, scored using affinity δG (S; Kcal/mol) scoring system.
Results and discussion
Until now, various reports have highlighted the isolation, identification, and biological properties of different phytoconstituents produced by medicinal plants, although few studies have referred to those provided by their endophytic fungi. Three different fungal strain coded as SA1, SA2, and SA3 were isolated and identified morphologically in additionally to phylogenetic analysis from sterilized seeds of Nigella sativa plant their names (Aspergillus sp. SA1, Aspergillus sp. SA2, and Aspergillus sp. SA3). Strain SA1 shared with 99.79% highest ITS sequence similarity with Aspergillus fasciculatus CBS 110.55 (NR_138285.1) (Aspergillus section Flavi). Strains SA2 and SA3 (identical ITS sequences) shared with 99.31% highest ITS sequence similarity to Aspergillus awamori ATCC 16,877 (NR_077143.1) and Aspergillus foetidus CBS 121.28 (NR_163668.1) (Aspergillus section Niger). The phylogenetic placement is shown in Fig. 1. Beside identical ITS sequences strains SA2 and SA3 also shared identical genomic fingerprint patterns (Fig. 2) which indicated genetic clonality or at least a close genetic relationship of the two strains. Strain SA1 showed fitting the phylogenetic differences based on the ITS sequences different genomic fingerprint patterns compared to strains SA2 and SA3.
Phylogenetic placement of the three studied fungi Aspergillus sp. SA1, Aspergillus sp. SA2 and Aspergillus sp. SA3 based on the fungal ITS sequence region ITS1–5.8 S – ITS2. The tree was constructed with the maximum-likelihood method in MEGA11. In total 39 sequences and 620 nucleotide positions were considered by the analysis. Numbers ad nodes represent bootstrap values of 70% and above. Two Penicillium sp. strains were used as outgroup. Bar: 0.02 substitutions per nucleotide position
Comparative genomic fingerprint analysis of strains Aspergillus sp. SA1, Aspergillus sp. SA2 and Aspergillus sp. SA3. Analyses were performed with primers (GTG)5, GACA and NS3. Depicted are fingerprint patterns resolved on a 1.4% agarose gel after staining with ethidium bromide. NTC: No template control; M: 100 bp DNA marker
The three strains were fermented using different media to investigate the chemical diversity of Aspergillus sp. SA1, Aspergillus sp. SA2, and Aspergillus sp. SA3.
The crude culture extracts were subjected to high-resolution mass spectrometry (HR-MS) analysis. Different metabolites produced by the three strains under different culture conditions were explored by LC‒MS-based metabolomics analysis as shown in the total ion chromatograms Fig. 3. Metabolic profiling was carried out for all extracts of Aspergillus sp. SA1, Aspergillus sp. SA2, and Aspergillus sp. SA3 results in annotation of 26 compounds (1–7), (8–17) and (18–26) respectively, as shown in Table 1; Fig. 4. Employing macros and algorithms that coupled MZmine with online and internal databases, such as MarinLit, DNP, and METLIN, as well as the comparison with the given literature data, the compounds were annotated. Many other chemical classes of metabolites were dereplicated such as polyketides, benzenoids, quinones, alcohols, phenols, and alkaloids. Identified compounds from Aspergillus sp. SA1 were shown in Table 1; Fig. 4 including Dibutyl phthalate (2) that was dereplicated from the mass ion peak at m/z 278.150 in agreement with the molecular formula C16H22O4, was previously isolated for the first time from this plant source have significant activity against Klebsiella pneumonia and Pseudomonas aeruginosa. The in vitro antibacterial evaluation of dibutyl phthalate forms a primary platform for further phytochemical and pharmacological studies [36]. Moreover, the mass ion peak at m/z 330.239, consistent with the molecular formula C18H34O5, was also identified as Penicitide B (3); Penicitide B as antifungal agent, [37, 38]. In the same vein, the mass ion peak at m/z 340.239, consistent with the molecular formula C23H32O2 was annotated as Plastoquinone-3 (4); was detected in spinach chloroplasts [39]. Similarly, Janthinolide A (6), which showed the molecular formula C23H32N2O7 and was annotated from the mass ion peak at m/z 448.219, these compound isolated from the coral endophytic Penicillium janthinellum [40], exhibited antifungal properties [41]. Another compound with the molecular formula C24H30O6 was identified as Austinoneol A (7) based on the observed mass ion peak at m/z 414.203, was previously isolated from Penicillium sp. [42]. On the other hand, metabolic profiling of the crude extracts of Aspergillus sp. (SA2) revealed a variety of metabolites as shown in (Table 1; Fig. 3), of which the mass ion peak at m/z 178.062 in consonance with the molecular formula C10H10O3 was also annotated as R-Mellein (8). This compound belongs to the family of pentaketides and have a widely distributed dihydroisocoumarin derivative in fungi [43], was first isolated in 1933 from Aspergillus melleus and exhibited a strong fungicidal agent [44]. Moreover, the mass ion peak at m/z 188.104, consistent with the molecular formula C9H16O4, was also identified as Aspinonene (9). This compound belongs to pentaketides previously purified from Aspergillus ochraceus (DSM-7428) in 1997 pentaketides [45]. When screened for activity towards MRSA aspinonene displayed mild inhibitory properties. Another new alkaloid from the culture broth of Aspergillus ochraceus with the molecular formula C17H13N3O3 was characterized as circumdatin G (11) based on the observed mass ion peak at m/z 307.095. This compound showed biological activity it inhibit the final stage of polyprotein processing during hepatitis C virus replication [46]. Moreover, an alkaloid with the molecular formula C17H13N3O4 was characterized as 2-hydroxycircumdatin C (12) based on the observed mass ion peak at m/z 323.090. This compound was also formerly identified from a natural source for the first time [47]. This compound isolated from A. ochraceus and showed notable antioxidant activity. The mass ion peak at m/z 445.199 in consonance with the molecular formula C26H27N3O4 was also annotated as avrainvillamide (13), was first isolated in 2000 from the marine fungus Aspergillus sp. [48], showed anti-insecticidal and antibacterial properties. This compound was also have antibiotic activity that inhibits the growth of multi-drug-resistant Staphylococcus aureus, Streptococcus pyogenes, and Enterococcus faecalis [49]. Additionally, notoamide B (14), dereplicated from the mass ion peak at m/z 447.215 and the corresponding molecular formula C26H29N3O4, also was previously identified as a metabolite of Aspergillus sp., showing stronger antibacterial activity against E. coli and P. aeruginosa also have potent insecticidal activities against H. armigera [50]. Furthermore, The mass ion peak at m/z 495.237 in consonance with the molecular formula C27H33N3O6 was also annotated as Spirotryprostatin C (16), was isolated from Aspergillus fumigatus [51]. This compound showed significant antibacterial activity against certain microbial pathogens, in which the highest antibacterial activity against Escherichia coli, Acinetobacter baumannii, Pseudomonas aeruginosa, Klebsiella pneumoniae, methicillin-resistant Staphylococcus aureus (MRSA), and Enterococcus faecalis [52]. The mass ion peak at m/z 514.258 in consonance with the molecular formula C30H34N4O4 was also annotated as Novofumigatamide (17), which was formerly identified from Aspergillus novofumigatus CBS11520 as the new Aspergillus sp. [53]. In the same vein, metabolic profiling of the crude extracts of Aspergillus sp.(SA3) revealed a moderate number of metabolites (Table 1; Fig. 3), peak at m/z 270.052, consistent with the molecular formula C15H10O5 was annotated as Emodin (19); This compound is an active ingredient of herbal medicine, Emodin (1,3,8-trihydroxy-6-methylanthraquinone) also has been reported to exhibit anti-inflammatory properties by reduction of cytokine production in human T-lymphocytes and endothelial cells [54]. This anthraquinone have antibacterial effects against Escherichia coli were proposed to be mediated through inhibition of respiration-driven solute transport in membrane [55]. Likewise, the mass ion peak at m/z 300.062 was annotated as Sydowinin A (20); a dimeric isocoumarin with the molecular formula C16H12O6, Sydowinin A is commonly produced by from Aspergillus sydowi [56]. Previously reported analogues showed antibacterial activities against common bacteria Bacillus subtilis and Escherichia coli [57] as well as against the Gram-positive bacterial strains Mycobacterium smegmatis ATCC 607 and Staphylococcus aureus ATCC 25,923, Gram-negative Pseudomonas aeruginosa ATCC 9027, and fungus Candida albicans ATCC10231 [58]. The mass ion peak at m/z 340.239 in consonance with the molecular formula C23H32O2 was also annotated as GERI-BP002-A (22), which was isolated from culture broth of Aspergillus fumigatus F93 by acetone extraction. These novel compound have inhibited ACAT activity by 50% at the concentration of 50 µM in an enzyme assay system using rat liver, GERI-BP002-A is an attractive target for treatment of hypercholesterolemia and atherosclerosis [59]. Additionally, (-)-Averantin (23), dereplicated from the mass ion peak at m/z 372.120 and the corresponding molecular formula C20H20O7, was also formerly isolated from Aspergillus versicolor, a sponge-derived fungus showing antibacterial activity against several clinical isolates of Gram + strains [60] and exhibited antifungal activities [61]. Moreover, the mass ion peak at m/z 377.137, consistent with the molecular formula C21H19N3O4, was also identified as Circumdatin J (24); benzodiazepine alkaloids isolated from the fungal strain Aspergillus ostianus IMBC-NMTP03, which also revealed wide antimicrobial potential against Enterococcus faecalis and Candida albicans [62]. The mass ion peak at m/z 393.132 in consonance with the molecular formula C21H19N3O5 was also annotated as circumdatin D (25), which was originally reported from Aspergillus ochraceus [62], this compound significantly inhibit Enterococcus faecalis growth and this compound showed antimicrobial effects against the yeast Candida albicans [62]. Finally, the mass ion peak at m/z 435.216, consistent with the molecular formula C25H29N3O4, was also identified as Notoamide L (26); this compound was isolated from Aspergillus species [63]. This provoked us to study the antibacterial potential of the total ethyl acetate extracts of the three endophytic fungi isolated from Nigella sativa seeds. Penicillium sp. SA1, Aspergillus sp. SA2, and Aspergillus sp. SA3 against five different pathogenic bacteria Staphylococcus aureus (ATCC 9144), Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27,853), Klebsiella pneumoniae (ATCC 13883), MRSA (ATCC 33591), and human pathogen Candida albicans (ATCC 10,231) using the quantitative antibacterial assay by IC50. Overall, the tested samples revealed varying in vitro growth inhibitory potencies against Staphylococcus aureus, Pseudomonas aeruginosa, Klebsiella pneumoniae, MRSA pathogenic bacteria, showing IC50 values in the range of 0.7‒27.3 µg/mL. As shown in Table 2, the extract of Penicillium sp. SA1 cultured on PDA media exhibited the highest antibacterial activity against Pseudomonas aeruginosa and Staphylococcus aureus, with IC50 value of 0.77 and 7.18 µg/mL, respectively, although it has weak activity against Klebsiella pneumoniae, Escherichia coli. Likewise, the extract of Aspergillus sp. SA2 cultured on the PDA medium, showed the highest activities against Pseudomonas aeruginosa and Klebsiella pneumonia, with IC50 values of 2.5, 27.3 µg/mL but has weak activity against Staphylococcus aureus, Escherichia coli and MRSA. In contrast, the extract of Aspergillus sp. SA3 cultured on PDA medium show highest activity against MRSA, Klebsiella pneumonia, Staphylococcus aureus and Pseudomonas aeruginosa with IC50 values of 2.5, 2.7, 4.1 and 5.4 µg/mL respectively, while it has weak activity with Escherichia coli. However, all the above-mentioned extracts showed higher antimicrobial and antifungal potential as compared to the positive control, Ciprofloxacin that revealed IC50 values of 1.2, 11.9, 16.3, 19.6 and 25.7 µg/mL in the case of Pseudomonas aeruginosa, Staphylococcus aureus, Escherichia coli, Klebsiella pneumonia and MRSA, respectively (Table 2).
Based on promising results obtained with various fungal extracts on six bacterial and fungal species, we decided to explore their cellular targets virtually using Molecular Orbital Environment (MOE®) Software. Molecular docking simulations were performed within active sites of 2 crystal structures obtained from Protein Data Bank (RSCB PDB): Fungal sterol 14α-demethylase (CYP51: a member of Cytochrome P450 superfamily) and bacterial DNA gyrase (bacterial topoisomerase II). Interestingly, most of three extracts’ compounds showed better docking score (S; Kcal/mol) than co-crystallized ligand; Fluconazole (as listed in Table 3).
In addition, these compounds exhibited close contact interactions with amino acid residues lining active site of CYP51, which indicated by their low RMSD values. Compounds 7, 14 and 25 were further explored for their in-site interactions, Fig. 5. The three compounds showed good overlapping with Fluconazole within CYP51 active site, also they showed number of H-bond and H-Bi interactions with various amino acid residues lining active site. Taking the average of docking score (S) for all compounds of each extract, we found that: SA1 (Compounds 1–7) showed the highest docking score with lowest RMSD value over other two extracts, which indeed matches its biological effect on Candida albicans isolates and this is a strong point for further investigation of such metabolites. On the other hand, working on the molecular docking simulations within bacterial DNA gyrase active site showed the following interesting observations: generally, molecules of the three extracts showed weak to moderate docking score (S) and RMSD (Å) values, as listed in Table 3). Furtherly, exploration of the binding interactions of Circumdatin J (Compound 24) within DNA gyrase (Fig. 6) with various amino acid residues lining active site, Fig. 5. As seen in Fig. 5a that compound 24 showed many interactions within DNA gyrase active site especially with Arg 482 residue which is one of crucial interactions shown to cause marked inhibition of DNA gyrase enzyme (www.pnas.org/cgi/doi/10.1073/pnas.1525047113.). Additionally, close view of DNA gyrase pocket (Fig. 5b) showed close distance of compound 24 to various amino acid residues lining active site. Finally, electrostatic potential map of DNA gyrase active site (Fig. 5c) showed perfect alignment of various functional groups of compounds 24 with different potential zones of H-donor, acceptor and Van der Waals interactions. Docking poses of ligands with best docking score and/or lowest RMSD values were inspected for their binding interactions for compounds 7, 14, 24, and 25. The above-mentioned results showed the potent effect of the three endophytic fungi (Aspergillus sp. SA1, Aspergillus sp. SA2, and Aspergillus sp. SA3) isolated from Nigella sativa seeds, as a powerful source of natural compounds with antibacterial activity against Gram-negative and Gram-positive bacteria. Especially, compounds identified from Aspergillus sp. SA3 showed the best average docking score within all the three investigated strains, and this provoked us in the upcoming work, to isolate the most active compounds to evaluate their antimicrobial and antifungal potential via in vitro studies.
a Schematic 2D interaction diagram of Circumatidin J (compound 24) within bacterial DNA gyrase (PDB ID: 5BTC) showing both H-bonding, Bi-Bi, and H-Bi interactions; b 3D representation of compound 24 within pocket of DNA gyrase active site showing its close distance with amino acid residues; c Electrostatic potential map of DNA gyrase active site and compound 24 functional groups impeded correctly within different potential zones of active site
Conclusion
Out of twenty isolated colonies, three isolates with distinct morphology were selected for further work and named SA1, SA2 and SA3 respectively. The three most active endophytic fungi were isolated from Nigella sativa seeds, Aspergillus sp. SA1, Aspergillus sp. SA2, and Aspergillus sp. SA3. These endophytic fungi have been shown to be a powerful source of natural compounds with biological activities. Particularly, when it was fermented using PDA culture medium, which could be attributed to its bioactive metabolites, of which twenty-six compounds were identified by LC-HR-ESI-MS belonging to various chemical classes. Crude extracts of endophytic Aspergillus sp. SA1, Aspergillus sp. SA2, and Aspergillus sp. SA3 have shown promising antibacterial activity against Gram-negative and Gram-positive bacteria. Collectively, compounds identified from Aspergillus sp. SA3 showed the best average docking score within all the three investigated strains. This could be used as explanatory for remarkable potency of SA3 extract on the five different bacterial species used in this study
Availability of data and materials
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
References
Vaz AB, et al. Antimicrobial activity of endophytic fungi associated with Orchidaceae in Brazil. Can J Microbiol. 2009;55(12):1381–91.
Card S, et al. Deciphering endophyte behaviour: the link between endophyte biology and efficacious biological control agents. FEMS Microbiol Ecol. 2016;92(8): fiw114.
Kumar A, Antony AR, Kannan VR. Exploration of endophytic microorganisms from selected medicinal plants and their control potential to multi drug resistant pathogens. J Med Plants Stud. 2015;3(2):49–57.
Manzoor T, et al. Extracellular vesicles derived from mesenchymal stem cells — a novel therapeutic tool in infectious Diseases. Inflamm Regeneration. 2023;43(1):17.
Lee DS et al. Marine bacteria are an attractive source to overcome the problems of antibiotic-resistant Staphylococcus aureus. 1st Ed. Mar Microbiol. 2013. p. 83–96.
Kodzius R, Gojobori T. Marine metagenomics as a source for bioprospecting. Mar Genom. 2015;24:21–30.
Demain AL. Pharmaceutically active secondary metabolites of microorganisms. Appl Microbiol Biotechnol. 1999;52:455–63.
Keller NP, Turner G, Bennett JW. Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol. 2005;3(12):937–47.
Strobel G. Harnessing endophytes for industrial microbiology. Curr Opin Microbiol. 2006;9(3):240–4.
Christina A, Christapher V, Bhore SJ. Endophytic bacteria as a source of novel antibiotics: an overview. Pharmacogn Rev. 2013;7(13):11.
Nurunnabi TR, et al. Molecular identification and antimicrobial activity of endophytic fungi isolated from Heritiera fomes (Buch.-Ham), a mangrove plant of the Sundarbans. Beni-Suef Univ J Basic Appl Sci. 2020;9(1):1–10.
Zhang X-g, et al. A new mixed inhibitor of adenosine deaminase produced by endophytic Cochliobolus sp. from medicinal plant seeds. Folia Microbiol. 2020;65:293–302.
Pharamat T, et al. Antimicrobial and anticancer activities of endophytic fungi from Mitrajyna javanica koord and Val. Afr J Microbiol Res. 2013;7(49):5565–72.
Ludwig-Müller J. Plants and endophytes: equal partners in secondary metabolite production? Biotechnol Lett. 2015;37(7):1325–34.
Bakathir HA, Abbas NA. Detection of the antibacterial effect of nigella sativa ground seedswith water. Afr J Tradit Complement Altern Med. 2011;8(2):159–64.
Gopane B, et al. Community diversity and stress tolerance of culturable endophytic fungi from black seed (Nigella sativa L). South Afr J Bot. 2021;137:272–7.
Shareef M, Khaliq T, Faisal M, Majeed W, Shahid M, Muhammad J. Comparative anti-bacterial activities of Nigella sativa and lincomycin in the gut of broiler chicks. 2017.
Chetana S, et al. Antibacterial activity of extract of seeds of Nigella sativa Linn. Pharmacol Online. 2009;2:823–82.
Pavithra N, Sathish L, Ananda K. Antimicrobial and enzyme activity of endophytic fungi isolated from Tulsi. J Pharm Biomedical Sci. 2012;16(16):2014.
Abdelmohsen UR, et al. Antioxidant and anti-protease activities of diazepinomicin from the sponge-associated Micromonospora strain RV115. Mar Drugs. 2012;10(10):2208–21.
Sayed AM, et al. Metabolomic profiling and antioxidant potential of three fungal endophytes derived from Artemisia annua and Medicago sativa. Nat Prod Res. 2022;36(9):2404–8.
May LA, Smiley B, G.J.C.J.o M. Comparative denaturing gradient gel electrophoresis analysis of fungal communities associated with whole plant corn silage. Université de Lorraine; 2001:47(9);829–41.
Fliegerova K, Mrazek J, Voigt KJFm. Differ Anaerob Polycentric fungi rDNA PCR-RFLP. 2006;51(4):273.
Kumar S, et al. MEGA7: molecular evolutionary genetics analysis version 70 for bigger datasets. 2016;33(7):1870–4.
Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of Progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80.
Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–20.
Glaeser SP, et al. Niabella hirudinis and Niabella drilacis sp nov, isolated from the medicinal leech Hirudo Verbana. 2013;63(Pt_9):3487–93.
Alvarez-Perez S et al. Characterization of multiple isolates of aspergillus fumigatus from patients: genotype, mating type and invasiveness. Med Mycol. 2009:47(6);601–8.
Hisham Shady N, et al. Metabolomic profiling and cytotoxic potential of three endophytic fungi of the genera aspergillus, Penicillium and Fusarium isolated from Nigella sativa seeds assisted with docking studies. Nat Prod Res. 2023;37(17):2905–10.
Alhadrami HA, et al. A metabolomic approach to target antimalarial metabolites in the Artemisia annua fungal endophytes. Sci Rep. 2021;11(1):2770.
Shady NH, et al. Mechanistic Wound Healing and Antioxidant Potential of Moringa oleifera Seeds Extract Supported by Metabolic Profiling, In Silico Network Design Molecular Docking, and In Vivo Studies. Antioxidants (Basel). 2022;11(9):1743.
Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3(2):163–75.
Vuuren Sv, Viljoen AM. Antimicrobial activity of limonene enantiomers and 1, 8-cineole alone and in combination. Flavour Fragr J. 2007;22(6):540–4.
Arshad M, et al. Synthesis and characterization of Zn doped WO3 nanoparticles: photocatalytic, antifungal and antibacterial activities evaluation. Mater Res Express. 2020;7(1):015407.
Elaasser MM, Abdel-Aziz MM, El-Kassas RA. Antioxidant, antimicrobial, antiviral and antitumor activities of pyranone derivative obtained from aspergillus candidus. J Microbiol Biotech Res. 2011;1(4):5–17.
Khatiwora E, et al. Antibacterial activity of Dibutyl Phthalate: a secondary metabolite isolated from Ipomoea carnea stem. J Pharm Res. 2012;5(1):150–2.
Aw Y-K, et al. Newly isolated Paenibacillus tyrfis sp. nov., from Malaysian tropical peat swamp soil with broad spectrum antimicrobial activity. Front Microbiol. 2016;7:219.
Gao S-S, et al. Secondary metabolites from a marine-derived endophytic fungus Penicillium chrysogenum QEN-24S. Mar Drugs. 2010;9(1):59–70.
Szymańska R, Kruk J. Novel and rare prenyllipids–occurrence and biological activity. Plant Physiol Biochem. 2018;122:1–9.
Bandeen-Roche K, et al. Phenotype of frailty: characterization in the women’s health and aging studies. J Gerontol A Biol Sci Med Sci. 2006;61(3):262–6.
Nicault, Matthieu Prospecting and induction of antimicrobial molecules by co-culture between bacteria (Streptomyces) and fungi (Basidiomycota) and fungi (Basidiomycetes). Université de Lorraine; 2020.
Lo H-C, et al. Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in aspergillus nidulans. J Am Chem Soc. 2012;134(10):4709–20.
Cabras A, et al. Occurrence, isolation and biological activity of phytotoxic metabolites produced in vitro by Sphaeropsis Sapinea, pathogenic fungus of Pinus radiata. Eur J Plant Pathol. 2006;115(2):187–93.
Nishikawa H. Biochemistry of filamentous Fungi III a metabolic product of aspergillus melleus Yukawa. Part II. Bull Agricultural Chem Soc Japan. 1933;9(10–12):148–51.
Fuchser J, Zeeck A. Secondary metabolites by Chemical Screening, 34.–Aspinolides and Aspinonene/Aspyrone co-metabolites, New Pentaketides produced by aspergillus ochraceus. Liebigs Ann. 1997;1997(1):87–95.
Dai J-R, et al. Circumdatin G, a new alkaloid from the fungus aspergillus ochraceus. J Nat Prod. 2001;64(1):125–6.
Cui CM, et al. Benzodiazepine alkaloids from marine-derived endophytic fungus aspergillus ochraceus. Helv Chim Acta. 2009;92(7):1366–70.
Mukherjee H. On the Biological activity of the natural product (+)-Avrainvillamide. Harvard University, ProQuest Dissertations Publishing; 2015. p. 3739011.
Finefield JM, et al. Fungal origins of the bicyclo [2.2. 2] diazaoctane ring system of prenylated indole alkaloids. J Nat Prod. 2012;75(4):812–33.
Zhang P, et al. Angularly prenylated indole alkaloids with antimicrobial and insecticidal activities from an endophytic fungus fusarium sambucinum TE-6L. J Agric Food Chem. 2019;67(43):11994–2001.
Wang F, et al. Seven new prenylated indole diketopiperazine alkaloids from holothurian-derived fungus aspergillus fumigatus. Tetrahedron. 2008;64(34):7986–91.
Lin S, et al. Structurally diverse and bioactive alkaloids from an insect-derived fungus Neosartorya fischeri. Phytochemistry. 2020;175: 112374.
Ishikawa K, et al. Quinazolinobenzodiazepine derivatives, novobenzomalvins A–C: fibronectin expression regulators from Aspergillus Novofumigatus. Sci Pharm. 2011;79(4):937–50.
Alves DS, et al. Membrane-related effects underlying the biological activity of the anthraquinones emodin and barbalCitation details for reference/s [70] is/are incomplete. Please supply the volume of this/these reference/s. Otherwise, kindly advise us on how to proceed.oin. Biochem Pharmacol. 2004;68(3):549–61.
Ubbink-Kok T, Anderson JA, Konings W. Inhibition of electron transfer and uncoupling effects by emodin and emodinanthrone in Escherichia coli. Antimicrob Agents Chemother. 1986;30(1):147–51.
Hamasaki T, Sato Y, Hatsuda Y. Structure of sydowinin A, sydowinin B, and sydowinol, metabolites from Aspergillus Sydowi. Agric Biol Chem. 1975;39(12):2341–5.
Yao Q, et al. Cytotoxic polyketides from the deep-sea-derived fungus engyodontium album DFFSCS021. Mar Drugs. 2014;12(12):5902–15.
Bao J, et al. New chromones from a marine-derived fungus, Arthrinium sp.,and their biological activity. Molecules. 2018;23(8):1982.
Kim Y-K, et al. GERI-BP002-A, novel inhibitor of acyl-CoA: cholesterol acyltransferase produced by aspergillus fumigatus F93. J Antibiot. 1996;49(1):31–6.
Masi M, Evidente A. Fungal bioactive anthraquinones and analogues. Toxins. 2020;12(11): 714.
Liu K, et al. The antifungal metabolites obtained from the rhizospheric aspergillus sp. YIM PH30001 against pathogenic fungi of Panax notoginseng. Nat Prod Res. 2014;28(24):2334–7.
Quang TH, et al. Cytotoxic and antimicrobial benzodiazepine and phenolic metabolites from Aspergillus Ostianus IMBC-NMTP03. Vietnam J Chem. 2021;59(5):660–6.
Youssef FS, Alshammari E, Ashour ML. Bioactive alkaloids from genus aspergillus: mechanistic interpretation of their antimicrobial and potential SARS-CoV-2 inhibitory activity using molecular modelling. Int J Mol Sci. 2021;22(4): 1866.
Burruano S, et al. Naphthalenone polyketides produced by Neofusicoccum Parvum, a fungus associated with grapevine Botryosphaeria dieback. Phytopathologia Mediterranea. 2016:55(2):197–206.
Yusoff YM, et al. Metabolomic profiling of Malaysian and New Zealand honey using concatenated NMR and HRMS datasets. Metabolites. 2022;12(1): 85.
Nakada T, Yamamura S. Three new metabolites of hybrid strain KO 0231, derived from Penicillium citreo-viride IFO 6200 and 4692. Tetrahedron. 2000;56(17):2595–602.
Xue C, et al. Janthinolide AB, two new 2, 5-piperazinedione derivatives from the endophytic penicillium janthinellum isolated from the soft coral Dendronephthya Sp. Pharmazie. 2006;61(12):1041–4.
Wen H, et al. Structurally diverse meroterpenoids from a marine-derived aspergillus sp. fungus. J Nat Prod. 2019;83(1):99–104.
Reveglia P, Masi M, Evidente A. Melleins—Intriguing Nat Compd. Biomolecules. 2020;10(5):772.
León A, et al. Phthalides: Distribution in nature, chemical reactivity, synthesis, and biological activity. Prog Chem Org Nat Prod. 2017:104:127–246. Springer.
Tsukamoto S, et al. Notoamides F – K, prenylated indole alkaloids isolated from a marine-derived aspergillus sp. J Nat Prod. 2008;71(12):2064–7.
Ma Y-M, et al. Structural diversity and biological activities of indole diketopiperazine alkaloids from fungi. J Agric Food Chem. 2016;64(35):6659–71.
Ryoo I-J, et al. Reticulone, a novel free radical scavenger produced by aspergillus sp. J Microbiol Biotechnol. 2009;19(12):1573–5.
Omolo JO, et al. New Variotin analogues from Aspergillus v iridi-nutans. J Nat Prod. 2000;63(7):975–7.
López-Gresa MP, et al. Circumdatin H, a new inhibitor of mitochondrial NADH oxidase, from Aspergillus Ochraceus. J Antibiot. 2005;58(6):416–9.
Acknowledgements
We thank Deraya University for laboratory facilities.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
1-Nourhan Hisham Shady, Sara Khalid Sobhy, Yaser A. Mostafa, Ramadan Yahia, Stefanie P Glaeser, Peter Kämpfer, Mo’men H. El-Katatny, Usama Ramadan Abdelmohsen wrote the main manuscript text. 2-Stefanie P Glaeser, Peter Kämpfer prepared Figs. 1 and 2. 3-Nourhan Hisham Shady, Usama Ramadan Abdelmohsen prepared Figs. 3 and 4. 4-Yaser A. Mostafa prepared Figs. 5 and 6. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Shady, N.H., Sobhy, S.K., Mostafa, Y.A. et al. Phytochemical analysis and anti-infective potential of fungal endophytes isolated from Nigella sativa seeds. BMC Microbiol 23, 343 (2023). https://doi.org/10.1186/s12866-023-03085-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-023-03085-4