Abstract
Background
Maternally inherited bacterial symbionts are extremely widespread in insects. They owe their success to their ability to promote their own transmission through various manipulations of their hosts’ life-histories. Many symbionts however very often go undetected. Consequently, we have only a restricted idea of the true symbiont diversity in insects, which may hinder our understanding of even bigger questions in the field such as the evolution or establishment of symbiosis.
Results
In this study, we screened publicly available Lepidoptera genomic material for two of the most common insect endosymbionts, namely Wolbachia and Spiroplasma, in 1904 entries, encompassing 106 distinct species. We compared the performance of two screening software, Kraken2 and MetaPhlAn2, to identify the bacterial infections and using a baiting approach we reconstruct endosymbiont genome assemblies. Of the 106 species screened, 20 (19%) and nine (8.5%) were found to be infected with either Wolbachia or Spiroplasma, respectively. Construction of partial symbiotic genomes and phylogenetic analyses suggested the Wolbachia strains from the supergroup B were the most prevalent type of symbionts, while Spiroplasma infections were scarce in the Lepidoptera species screened here.
Conclusions
Our results indicate that many of the host-symbiont associations remain largely unexplored, with the majority of associations we identify never being recorded before. This highlights the usefulness of public databases to explore the hidden diversity of symbiotic entities, allowing the development of hypotheses regarding host-symbiont associations. The ever-expanding genomic databases provide a diverse databank from which one can characterize and explore the true diversity of symbiotic entities.
Similar content being viewed by others
Background
Facultative endosymbiotic bacteria are extremely common in insects. Reports suggest that at least 40% of all insects are infected by the facultative endosymbiotic bacterium Wolbachia [1, 2], while up to 10% of insect species (and up to 23% of Aranaea species) carry Spiroplasma, another facultative endosymbiotic bacterium [3,4,5]. These two symbionts owe their success to their abilities to affect their host biology and promote their own transmission to the next generation of hosts. One such ability revolves around the manipulation of the hosts reproductive system, this occurs in a diverse range of hosts [6,7,8], and more specifically in the butterflies Hypolimnas bolina for Wolbachia [9, 10] and Danaus chrysippus for Spiroplasma [11]; or play defensive roles against diverse parasites and pathogens of their host, including viruses, other bacteria, or parasitoids [12]. Additionally, both of these maternally inherited symbionts have been suggested to occasionally transfer horizontally between host species [13,14,15,16]. Studies have shown that divergent species sharing the same diet [13, 14, 17, 18], or the same parasites [16], are also prone to share similar symbiotic strains. Hybridization between closely related host species may also support such horizontal transfers of the symbionts through the introgressed matriline [15, 19]. Altogether, the diversity of phenotypes associated to these symbionts, and their versatile transmission modes, have made host-symbiont associations excellent study systems for various eco-evolutionary processes. Yet, many host-symbiont interactions remain un-noticed. Thus, we lack a true understanding of the diversity and origin of these symbionts in insects, which in turn challenges the comprehensive study of the evolution of these microbial symbioses.
Most phylogenetic studies on Wolbachia and Spiroplasma are based on a small set of markers. Such sets often only include some or all of the five Multi Locus Sequence Typing (MLST) markers [20] and the wsp gene [21] for Wolbachia; and the data is even more restricted for Spiroplasma, as no MLST markers are yet available for this symbiont. Although broadly used for symbiont screening, and strain characterization, these markers have been criticized for being highly conserved, and thus for being inadequate for depicting the true strain diversity [22] and evolutionary rate of each symbiont. Instead, recent studies advocate for the use of whole genome data [22, 23], but the production of this genomic data is not without difficulties. Sequencing and assembling the genomes of isolated endosymbionts remain costly and methodologically challenging. This is because it is still not always possible to culture and sequence symbionts in isolation from their hosts [24, 25]. Consequently, although a variety of whole genome sequences are available for both Wolbachia and Spiroplasma, they are still unlikely representative of the true strain diversity that may exist in nature for these bacteria. Furthermore, by only targeting (I) host species that do not belong to the same natural species communities or environments, and are not (or little) interacting in nature, or (II) species that do not share direct phylogenetic relationships (but see [26]), it will remain difficult for the field to infer any major eco-evolutionary event that may shape symbiosis, including horizontal transfer events of the symbionts between host species. As a multi-strain genome-sequencing project targeting all symbionts of interacting/related host species would represent a considerable financial and time investment, such important genomic material is unlikely to become available in the near future. Until then, other methods that allow the field to accumulate genomic data from a wider diversity of symbiotic strains can be, and have been, considered [27,28,29].
With the constant development of sequencing techniques, the field of genomic diversity is regularly acquiring new genomic material from a wide array of species [30, 31]. For the order Lepidoptera, we have access to genomic material from most families, and sometimes even over 40 species sequenced per family (e.g. 40 genomes of Nymphalidae, mostly due to the intensive genomic work from the Heliconius Genome Consortium [32]. The deposited sequence read archives (SRAs) contain the raw host genomic material, but also reads from various other entities originally considered as ‘trash’ or ‘contaminants’ to many, and often not analyzed nor discussed. This non-target material however offers opportunities for a broad screening of DNA material from diverse hidden endosymbionts, and for building the partial to complete assemblies of various symbiotic microorganisms without having to sequence the symbionts independently of their hosts [27]. Here, we screened for genomic material from Wolbachia and Spiroplasma, two common insect endosymbiotic bacteria, in 1094 SRAs files/samples from 106 unique Lepidopteran species. We described symbiotic infections new to the literature [6] and characterized strain diversity using a phylogenomic rather than a MLST-based phylogenetic approach.
Results
Identification of reads originating from the endosymbionts Wolbachia and Spiroplasma
Using MetaPhlAn2 we identified 58 of the 1094 tested SRAs as being infected with Wolbachia (with a threshold > 1000 reads), corresponding to 55 individual samples and 16 species (15.1% of the total 106 species screened here). While only six samples were identified as containing Spiroplasma reads, representing five species (4.7%). In comparison, Kraken2 (using our custom databases) identified a larger number of SRAs positive for these same infections. A total of 70 SRAs were identified as containing Wolbachia, representing 64 biosamples and 20 host species (19%), with the majority also having reads identified as belonging to the Wolbachia-associated phage WO (Table 1). While 23 SRAs tested positive for Spiroplasma, corresponding to 16 individual samples and nine host species (8.5%). Tables 1 and 2 contain a list of all biosamples and SRAs identified as infected in our screen.
Neither software identified any sample as being infected by both bacterial symbionts simultaneously. Generally speaking, the majority of samples identified in the MetaPhlAn2 analysis were also identified using Kraken2 (with the exception of two SRAs; Fig. S1). The Wolbachia analysis with MetaPhlAn2 identified two SRAs that were not positive in the Kraken2 analysis (Sample number 47 - SRR8549338 and number 64 - SRR6505226). A closer look at these two samples showed that while MetaPhlAn2 identified 1152 and 1727 reads as Spiroplasma, Kraken2 only identified 60 and 620 for each sample, respectively and showed no presence of the Wolbachia-associated phage WO. Due to the small number of Kraken2 identified reads, and phage absence (whose presence has been associated with strong support for Wolbachia infection [28]) these samples were not taken forward for further analysis, and are likely to represent false positives.
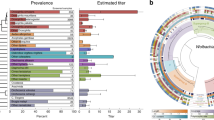
Overall, the incidence levels for both symbionts in our dataset are slightly lower but comparable to those suggested by the available literature. Wolbachia infects 40–80% of all arthropod species [1, 33, 34], and Spiroplasma is expected in only 5–30% of all terrestrial arthropod species [5, 35]. Nine of the Lepidoptera species positive here for Wolbachia or Spiroplasma infection were previously reported to carry the infections in a comprehensive review on symbiont infection in Lepidoptera by Duplouy & Hornett [6]. Wolbachia was indeed previously reported in eight of these species, including Heliconius erato (however subspecies H. erato chesstertonii only [36], which is not included in our sample), Pararge aegeria [5], Parnassius apollo [37], Polygonia c-album [38], Operophtera brumata [39], Plutella australiana [40], P. xylostella [41, 42], and Tuta absoluta [43]; while Spiroplasma was already previously detected, and intensively studied, in the African monarch D. chrysippus [11, 44]. These results suggest that most of the host-symbiont associations detected here have yet to be described and studied in their natural habitats.
Identification of potential contamination and parasitoids
As we are screening pre-existing genomic data from a variety of tissue types, ranging from specific tissues to whole bodies, the possibility exists that the Wolbachia and Spiroplasma infections might be those of parasites, parasitoids or host plants of the Lepidoptera, as opposed to the targeted Lepidoptera species themselves. A complete summary of contaminant groups is given in File S1. Within, the samples positive for Spiroplasma, two (Sample Numbers: 140, 131) had 0.1% of the total reads assigned to Hymenoptera, and a further three (Sample Numbers: 130, 136, 145) were positive for the plant phyla Streptophyta, with Sample Number 145 having ~ 10% of reads assigned to a potential host plant, Vigna unguiculata. In comparison, of the 64 biosamples identified as positive for Wolbachia, two (Sample Numbers: 2, 30) had 0.1% of reads assigned to Hymenoptera or 0.3% to Coleoptera, respectively (See Table 3). A further three samples (Sample Numbers: 2, 37, 45) were positive for the plant phyla Stretophyta, with up to 1% of reads being assigned to the phylum.
Wolbachia
Two to three rounds of baiting of Wolbachia originating reads with mirabait revealed that on average 6.35% of the total number of reads from each Wolbachia-infected SRA are of the symbiont (range: 0.002–54.26%, File S2). The resulting extracted reads were used to construct 64 partial assemblies, with 18 being < 0.5 Mbp in size. On average, the assemblies consisted of a total size of 0.9 Mbp spread across 245 contigs. Complete assembly statistics can be found in File S2. Identification of BUSCO genes found on average 87 Complete and Single copy orthologues (Range: 0–131, File S2), in comparison 121 genes (out of 148 total) were identified from the reference genomes (File S3). The samples that lacked the identification of any BUSCO genes were those of < 0.1 Mbp in size, which is expected due to their incomplete and fragmented nature. Noticeably, 12 of the assemblies lacking gene identification belonged to Spodoptera litura. Following manual curation of the dataset a total of 69 Wolbachia strains (21 references and 48 samples) and 133 genes were included in the phylogenetic analysis. Concatenation of the BUSCO genes sequences resulted in a final alignment of 141,346 bp.
The MLST and wsp genes were identified for 68 strains (47 samples and 21 references), with concatenation resulting in a final alignment of 2539 bp. In addition to the concatenated alignment, alignments for each of the five MLST and one wsp genes were carried forward. All five MLST genes were identified in 48/68 (36 samples and 12 references) strains, while the wsp gene was present in 15/68 (5 samples and 10 references) strains.
All the Wolbachia strains identified from our samples belong to the A- and B- supergroups; with the majority (44/48, 92%) belonging to the B-supergroup. Despite using a slightly different set of strains, similar tree configurations were obtained by using the concatenated BUSCO sequences or the concatenated sequences of the MLSTs and wsp genes (Figs. 1 and 2). Finally, the phylogenies based on only one single MLST gene or the wsp gene showed the same groupings of samples into either A- and B- supergroups, with generally fewer representative samples per phylogeny, and lower resolution among individual samples (Figs. S2, S3, S4, S5, S6 and S7.
Phylogenetic relationships between Wolbachia positive SRAs and reference genomes based on 133 BUSCO genes. Tree was rooted using the reference genomes of the C- D- and F-supergroups (black, N = 5 reference genomes). All samples characterized in this study (annotated with a sample number and host species) belong to either the A-supergroup (orange, N = 3 + 8 reference genomes) or the B-supergroup (purple, N = 45 + 8 reference genomes)
Phylogenetic relationships between Wolbachia positive SRAs and reference genomes based on five MLST and the wsp genes. Tree was rooted using reference genomes data from Wolbachia strains wBm and wClec-F from the D- and F-supergroup respectively (in black). All samples characterized here (annotated with a sample number and host species) belong to either the A-supergroup (orange, N = 4 + 5 reference strains) or the B-supergroup (purple, N = 44 + 5 reference strains)
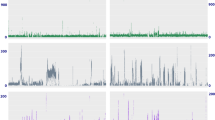
Interestingly, 14 SRAs from the unique bioproject: PRJNA344815 [45] were identified as being positive for Wolbachia under our criteria. However, only two produced relatively complete assemblies (> 1.2 Mbp in length) from which BUSCO and MLST genes could be extracted. To determine whether the remaining 12 SRAs potentially represented false positives or potentially host insertion of Wolbachia gene(s) within the host genome a mapping approach was taken. The mapping of the reads baited out during the second round of baiting with mirabait against the wPip genome (GCF_000073005.1) show for the two samples that produced good assemblies (Sample Number 42 - SRR5132433 and Number 36 - SRR5132404) high levels of coverage is seen along the entire wPip genome (Fig. 3A and B). In comparison, the remaining 12 SRAs showed low and sometimes patchy coverage along the reference genome (Fig. 3C - N). However, due to the distribution of reads along the reference genome these samples are thought to represent true infection, with low numbers of sequencing reads originating from Wolbachia, resulting in overall low coverage and hence the inability to generate reasonable assemblies for gene extraction and phylogenetic analysis. Nevertheless, this highlights that although it might not be possible to always assemble a symbiont genome from positive samples, the screening approach used here provides interesting testable hypotheses for further work.
Mapping of the 14 Wolbachia positive Spodoptera litura samples along the wPip genome. Reads identified as belonging to Wolbachia were mapped to the wPip Wolbachia reference genome (GCF_000073005.1). Coverage values shown on the vertical side of the figure. The graphs correspond to the following samples, (represented as sample number, as given in Table 1, − SRA Accession): A) 42 - SRR5132433; B) 36 - SRR5132404; C) 34 - SRR5132402; D) 33 - SRR5132396; E) 32 - SRR5132393; F) 31 - SRR5132392; G) 44 - SRR5132442; H) 43 - SRR5132437; I) 39 - SRR5132426; J) 38 - SRR5132419; K) 37 - SRR5132409; L) 35 - SRR5132403; M) 40 - SRR5132427; N) 41 - SRR5132432
Spiroplasma
Baiting of Spiroplasma reads identified that on average 2.38% of the total number of reads in infected SRA files belonged to the symbiont (Range: 0.0004–19.75%, File S4). A maximum of two baiting rounds was required for optimal sequence baiting of Spiroplasma. Of the 16 samples identified as being positive, only 15 produced assemblies. In the case of SAMN06758611 (Sample Number 145) none of the produced contigs blasted to any of the Spiroplasma references, and therefore this sample was discarded from further analysis.
Identification of BUSCO genes found on average 27 single copy complete orthologs (Range 0–47, File S4), in comparison 55 genes (File S3) were extracted from the reference genomes (148 BUSCO genes total). The samples that lacked identification of any BUSCO genes were those with assemblies of < 0.1 Mbp in size. Following manual curation of the dataset, a total of 55 Spiroplasma strains (42 references + 13 samples) and 63 genes were used for phylogenetic analysis. Concatenation resulted in a final alignment of 40,401 bp.
Based on the BUSCO phylogeny, all the Spiroplasma strains identified from our samples are similar to previously characterized strains (ie. reference genomes) (Fig. 4); Spiroplasma is here distributed among three clades: Clade I, or the Apis clade as described by Gasparich et al [46], contains most of the Spiroplasma-infected SRAs (7 in total), all of which group together with Spiroplasma strains originating from Hymenoptera, Diptera and Coleoptera hosts, including Spiroplasma apis, S. clarkia and S. sabaudiense; Clade II, or the Citri-Chrysopirola-Mirum clade [46], includes only a single SRA sample, which is grouped with Spiroplasma reference genomes from Diptera and Hymenoptera hosts (ie. S. mirum, S. chrysopirola and S. poulsonii); lastly, Clade III consists of five SRA samples grouped with the single Lepidoptera Spiroplasma reference (from Danus chrysippus), and a Hymenoptera reference. It remains unknown whether Clade III corresponds to the ixodetis clade described by Gasparich et al [46], as the species included in the ixodetis clade and our Clade III were reciprocally absent between the two studies.
Phylogenetic relationship between Spiroplasma positive SRAs and reference genomes based on 63 BUSCO genes. All samples characterized here are annotated with a sample number, followed by either the host species (for SRA screened samples) or the Spiroplasma species (for reference samples) belong to either the Clade I (blue), II (red) or III (green). All reference samples are coloured with a darker shade of the Clade colour
Discussion
By screening 1094 SRAs files from diverse Lepidoptera species for genomic material from Wolbachia and Spiroplasma symbionts, we isolated and characterized infections by either of these two symbiotic bacteria in 28 Lepidoptera species. For many of these host species, these infections had, to our knowledge, never been described [6]. Noticeably, some of these newly discovered infections were found in species with strong prior ecological and evolutionary knowledge. In particular, our screening work revealed an additional five Heliconius species (including six subspecies of H. erato) infected with Spiroplasma, and one species with a Wolbachia infection. Previous studies had identified Spiroplasma from H. clysonymus [47], H. doris [48], and H. aoedes [49], while Wolbachia was identified from H. cydno [47] and H. erato chestertonii and H. e. venus [36]. Additionally, the taxonomy browser in NCBI suggests an additional 16 species and subspecies of Heliconius carrying Spiroplasma (H. charithonia, H. clysonymus, H. cydno chioneus, H. demeter, H. e. notabilis, H. eratosignis, H. melpomene, H. m. amaryllis, H. m. meriana, H. pachinus, H. sara, H. telesiphe, H. timareta, H. t. timareta, H. wallacei, and H. xanthocles). Altogether, these results suggest that endosymbiotic bacteria commonly infect the Heliconius butterfly clade. What however remains surprising is that despite more than 350 studies (Pubmed Dec 2021) using Heliconius butterflies as study organisms, one has yet to experimentally test the role of these symbionts in these species. With Spiroplasma and Wolbachia infecting Heliconius species from different clades across the Heliconius phylogeny [50], one could for example wonder whether these symbionts have played a role in the speciation and diversification of this species rich insect genus.
Despite our efforts to optimize the detection of symbiotic infections and the extraction of their genomic material from the SRA samples (not specifically aimed at metagenomics analysis), we could only produce the partial genomic assemblies of 64 Wolbachia strains and 15 Spiroplasma strains. Three hypotheses can explain the incompleteness of our assemblies: (a) low sequencing data quality and/or incomplete sequencing of the entire symbionts genomic chromosomes; (b) contamination with DNA from another samples; (c) insertion of some genomic material from the symbiont(s) within the genome of the host. Methodological boundaries to the detection and construction of full symbiotic genomes are several folds. The use of Kraken2 for screening the SRAs yields more positive results than the screening using the default MetaPhlAn2, leading to a wider range of genomic assemblies. This is most likely due to the use of a customized reference database with either Wolbachia or Spiroplasma reference genomes in Kraken2. Nevertheless, both software provided useful results for screening of endosymbionts. Kraken is considered to have good performance metrics in terms of accuracy and abundance profiles [51], but the main advantage to using Kraken2 is the ability to create custom databases, however it requires large amounts of memory (> 100 Gb). In cases where high amounts of memory are unavailable, MetaPhlAn2 is a good alternative and has low computational requirements and fast classification speed [51], however the main limiting flaw is its’ database and the inability to create custom databases, which for us resulted in fewer ‘infected’ samples.
Kraken2 is also likely to produce some false negative results, simply because the SRA data we screened was not optimized for the sequencing and analyses of metagenomes. For example, the tissue type used for extraction, is likely to affect if or how much endosymbiotic bacteria ends up in the final DNA library. An arbitrary cut off of 1000 reads was chosen as we considered fewer reads than this would result in insufficient data to yield a useful assembly result, therefore samples with slightly lower than 1000 Wolbachia or Spiroplasma reads could also potentially represent ‘true’ positives that may warrant further investigation. Our protocol was optimized to make sure each step significantly improved the quality of our final assemblies in a time-efficient manner.
Additionally, the endosymbiont genomes we constructed might potentially have originated from contaminant parasites, or associated host plant, as opposed to the targeted Lepidoptera. Since the data investigated in this study has been obtained from publically available genomic data, we have no control over the tissue types used for extraction, or the sample preparation. To investigate potential contamination, we took a conservative approach and listed all samples for which > 0.1% of the reads belonged to potential ‘contaminant’ taxa (ie. parasitoid, parasite, or host plant). Although it should be noted that there is no consensus on which thresholds should be used with metagenomic taxonomic classifiers [51, 52], with thresholds being considered on a project specific basis, we believe that the endosymbiont genomes presented in this study originate from the Lepidoptera host targeted by the sequencing project, with possibly one notable exception. The sample number 145 includes 10% of reads assigned to the plant Vigna unguiculata. However, the assembly produced from the baited Spiroplasma reads from this sample was discarded from downstream analysis, due to none of the assembled contigs blasting to any Sprioplasma assemblies from our reference database. Nonetheless, the true infection status of many of the species screened for infection here remains to be confirmed through screening of fresh wild samples. Similarly, the true role of these infections will only be fully tested through ecological studies in the hosts respective natural habitats.
Despite the shortcomings, the 48 Wolbachia and 13 Spiroplasma partial genomic assemblies (out of 64 and 15 produced in this study, respectively) included enough target genes to support phylogenomic analyses of the symbiotic strains. The Wolbachia phylogenetic trees built either using the MLST sequences only, or using the BUSCO genes sequences, both similarly divided the strains within the A- and B-supergroups, with a higher number of strains belonging to the B-supergroup. This observation is not new, as Lepidoptera have been described as hosts to a greater number of B-supergroup Wolbachia compared to A-supergroup Wolbachia [44, 53,54,55,56]. Additionally, the phylogenomic approach using the BUSCO genes revealed higher Wolbachia strain diversity than did the MLST-based phylogenetic analyses. This supports the idea that the MLST markers are unsuited for fine-scale strain differentiation in this bacterial clade [22, 23]. Nonetheless, even when using a whole-genome typing method such as the BUSCO genes, the resulting phylogenomic tree highlighted very little divergence within the B-supergroup Wolbachia strains, with divergent Lepidoptera species carrying closely related strains. Similarly, although the newly characterized Spiroplasma strains belong to three divergent clades, few strains from highly divergent host species show very little genetic differences. Both Wolbachia and Spiroplasma are vertically transmitted in insects [57, 58], but hybrid introgression and other shared host-resource have been proposed as platforms for the horizontal transfer of these microbial symbionts [14, 58]. As many host species are from different geographic regions and evolved in different environments, it remained impossible to identify which eco-evolutionary routes may have supported the transfer of these symbionts between host species [59].
Additionally, while large, almost complete assemblies of symbiotic chromosome are often evidence for true natural infections, partial symbiont assemblies that are much shorter than 500,000 bp long often require further investigation. As discussed above, we are confident that our quality criteria across our methodology allowed us to remove many potential false positives. In our dataset, however, few SRA samples clearly included genomic material of Wolbachia origin, which supported the construction of small assemblies, from which none of the BUSCO or Wolbachia MLST genes were retrieved. This was the case for 12 of the 14 positive SRAs from the moth species Spodoptera litura (Noctuidae, Fabricius 1775). Closer investigation into these SRAs showed that mapping of the baited reads along the wPip reference genome had low and sometimes patchy coverage. Therefore, we conclude that these are likely to represent true positives, but with insufficient coverage of the Wolbachia genome to promote an adequate assembly for gene extraction and phylogenetic analysis, using the approach taken here. Alternative gene identification methods, such as programs designed to identify exons from fragmented genomes [60] could represent an alternative approach to the identification of the corresponding BUSCO genes.
Conclusions
This study highlights the usefulness of existing genomic data to investigate the true diversity of endosymbiotic bacteria. Here we present two methods to detect the presence of endosymbionts, such as Wolbachia and Spiroplasma. Generally speaking, provided large amounts of memory are available for computation, Kraken2 identified more samples as containing Wolbachia and Spiroplasma compared to MetaPhlAn2. We also successfully produced partial endosymbiont genomes that can be mined for phylogenetically informative genes, to better understand their evolutionary histories. The deep analysis of such ‘once hidden symbiont’ genomic material from a wider diversity of hosts and environments, than currently available, will benefit the studies of different eco-evolutionary events associated to the evolution and establishment of bacterial symbioses in arthropods, including radiation, horizontal transfers, and lateral gene transfers.
Material & methods
Dataset construction
All samples included in this study are available in the NCBI Sequence Read Archive (SRA). To identify samples for screening, all accession numbers that matched the criteria of Lepidoptera genomic DNA were sent to the NCBI run selector (as of September 2020). Within the run selector, the following criteria were used for sample selection: (i) ran on the illumina platform, (ii) had a WGS assay type, and (iii) paired library layout. This resulted in a total of 1094 samples for analysis, covering 106 species (File S5). We used prefetch and Fasterq-dump v. 2.9.6 from the SRA Toolkit (NCBI SRA) to download the reads from each accession.
Taxonomic assignment
Reads were assigned taxonomic labels with Kraken2 [61] and MetaPhlAn 2.0 [62]. Kraken2 assigns taxonomic labels using a k-mer based search, whereby each k-mer within a query is matched to the lowest common ancestor of genomes in the database containing the given k-mer, this information is then used by the classification algorithm to infer the taxonomic classification. In comparison, MetaPhlAn2 uses clade-specific marker genes (identified from across ~ 17,000 reference genomes) to determine the taxonomic composition of the input dataset. Kraken2 was run using a confidence threshold of 0.05, a mpa style output and a custom database which contained; (i) the standard kraken database, (ii) Refseq viral database, (iii) Refseq plasmid database, (iv) Refseq bacteria database, (v) Univec core database and (vi) available Lepidoptera sequences (downloaded as of April 2020). A full list of taxa included in the database is given in File S6. MetaPhlAn was run using the analysis type rel_ab_w_read_stats, which provides the relative abundance and an estimate of read numbers originating from each clade. The resulting outputs were screened for lines matching Wolbachia or Spiroplasma. Based on the Kraken2 results, datasets that contained > 1000 hits to either Wolbachia or Spiroplasma were taken forward for further analysis. Overlap between each analysis was inferred and Venn diagrams constructed with Jvenn [63].
To rule out possible sources of contamination with material from parasites, parasitoids or host plants, we screened the Kraken2 results for the presence of any Insecta Orders, Arthropod classes, Nematoda, Platyhelminthes and Plant Phyla. Any contaminant groups with > 1000 reads assigned are listed in File S1. The conservative limit of 1000 reads was used as a pre-screen for contaminants. However, due Kraken2 often reporting false positives in relation to low abundance taxa [51, 52], only samples whereby more than 0.1% of the total reads were assigned to ‘contaminants’ are suspected of being contaminated.
Wolbachia and Spiroplasma reference database construction
To identify reads originating from either Wolbachia or Spiroplasma a reference dataset containing genomes from either Wolbachia or Spiroplasma were constructed. A total of 21 Wolbachia and 42 Spiroplasma genomes were downloaded from NCBI (September 2020), respectively. For the Wolbachia reference database, genomes chosen to represent strains found in the supergroups A, B, C, D and F, with a single representative per stain being included. Spiroplasma genomes originating from insect hosts were included in the Spiroplasma database. A complete list of the genomes included in each database is shown in File S3.
Identification and assembly of Wolbachia and Spiroplasma reads
Independent SRA experiments or runs originating from the same Biosamples that met our screening criteria were combined into a single dataset. To identify reads originating from the endosymbiont, a modified version of Pascar and Chandler [29] method was used. For each sample, reads were extracted from the full dataset that matched at least one kmer to the respective reference dataset using the mirabait tool from MIRA 4.0.2 [64] using a kmer value of 31. The extracted reads were assembled using SPAdes 3.13.1 [65], with the baited reads being considered single end and kmer values of 21, 33 and 55. The resulting contigs were then blasted back to their respective endosymbiont database using standalone blast 2.0.0+ (megablast, evalue threshold of e-10, 70% minimum percentage identity), contigs lacking significant blast hits were removed. The remaining contigs were used as the reference for the second round of baiting, followed by reassembly, and blast search. This process was repeated until a < 5% increase in the number of reads baited was observed, for this dataset no significant increase was seen after three baiting rounds. The assemblies produced at the end of round of either round two or three were carried forward for downstream analysis. All final assemblies are available at Zenodo: https://doi.org/10.5281/zenodo.6517359
Gene identification, alignment and phylogenetic reconstruction
The following steps were carried out for each symbiont dataset independently. The assemblies resulting from the final round of baiting were used to identify single copy bacterial genes with BUSCO v 3.0.2 [66, 67] in genome mode, utilizing the bacteria odb9 database. Individual genes were aligned based on BUSCO IDs using MAFFT 7.407 [68], using the auto option which chooses the best alignment method based on the data. The resulting gene alignments were manually screened and curated using Geneious Prime® 2020.2.4 (http://www.geneious.com). This step was carried out to ensure correct orthology and alignment. Genes identified in < 10 samples were removed from the final dataset. This resulted in a final dataset of 133 Wolbachia and 63 Spiroplasma genes.
For all Wolbachia assemblies, in addition to the BUSCO genes, the five MLST genes (CoxA, FbpA, FtsZ, Gatb, HcpA) and the wsp gene were identified using a blast approach and extracted when present. A reference set for each gene was obtained from GenBank (See File S5 for a complete list) with representative strains of the Wolbachia A-, B-, F- and D-supergroups. A blast search was carried out against these references using Genious Prime® 2020.2.4, and the corresponding region were extracted from our assemblies. Individual gene alignments were produced using the pairwise Geneious Alignment, default options, in Geneious Prime. Alignments were manually screened to check for correct alignment.
For the phylogenetic analysis of Spiroplasma samples, all BUSCO genes were concatenated in alphabetical gene order, resulting in an alignment of 40,401 bp. The following datasets were constructed for the analysis of Wolbachia samples: (I) Concatenated BUSCO genes (141,346 bp), (II) Concatenated MLST + wsp genes (2539 bp), (III – VIII) Individual MLST and wsp genes. Alignments are available from Zenodo doi: https://doi.org/10.5281/zenodo.6517359 All concatenated alignments were double-checked for misaligned regions using AliView [69].
Phylogenetic reconstruction was carried out for each dataset with CIPRES v.3.3 [70] using RAxML-HPC2 on XSEDE [71] with the Gamma+I parameter. Tree visualization and figures were produced with FigTree (http://tree.bio.ed.ac.uk/software/figtree/) and ITOL [72, 73] using the bipartitions output trees produced by RAxML.
A closer look at the Spodoptera litura samples
Since 12 of the 14 SRAs belonging to Spodoptera litura produced poor assemblies, for which no BUSCO or MLST genes could be identified, we wanted to further investigate their composition. To determine if the samples potentially represented false positives or host insertions of Wolbachia genes rather than true infections a mapping analysis was carried out. Reads baited during the second round of Mirabait were mapped to the wPip reference genome (GCF_000073005.1) with bowtie2 v.2.4.1 [74], using the sensitive local option. The resulting sam files were converted to sorted bam with samtools v1.10 [75]. Coverage information was obtained using samtools depth, and the resulting graphs plotted with the ggplot package [76] in R.
Availability of data and materials
The raw data analyzed were obtained from the NCBI SRA archive, all SRA numbers are given in File S5. The resulting symbiont assemblies and final gene alignments are available from Zenodo doi: https://doi.org/10.5281/zenodo.6517359
References
Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia? - a statistical analysis of current data. FEMS Microbiol Lett. 2008;281:215–20.
Detcharoen M, Schilling MP, Arthofer W, Schlick-Steiner BC, Steiner FM. Differential gene expression in Drosophila melanogaster and D. nigrosparsa infected with the same Wolbachia strain. Sci Rep. 2021;11:1–9.
Duron O, Bouchon D, Boutin S, Bellamy L, Zhou L, Engelstädter J, et al. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 2008;6:1–12.
Cisak E, Wójcik-Fatla A, Zając V, Sawczyn A, Sroka J, Dutkiewicz J. Spiroplasma - an emerging arthropod-borne pathogen? Ann Agric Environ Med. 2015;22:589–93.
Russell JA, Funaro CF, Giraldo YM, Goldman-Huertas B, Suh D, Kronauer DJC, et al. A veritable menagerie of heritable Bacteria from ants, butterflies, and beyond: broad molecular surveys and a systematic review. PLoS One. 2012;7:e51027.
Duplouy A, Hornett EA. Uncovering the hidden players in Lepidoptera biology: the heritable microbial endosymbionts. PeerJ. 2018;6:e4629.
Hassan SSM, Idris E. Male-killing in African butterflies. Trends Evol Biol. 2013;5:7–14.
Correa CC, Ballard JWO. Wolbachia associations with insects: winning or losing against a master manipulator. Front Ecol Evol. 2016;3:153.
Dyson EA, Kamath MK, Hurst GDD. Wolbachia infection associated with all-female broods in Hypolimnas bolina (Lepidoptera: Nymphalidae): evidence for horizontal transmission of a butterfly male killer. Heredity (Edinb). 2002;88:166–71.
Duplouy A, Hurst GDD, O’Neill SL, Charlat S. Rapid spread of male-killing wolbachia in the butterfly Hypolimnas bolina. J Evol Biol. 2010;23:231–5.
Jiggins FM, Hurst GDD, Jiggins CD, Schulenburg JHGVD, Majerus MEN. The butterfly Danaus chrysippus is infected by a male-killing Spiroplasma bacterium. Parasitology. 2000;120:439–46.
Hamilton PT, Perlman SJ. Host defense via Symbiosis in Drosophila. PLoS Pathog. 2013;9:1–4.
Stahlhut JK, Desjardins CA, Clark ME, Baldo L, Russell JA, Werren JH, et al. The mushroom habitat as an ecological arena for global exchange of Wolbachia. Mol Ecol. 2010;19:1940–52.
Sintupachee S, Milne JR, Poonchaisri S, Baimai V, Kittayapong P. Closely related Wolbachia strains within the pumpkin arthropod community and the potential for horizontal transmission via the plant. Microb Ecol. 2006;51:294–301.
Raychoudhury R, Baldo L, Oliveira DCSG, Werren JH. Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution (N Y). 2009;63:165–83.
Vavre F, Fleury F, Lepetit D, Fouillet P, Boulétreau M. Phylogenetic evidence for horizontal transmission of Wolbachia in host- parasitoid associations. Mol Biol Evol. 1999;16:1711–23.
Ahmed MZ, Breinholt JW, Kawahara AY. Evidence for common horizontal transmission of Wolbachia among butterflies and moths. BMC Evol Biol. 2016;16:1–16.
Chrostek E, Pelz-Stelinski K, Hurst GDD, Hughes GL. Horizontal transmission of intracellular insect symbionts via plants. Front Microbiol. 2017;8:28.
Deng J, Assandri G, Chauhan P, Futahashi R, Galimberti A, Hansson B, et al. Wolbachia-driven selective sweep in a range expanding insect species. BMC Ecol Evol. 2021;21:1–17.
Baldo L, Hotopp JCD, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72:7098–110.
Zhou W, Rousset F, O’Neill S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc R Soc B Biol Sci. 1998;265:509–15.
Bleidorn C, Gerth M. A critical re-evaluation of multilocus sequence typing (MLST) efforts in Wolbachia. FEMS Microbiol Ecol. 2018;94:163.
Gerth M, Gansauge MT, Weigert A, Bleidorn C. Phylogenomic analyses uncover origin and spread of the Wolbachia pandemic. Nat Commun. 2014;5:1–7.
Whitcomb RF, Tully JG, McCawley P, Rose DL. Application of the growth inhibition test to Spiroplasma taxonomy. Int J Syst Bacteriol. 1982;32:387–94.
Dobson SL, Marsland EJ, Veneti Z, Bourtzis K, O’Neill SL. Characterization of Wolbachia host cell range via the in vitro establishment of infections. Appl Environ Microbiol. 2002;68:656–60.
Gerth M, Röthe J, Bleidorn C. Tracing horizontal Wolbachia movements among bees (Anthophila): a combined approach using multilocus sequence typing data and host phylogeny. Mol Ecol. 2013;22:6149–62.
Scholz M, Albanese D, Tuohy K, Donati C, Segata N, Rota-Stabelli O. Large scale genome reconstructions illuminate Wolbachia evolution. Nat Commun. 2020;11:1–11.
Ghanavi HR, Twort VG, Duplouy A. Exploring bycatch diversity of organisms in whole genome sequencing of Erebidae moths (Lepidoptera). Sci Rep. 2021;11:1–8.
Pascar J, Chandler CH. A bioinformatics approach to identifying Wolbachia infections in arthropods. PeerJ. 2018;2018:e5486.
Giani AM, Gallo GR, Gianfranceschi L, Formenti G. Long walk to genomics: history and current approaches to genome sequencing and assembly. Comput Struct Biotechnol J. 2020;18:9–19.
Lewin HA, Robinson GE, Kress WJ, Baker WJ, Coddington J, Crandall KA, et al. Earth BioGenome project: sequencing life for the future of life. Proc Natl Acad Sci U S A. 2018;115:4325–33.
The Heliconius Genome Consortium. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–8.
Weinert LA, Araujo-Jnr EV, Ahmed MZ, Welch JJ. The incidence of bacterial endosymbionts in terrestrial arthropods. Proc R Soc B Biol Sci. 2015;282:20150249.
Sazama EJ, Bosch MJ, Shouldis CS, Ouellette SP, Wesner JS. Incidence of Wolbachia in aquatic insects. Ecol Evol. 2017;7:1165–9.
Kautz S, Rubin BER, Moreau CS. Bacterial infections across the ants: frequency and prevalence of Wolbachia, Spiroplasma, and Asaia. Psyche (London). 2013. https://doi.org/10.1155/2013/936341.
Muñoz AG, Baxter SW, Linares M, Jiggins CD. Deep mitochondrial divergence within a Heliconius butterfly species is not explained by cryptic speciation or endosymbiotic bacteria. BMC Evol Biol. 2011;11:1–11.
Łukasiewicz K, Sanak M, Węgrzyn G. A lack of Wolbachia-specific DNA in samples from apollo butterfly (Parnassius apollo, Lepidoptera: Papilionidae) individuals with deformed or reduced wings. J Appl Genet. 2016;57:271–4.
Kodandaramaiah U, Weingartner E, Janz N, Dalén L, Nylin S. Population structure in relation to host-plant ecology and Wolbachia infestation in the comma butterfly. J Evol Biol. 2011;24:2173–85.
Derks MFL, Smit S, Salis L, Schijlen E, Bossers A, Mateman C, et al. The genome of winter moth (Operophtera brumata) provides a genomic perspective on sexual dimorphism and phenology. Genome Biol Evol. 2015;7:2321–32.
Ward CM, Baxter SW. Draft genome assembly of a Wolbachia endosymbiont of Plutella Australiana. Genome Announc. 2017;5:e01134–17.
Jeyaprakash A, Hoy MA. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol Biol. 2000;9:393–405.
Delgado AM, Cook JM. Effects of a sex-ratio distorting endosymbiont on mtDNA variation in a global insect pest. BMC Evol Biol. 2009;9:1–10.
Škaljac M, Kostanjšek R, Žanić K. The presence of Wolbachia in Tuta absoluta (Lepidoptera: Gelechiidae) populations from coastal Croatia and Montenegro. Afr Entomol. 2012;20:191–4.
Salunke BK, Salunkhe RC, Dhotre DP, Walujkar SA, Khandagale AB, Chaudhari R, et al. Determination of Wolbachia diversity in butterflies from Western Ghats, India, by a multigene approach. Appl Environ Microbiol. 2012;78:4458–67.
Liu J, Li S, Li W, Peng L, Chen Z, Xiao Y, et al. Genome-wide annotation and comparative analysis of cuticular protein genes in the noctuid pest Spodoptera litura. Insect Biochem Mol Biol. 2019;110:90–7.
Gasparich GE, Whitcomb RF, Dodge D, French FE, Glass J, Williamson DL. The genus Spiroplasma and its non-helical descendants: phylogenetic classification, correlation with phenotype and roots of the mycoplasma mycoides clade. Int J Syst Evol Microbiol. 2004;54:893–918.
Salazar Sastoque MP. Exploring the abdominal microbiome of two Heliconius species in the central Colombian Andes. Bogotá: Universidad del Rosario; 2021.
Van Schooten B, Godoy-Vitorino F, McMillan WO, Papa R. Conserved microbiota among young Heliconius butterfly species. PeerJ. 2018;6:e5502.
Hammer TJ, Dickerson JC, McMillan WO, Fierer N. Heliconius butterflies host characteristic and Phylogenetically structured adult-stage microbiomes. Appl Environ Microbiol. 2020;86:e02007–20.
Kozak KM, Wahlberg N, Neild AFE, Dasmahapatra KK, Mallet J, Jiggins CD. Multilocus species trees show the recent adaptive radiation of the mimetic Heliconius butterflies. Syst Biol. 2015;64:505–24.
Ye SH, Siddle KJ, Park DJ, Sabeti PC. Benchmarking Metagenomics tools for taxonomic classification. Cell. 2019;178:779–94.
McIntyre ABR, Ounit R, Afshinnekoo E, Prill RJ, Hénaff E, Alexander N, et al. Comprehensive benchmarking and ensemble approaches for metagenomic classifiers. Genome Biol. 2017;18:1–19.
Duplouy A, Brattström O. Wolbachia in the genus Bicyclus: a forgotten player. Microb Ecol. 2018;75:255–63.
Tagami Y, Miura K. Distribution and prevalence of Wolbachia in Japanese populations of Lepidoptera. Insect Mol Biol. 2004;13:359–64.
Russell JA, Goldman-Huertas B, Moreau CS, Baldo L, Stahlhut JK, Werren JH, et al. Specialization and geographic isolation among Wolbachia symbionts from ants and lycaenid butterflies. Evolution (N Y). 2009;63:624–40.
Ilinsky Y, Kosterin OE. Molecular diversity of Wolbachia in Lepidoptera: prevalent allelic content and high recombination of MLST genes. Mol Phylogenet Evol. 2017;109:164–79.
Newton ILG, Savytskyy O, Sheehan KB. Wolbachia utilize host actin for efficient maternal transmission in Drosophila melanogaster. PLoS Pathog. 2015;11:e1004798.
Haselkorn TS, Markow TA, Moran NA. Multiple introductions of the Spiroplasma bacterial endosymbiont into Drosophila. Mol Ecol. 2009;18:1294–305.
Duplouy A, Pranter R, Warren-Gash H, Tropek R, Wahlberg N. Towards unravelling Wolbachia global exchange: a contribution from the Bicyclus and Mylothris butterflies in the Afrotropics. BMC Microbiol. 2020;20:1–9.
Neethiraj R, Hornett EA, Hill JA, Wheat CW. Investigating the genomic basis of discrete phenotypes using a Pool-Seq-only approach: new insights into the genetics underlying colour variation in diverse taxa. Mol Ecol. 2017;26:4990–5002.
Wood DE, Lu J, Langmead B. Improved metagenomic analysis with kraken 2. Genome Biol. 2019;20:1–13.
Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–4.
Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. Jvenn: An interactive Venn diagram viewer. BMC Bioinformatics. 2014;15:1–7.
Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WEG, Wetter T, et al. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–59.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77.
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–2.
Waterhouse RM, Seppey M, Simao FA, Manni M, Ioannidis P, Klioutchnikov G, et al. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 2018;35:543–8.
Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66.
Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30:3276–8.
Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES science gateway for inference of large phylogenetic trees. 2010 Gatew Comput Environ Work GCE. 2010;2010:1–8.
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3.
Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–9.
Letunic I, Bork P. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–8.
Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–9.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Hadley W. ggplot2: elegant graphics for data analysis - Hadley Wickham - Google Books; 2016.
Acknowledgements
Thanks to Dr. M. Gerth and Dr. F. Valerio for discussion on the phylogenomics analyses. Thank you to two anonymous reviewers for helpful comments on a version of this manuscript.
Funding
AD was funded by Marie Skłodowska-Curie Individual fellowship (#790531, Host Sweet Home), and by the Academy of Finland research fellowship (#321543). Open access funded by Helsinki University Library.
Author information
Authors and Affiliations
Contributions
AD designed the study, all authors collected and analyzed the data, all authors contributed to the manuscript, and read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 7. Figure S1:
Venn diagram representing the overlap between samples identified as having > 1000 reads associated with either A) Wolbachia or B) Spiroplasma. MetaPhlan results are represented in Green and Kraken in Blue.
Additional file 8. Figure S2:
Phylogenetic relationships between Wolbachia positive SRAs and reference sequences for the MLST gene coxA.
Additional file 9. Figure S3:
Phylogenetic relationships between Wolbachia positive SRAs and reference sequences for the MLST gene fbpa.
Additional file 10. Figure S4:
Phylogenetic relationships between Wolbachia positive SRAs and reference sequences for the MLST gene ftsz.
Additional file 11. Figure S5:
Phylogenetic relationships between Wolbachia positive SRAs and reference sequences for the MLST gene GatB.
Additional file 12. Figure S6:
Phylogenetic relationships between Wolbachia positive SRAs and reference sequences for the MLST gene hcpa.
Additional file 13. Figure S7:
Phylogenetic relationships between Wolbachia positive SRAs and reference sequences for the wsp gene.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Twort, V.G., Blande, D. & Duplouy, A. One’s trash is someone else’s treasure: sequence read archives from Lepidoptera genomes provide material for genome reconstruction of their endosymbionts. BMC Microbiol 22, 209 (2022). https://doi.org/10.1186/s12866-022-02602-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-022-02602-1