Abstract
Background
Pseudomonas aeruginosa is a common pathogen in Hospitalized patients, and its various resistance mechanisms contribute to patient morbidity and mortality. The main aims of the present study were to assess the susceptibility of biofilm-producing and non-producing P. aeruginosa isolates to the five commonly used Hospital disinfectants, to evaluate the synergistic effect of selected disinfectants and Ethylene-diamine-tetra acetic acid (EDTA), and the effect of exposure to sub-inhibitory concentrations of Sodium hypochlorite on antimicrobial susceptibility test.
Results
The results showed that sodium hypochlorite 5% and Ethanol 70% were the most and least effective disinfectants against P. aeruginosa, respectively. The addition of EDTA significantly increased the effectiveness of the selected disinfectants. The changes in the antibiotic-resistance profiles after exposure to sub-inhibitory concentrations of disinfectants were observed for different classes of antibiotics (Carbapenems, Aminoglycosides, Cephalosporins, Fluoroquinolones). As well as near the all isolates harbored efflux pump genes and 117 (97.5%) of isolates produced biofilm.
Conclusion
In the current study, the mixture of disinfectant and EDTA were the most suitable selection to disinfect Hospital surfaces and instruments. Also, it was clear that exposure to sub-inhibitory concentrations of Sodium hypochlorite results in resistance to some antibiotics in P. aeruginosa species. Strong and intermediate biofilm formers belonged to MDR/XDR strains. Future studies should include more complex microbial communities residing in the Hospitals, and more disinfectants use in Hospitals.
Similar content being viewed by others
Background
P. aeruginosa is a gram-negative bacilli and is known as human opportunistic pathogen, especially for high-risk patients, including burn wounds, immunocompromised patients, and cystic fibrosis [1, 2]. It is a member of the ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter species) the leading cause of nosocomial infections throughout the world [3]. Also, P. aeruginosa is one of the bacteria that the World Health Organization (WHO) named as antibiotic‐resistant “priority pathogens” and has acquired and expanded different kinds of resistance mechanisms [3,4,5]. P. aeruginosa infections are difficult to treat because of their ability to acquire resistance to antibiotics [6]. P. aeruginosa possesses both cell-associated (lipopolysaccharide, flagella, alginate/biofilm pili, lectins) and extracellular (cytotoxin, proteases, hemolysins, siderophores, pyocyanin, exoenzyme S, exotoxin A, exoenzyme U, etc.) virulence factors [7]. Multi-drug-resistant (MDR) or Extensively drug-resistant (XDR) strains, in the nosocomial base, are a global threat to health care systems and vulnerable patients, and they have been reported to cause a large number of Hospital incidence in high-risk patients such as patients admitted to intensive care units (ICUs) [1]. As a general rule, MDR is defined as acquired non-susceptibility to at least one agent in ≥ 3 antimicrobial categories and XDR is referred as non-susceptibility to at least one agent in all categories but sensitive to ≤ 2 antimicrobial categories [2]. Over the years, selective pressure by administration of different classes of antibiotics has resulted in micro-organisms bearing additional types of resistance mechanisms that led to MDR (enzymatic mechanisms of drug modification, enhanced efflux pump expression, novel penicillin-binding proteins (PBPs), mutated drug targets, and altered membrane permeability) [8]. Bacterial resistance to antibiotics is due to acquired or intrinsic mechanisms [8]. Inadequate surveillance, misuse of antibiotics, and excess administration of antibiotics in the livestock industry resulted in the appearance and spread of MDR/XDR bacteria all over the world [3, 9].
Today, resistance to antibiotics and disinfectants in various bacterial strains is a major public health problem in the world, with increasing growth worldwide. Reports of this have generally been based on the detection of antibiotic resistance in bacteria associated with nosocomial infections, but in recent years with the identification of MDR, strains in different countries, ways to spread and spread the relevant genes are important [4, 5].
One of the most important factors in the spread of nosocomial infections is the improper use of disinfectants. As none of the disinfectants are equally suitable for all different disinfection needs, studies are needed to determine the disinfectant effects of different disinfectants so that you can choose the right disinfectant [5,6,7].
Improper use of disinfectants, dilution in the environment after discharge, and biodegradation result in biocide concentration gradients. As a result, microorganisms are alternately exposed to non-lethal concentrations of disinfectants. Recent studies have shown that, when bacteria are exposed to non-lethal concentrations of disinfectants, it facilitates resistance to disinfectants and may also lead to resistance to other antimicrobials, such as antibiotics [10,11,12,13,14]. There is a growing concern that the widespread use of disinfectants has also been involved in antibiotic resistance [15, 16].
P. aeruginosa have mechanism of developing resistance to disinfectants and antibacterial agents. Among these, small multidrug resistance (SMR) proteins are located in the inner layer of the cytoplasmic membrane and are divided into three groups: SUG, SMP, and PSMR. They cause resistance to biocides and a number of antibiotics. The genes qacE and qacEΔ1 are located in the SMP subgroup and have been identified on plasmids and integrons of many gram-negative and gram-positive bacteria that are resistant to drugs. qacEΔ1 gene is a mutation of the qacE gene [17]. SUG genes are also located on the plasmid. The SMR family includes proton-dependent efflux pumps [18]. Qac genes is resulted in resistance against quaternary ammonium compounds, also these genes code for resistance to a broad spectrum of other cationic compounds such as biguanides, diamidines and intercalating dyes [19]. In P. aeruginosa the efflux pumps prevent the formation of lethal concentrations of toxic compounds by directing extracellular compounds (antibiotics, chlorhexidine, various drugs, etc.) out of the bacteria, and promotes the survival of the pathogen. Increased expression of efflux pump genes causes MDR isolates [20, 21]. Close association between resistance to antibiotics and biocides can be explained by the fact that a variety of antibiotic resistance genes are harbored by class 1 integrons (mobile genetic elements). Therefore, there is a global concern that the inappropriate use of disinfectants could select gram-negative antibiotic-resistant bacteria [22, 23].
P. aeruginosa use biofilm formation as another mechanism to resist disinfectants [24]. One of the purposes of the current study is to determine the correlation between antibiotic/detergent resistance and biofilm formation in P. aeruginosa. Treatment of P. aeruginosa infections is challenging, due to the acquired and intrinsic resistance of P. aeruginosa against a wide range of antibacterial agents [25, 26], biofilm formation, enzyme production suppression, and overexpression of efflux pumps, known as resistance mechanisms of this microorganism [27].
The present study identified resistance and sensitivity to common disinfectants in two steps: with or without Ethylene-diamine-tetra-acetic acid (EDTA). The disinfectants examined in the current study were Sodium hypochlorite 5%, Ethanol alcohol 70%, Sayasept- HP 2%, Chlorhexidine 2%, Dettol 4.8%. Based on the studies conducted in Hospitals in Iran, as well as surrounding countries, disinfectants were selected for this study. These disinfectants are also widely used worldwide for disinfection purposes. Detection of efflux pump genes (qac-E, qacE-Δ1, and sug-E1) by PCR technique was performed. P. aeruginosa strains were also evaluated in terms of biofilm production. Also, the relationship between resistance to disinfectant, and biofilm production was assessed. In the present study, the relationship between resistance to disinfectants, and antibiotics was investigated. The effect of exposure to sub-inhibitory concentrations of Sodium hypochlorite on antimicrobial susceptibility test was determined.
Results
A total of 120 (12.1%) P. aeruginosa strains were collected from 986 clinical specimens of Hospitalized patients. Out of 120 obtained isolates, 67 (55.8%) were from Urine, 11 (9.2%) of them from Broncho Alveolar Lavage (BAL), and 28 (23.3%), 14 (11.7%) of them were from blood and wound respectively (Fig. 1). In the current study, all isolates were identified by biochemical tests (Table 1).
Antimicrobial susceptibilities
The highest resistance rate was against Cefoxitin and Ampicillin/Sulbactam (100%) followed by Imipenem (45.8%) and Levofloxacin (33.3%). The highest susceptibility rate was related to Colistin 116 (96.7%), Piperacillin/Tazobactam 94 (78.3%), Piperacillin 93 (77.5%), Tobramycin 93 (77.5%), and Gentamicin 91 (75.8%) respectively (Table 2 and 3) (Fig. 2A).
Table 2 shows the percentage of antibiotic resistance and susceptibility by each antibiotic. A comparative table of the results of antibiotics susceptibility tests before and after exposure to Sodium hypochlorite is shown in Table 3. Base on susceptibility testing results, 65(61.7%) and 19(15.8%) isolated strains were categorized as MDR and XDR strain respectively.
The changes in the antibiotic-resistance profiles after exposure to sub-inhibitory concentrations of Sodium hypochlorite were observed for different classes of antibiotics (Table 3). Most of P. aeruginosa strains showed increased resistance to different kinds of classes of antibiotics, with respect to resistance Meropenem 13(10.8%), Ciprofloxacin 12 (10%), Tobramycin and Gentamicin 10 (8.3%), Imipenem and Cefepime 9 (7.5%), Amikacin 7 (5.9%), Ceftazidime and Levofloxacin 6 (5%) showed the most changes. As a result, the rate of MDR 16 (15.4%) and XDR 4 (3.4%) increased (Fig. 2B).
Determination of minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) of disinfectants
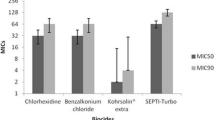
In this study the most effective disinfectants were Sodium hypochlorite 5%, Chloroxylenol (Dettol) 4.8%, Sayasept-HP 2%, Chlorhexidine 2%, and Ethanol 70%, respectively. Moreover, the disinfectant susceptibility testing in this study confirmed 1/32 concentration of Sodium hypochlorite and Dettol have lethal effect (MBC) on 120 (100%) of the P. aeruginosa isolates. On the other side, the less effective disinfectant was Ethanol 70% and 59(49.2%) of isolates did not show the lethal effect on 4.37% concentration of Ethanol (Table 4). Also results of the current study imply that higher concentration of disinfectants (higher MBC) needs to kill MDR/XDR isolates (Fig. 3 A-E).
Synergistic effect of selected disinfectants and Ethylene-diamine-tetra acetic acid (EDTA)
Adding EDTA increased the efficiency of all disinfectants included in this study. At the concentrations used, disinfectants without EDTA were efficient at higher concentrations in comparison with mixed EDTA and, when combined with EDTA, a reduction of concentration was observed in MBC and MIC. The effects of EDTA and disinfectants were additive. Ethanol 70%, Sodium hypochlorite 5%, Sayasept-HP 2%, respectively gave the best results when combined with EDTA, although additive effect of EDTA with Chloroxylenol (Dettol) 4.8%, and Chlorhexidine 2% were lower than that of other antiseptics (Table 4). A comparative diagram of MBC disinfectants at different concentrations before and after mixing with EDTA is shown in Figs. 4 and 5.
Biofilm assessment
In this part, 117 (97.5%) of clinical P. aeruginosa isolates were found to develop biofilm, and 45 (37.5%), 51 (42.5%), and 21 (17.5%) isolates were strong, intermediate, and weak biofilm formers respectively, compared to the reference strain. 37 (30.8%) and 28 (23.3%) of strong and intermediate biofilm formers belong to MDR/XDR strains. Also, in the case of strong biofilm-producing isolates, higher concentrations of disinfectants were used to kill the isolates (MBC). Our results indicate, the higher concentrations of biocides (MBC) should be provided to kill strong and moderate biofilm-producing isolates (Fig. 6 A-E).
Detection of efflux pump genes (qac-E, qacE-Δ1, sug-E1) by PCR technique
Genomic detection of qacE, qacΔE1, and sug-E1 showed that 111 (92.5%), 21 (17.5%), and 114 (95%) out of 120 P. aeruginosa isolates harbor the qacE, qacΔE1, and sug-E1 genes, respectively. Among the isolates carrying the qacEΔ1 gene, 16 isolates were MDR / XDR (Fig. 7).
Discussion
Since 1980, nosocomial infections, caused by P. aeruginosa have been classified as a big issue in Hospital, as a result of this problem, medical costs for health care systems have been high [28]. Many studies have shown that disinfectants and antibiotics efficacy is gradually reduced [29]. For this reason, one of the aim of this study was to assess the susceptibility of the isolates to the antibiotics and disinfectants. There are several reasons for the prevalence of disinfectants resistance: inaccurate concentration, inappropriate usage, and insufficient training to prepare and store Hospital disinfectants are among them [30]. Compared to many resistance surveys about antibiotics, the number of global research regarding biocides resistance is insufficient. Due to the clinical importance of P. aeruginosa, the efficacy of five Hospital disinfectants was assessed against clinical isolates of P. aeruginosa.
The effectiveness of Sodium hypochlorite in the active ingredient concentration of 0.078% at 37 °C for 18–24 h was determined as MBC for all the isolates included in this study, which is more effective between most concentrated active ingredients of different disinfectants in the present study. These results are consistent with another study performed on P. aeruginosa, and Sodium hypochlorite 0.5% has been shown to be more effective than Ethanol and Savlone [31]. The results in a study conducted in Brazil showed that Sodium hypochlorite is more effective than ammonium tetravalent compounds against bacteria. In our study, Sodium hypochlorite was more effective than Sayaspet, which is a fifth-generation of ammonium tetravalent compounds [32]. The results of other studies on the active ingredient Sodium hypochlorite confirm the results of our work [33,34,35]. Also, the results of the current study showed that eradicating of MDR/XDR isolates needs higher concentrations of disinfectants. This highlights the importance of using the suitable disinfectant at the right concentration to kill MDR/XDR bacteria. Given that bacteria are becoming resistant to disinfectants, new disinfectants with new compounds or mixtures must be considered to kill these isolates.
Currently, EDTA has been approved as an antimicrobial agent to reduce the risk of bacterial biofilm formation and colonization. EDTA is known as a metal chelator and disrupts the outer lipopolysaccharide layer of Gram-negative bacteria, and the membrane becomes more penetrable to disinfectants [36]. Another goal of the current study was to determine the synergistic effect of EDTA in combination with five other non-antibiotic antimicrobials. In the current survey, the addition of EDTA increased the efficacy of selected disinfectants significantly. The results showed that disinfectants are able to kill MDR/XDR isolates in lower concentrating with the mixture of the EDTA. A study reported that tetra-sodium EDTA 4% is able to eradicate pre-formed biofilms of clinical isolates [37]. Some surveys revealed that combined antibiotics are more effective compared to single antibiotics [38]. The efficacies of disinfectants currently are being investigated in order to decrease the rate of emerging resistance among clinically isolated bacteria. For decades, EDTA has been known as a potentiating and sensitizing agent. Several studies showed that the action of EDTA biofilm disrupting is due to its ability to cations sequestering (Mg2 + , Ca2 + , and Fe3 +), as a result, increases the effect of other antimicrobial agents [39,40,41]. The combination of antibiotics and other antimicrobial agents with disodium EDTA has been broadly studied [39, 42, 43]. A study was conducted on Candida and methicillin-resistant S. aureus (MRSA), which a combination of Ethanol (25%), EDTA (30 mg/mL), and Minocycline (3 mg/mL) synergistically eradicated pre-formed biofilms [39]. Another survey has been performed on common pathogens involved in canine otitis especially Pseudomonas, which revealed that the combination of Tris–EDTA with Chlorhexidine 0.15% has excellent synergistic activity against all isolates [44]. These trials propose that the combination of Chlorhexidine or Ethanol with EDTA does not compromise the activity of one another. However, standard EDTA or disodium EDTA is not a potent and practical antimicrobial agent, even when used at high-level concentrations, and is not able to kill bacteria. On the other side, some studies showed that tetra-sodium EDTA has broad-spectrum antimicrobial activity on its own [37, 41]. It was reported, tetra-sodium EDTA (40 mg/ mL) decreased biofilm colonization by P. aeruginosa, S. epidermidis, K. pneumoniae, C. albicans, and E. coli on catheter segments [41]. In another survey killing ability of tetra-sodium EDTA 4% against clinically relevant pathogens was reported, and 4% tetra-sodium EDTA kill clinically significant bacterial and fungal pathogens isolated. The tetra-sodium EDTA solution was able to kill all microorganisms tested, at a concentration of 4% or less, and in less than 24 h of exposure [37].
There are studies that are significant to attain a better understanding of the interaction between bacteria and biocides and emerging resistance and cross-resistance in bacteria [45, 46]. In this study, there was a significant difference between the results of the antibiogram before and after exposure to Sodium hypochlorite in most antibiotics. The reason for choosing Sodium hypochlorite to study the effect of sub-inhibitory on P. aeruginosa was that it is widely used in most countries in inappropriate concentrations. Our results showed that after exposure to Sodium hypochlorite 16 isolates from antibiotic-sensitive group, categorized as MDR, and among them 4 isolates became XDR. It is worth bearing in mind that, the in-use concentration of disinfectants, in most times is 1000 times greater than of their MIC, to gain a rapid killing of bacteria. Biocide at high-level concentration usually interacts with several targets in the bacterial cell. For this reason, bacteria hardly become resistant via adaptation or other mechanisms. However, bacterial are usually exposed to sub-inhibitory concentrations of biocides. It has been shown that bacteria exposed to sub-inhibitory concentrations of biocides result in increased resistance to biocides and antibiotics in bacteria [12,13,14, 47]. In a study conducted in 2017, bacteria-harboring biocide resistance genes were more, probable to harbor an antibiotic resistance gene in comparison with bacteria lacking biocide resistance genes [48]. In the current study, we analyzed the correlation between biocides and antibiotics. Positive connections were detected between exposing to the sub-inhibitory concentration of biocides and antibiotic resistance. Such relationships were extensively reported and often involved the up-regulation of efflux pumps [49].
Usage of disinfectants in Hospitals must be intently measured and re-evaluated due to the selection pressure effect of antimicrobials on the advent of resistant bacteria which could be spread to Hospitalized patients. For this reason, resistance is inducible after being exposed to the sub-inhibitory level of disinfectants (sodium hypochlorite), which results in an increase in the isolates resistant to some antibiotics. Therefore, it is noteworthy to evaluate some disinfectants and assess correlations with antibiotic resistance, which should be considered for disinfection practices. It is not correct to consider bacteria that grow in low concentrations of disinfectants as resistant to biocides. This must be determined as ‘increasing MIC value’ or reducing susceptibility, and as a result, it is important to evaluate the bactericidal concentration instead of the inhibitory concentration of disinfectants [50]. It should be noted that the results of different surveys and the methods employed must also be considered. However, opposed results show that there is not any correlation between antibiotic resistance and exposure to sub-lethal concentrations of biocides, which may be due to different bacterial species selected (Listeria monocytogenes) or the type of disinfectant selected, which indicates the importance of research on the induction of resistance in different bacterial species with different disinfectants [51]. It is not completely obvious that there is a correlation between biocide resistance and antibiotic resistance, and surveys still continue in this subject [52, 53]. In conclusion, it is clear that biocide concentration is a significant element in bacterial resistance induction. In hospitals, bacteria are exposed to a low concentration of biocide if the disinfectant prepares at low concentrations, and if the diluted disinfectant is kept for a long time [50].
A vital key used by P. aeruginosa to survive in harsh environments such as exposure to antibiotics agents is biofilm formation [54]. The National Institute of Heart, Blood, and Lung reported that up to 80% of all infections caused by bacterial are related to biofilm formation [55]. The results of our study showed that 117 (97.5%) isolates formed biofilm, which was similar to other studies [56]. The results of our study revealed that isolates that produced strong and intermediate biofilm are more resistance to antibiotics and disinfectants. Similarly, antibiotic resistance has increased by biofilm formation, resulting in higher antibiotic concentrations in MDR P. aeruginosa isolates infections [57]. A study indicated that the rate of P. aeruginosa biofilm formation from Iranian patients varied from 48.5% to 99.5%. Generally, the biofilm formation ratio was reported as 87.6%. As well, 27.4%, 30.2%, and 47.7% of P. aeruginosa isolates were weak, moderate, and strong biofilm producers, respectively [24]. Accordingly, our data were aligned with the results published in studies where 40–100% of P. aeruginosa isolates produce biofilm [58, 59]. Karami et al., reported 73% of both clinical and environmental isolates were biofilm producers [60]. Also, other studies have revealed the importance of biofilm formation by P. aeruginosa [61, 62]. In line with our study, it was reported that 58.6% of MDR isolates produce strong biofilm. These results revealed a significant correlation between biofilm formation and MDR isolates [63]. It should be noted that, in contrast to these findings, some studies from different parts of the world indicated a lower prevalence of biofilm formation, and there was no correlation between antibiotic resistance and biofilm-producing [64, 65]. This issue is possibly linked to other resistance mechanisms (efflux pumps, purines, chromosomal mutation, and plasmid acquisition) involved in antibacterial resistance [66].
Another aim of the present study was to investigate the prevalence of qacE∆1, qacE, and sug-E1 genes, and their relationship with resistance to antibiotics and biocides in P. aeruginosa. In the current study, the prevalence of efflux pump genes was very high, and due to the high prevalence of qacE and sug-E1 genes, no association was found between these genes and resistance to disinfectants and antibiotics. It should be noted that, 21 isolates carrying the qacE∆1 gene. Between them 16 isolates were MDR or XDR, that indicating an association between this gene and antibiotic resistance. The prevalence of qacE ∆1 and sug-E1 genes in our study were consistent with findings of other studies [67,68,69].
Conclusions
In the current study, most isolates produced strong and moderate biofilm. The results indicated that strong and moderate biofilm formation isolates need higher concentration (MIC and MBC) of disinfectant for killing. It should be noted that most of MDR/XDR isolates produced strong and moderate biofilm. The present study indicated that EDTA has a significant additive effect in increasing the lethality (MBC) and inhibitory (MIC) power of disinfectants. It is worth noting that EDTA, as a chelating agent of divalent compounds, destroys the biofilm. Therefore, it is suggested to use alternative compounds such as EDTA in combination with disinfectant to increase the potency of disinfectant by creating synergistic effects against MDR/XDR P. aeruginosa isolates. EDTA is also fully biodegradable, and has no toxic effects on humans, which is less harmful to the environment and human health than other disinfectants. Collectively, our results revealed that exposure to a sub-inhibitory concentration of Sodium hypochlorite can induce resistance to some antibiotics in P. aeruginosa. The results are important because the cross-link between exposure to the Sodium hypochlorite and antibiotic resistance was observed in at least nine antibiotics of the fourteen tested. Appropriate concentration of disinfectants is a critical key to the eradication of bacteria in Hospitals. We also strongly recommend better training on the correct use of disinfectants in Hospitals, since preserving the efficacy of disinfectants is crucial to maintaining hygiene levels in Hospitals and reducing the need for using antimicrobials. This study also showed that Sodium hypochlorite has high lethality and inhibitory power, and Ethanol alcohol has low lethality against isolates of this study. Future studies should include more complex microbial communities residing in the Hospitals, additional pseudomonas strains as well as other detergents typically used to clean and disinfect the Hospital surfaces and medical instruments. Based on this study appropriate use of disinfectants at proper concentrations for different species of bacteria should be addressed to avoid inducing resistance mechanisms in bacteria. Also, to the field of study using EDTA in combination with disinfectants should be addressed.
Materials and methods
Bacterial strains
A cross-sectional study was performed from April 2019 to July 2020, by approval of the Ethics Committee of Qazvin Medical University (IR.QUMS.REC.1398.156). A total of 120 samples of P. aeruginosa were isolated from 986 clinical specimens of Hospitalized patients. Isolates from urine, wound, blood cultures, and BAL were included in this study. All isolates were identified on the basis of cultural, biochemical, and morphological characteristics as per Bergey’s Manual of Systemic Bacteriology [70]. Standard laboratory methods such as growth on Cetrimide agar medium, Triple sugar iron agar, oxidase test, catalase test, Methyl red test, Voges Proskauer test, Citrate utilization, Urease test (Merck, Darmstadt, Germany), Motility test, grow at 42 °C, pigment production was performed to identify P. aeruginosa strain. For further investigation, all P. aeruginosa isolates cultured in trypticase soy broth (TSB) then were supplemented with 15% glycerol and were stored at − 20 °C. In the next stage, all isolates were confirmed by PCR method. All bacterial culture media were purchased from the Merck, Darmstadt, Germany.
Antibiotic susceptibility testing
Antibiotic susceptibility test (AST) was done using the disk diffusion method, based on the Clinical and Laboratory Standard Institute CLSI 2020 guideline [71] on Mueller–Hinton agar (Merck, Darmstadt, Germany).
Accordingly, a platinum loop was used to pick 3–5 pure P. aeruginosa colonies and transferred to a tube containing 5 ml sterile nutrient broth. The isolates were grown in nutrient broth and incubated at 370C until the turbidity was adjusted with the 0.5 McFarland standards (4–6 h). The suspension was evenly swabbed over the surface of Mueller Hinton agar. The inoculated plates were then incubated at 37 °C for 18–24 h. Diameters of the zone of inhibition around the discs were measured, and the isolates were categorized as resistant, intermediate, and sensitive according to the standardized table supplied by the CLSI2020. P. aeruginosa ATCC 27,853 was used as control.
Antibiotic disks (MAST Diagnostics, Merseyside, UK) tested were as follows: Piperacillin [30], Piperacillin/Tazobactam (PTZ, 100 μg/10 μg), Ceftazidime (CAZ, 30 μg), Levofloxacin (LEV, 5 μg), Amikacin (AN, 30 μg), Imipenem (IMI, 10 μg), Gentamicin (GM, 10 μg), Meropenem (MEM, 10 μg), Tobramycin (TOB, 10 μg), Ciprofloxacin (CIP, 5 μg), Cefepime (CPM, 30 µg). Cefoxitin (30 μg), Ampicillin/Sulbactam (10 μg /10 μg). P. aeruginosa (ATCC 27,853) and Escherichia coli (ATCC 25,922) was used as control. The MICs for Colistin were determined using the MIC (micro broth dilution) method, then incubated at 37 °C for 18–24 h. Colistin susceptibility was interpreted according to the CLSI 2020 clinical breakpoints (≥ 4 = resistance, ≤ 2 = intermediate) [71].
To determine the effect of exposure to sub-inhibitory concentrations of Sodium hypochlorite on antimicrobial susceptibility, an antibiogram was done after exposing to sub-inhibitory concentrations of Sodium hypochlorite, and results were compared with before exposing. For this goal, an antibiogram test was performed for bacteria that had grown in the highest concentration of Sodium hypochlorite. The test was performed in the same manner as described for determining the MIC and MBC. Briefly, 20 µL of the suspension in the MIC wells were aseptically transferred to the 5 ml sterile nutrient broth. The isolates were grown in nutrient broth and incubated at 370C until the turbidity was adjusted with the 0.5 McFarland standards (4–6 h). The suspension was evenly swabbed over the surface of Mueller Hinton agar. The inoculated plates were then incubated at 37 °C for 18–24 h. Diameters of the zone of inhibition around the discs were measured, and the isolates were categorized as resistant, intermediate, and sensitive according to the standardized table supplied by the CLSI2020. P. aeruginosa ATCC 27,853 was used as control.
Determination of disinfectants MICs and MBCs
In Iranian Hospitals, Ethanol 70%, Chlorhexidine 2%, Sodium hypochlorite 5%, Chloroxylenol (Dettol) 4.8%, Sayasept- HP 2% are the most applicable disinfectants (Table 5). The MICs and MBCs of the all mentioned disinfectants were assessed by broth micro-dilution method (micro titer assay, 96-well plate) [72]. Briefly, 100 μl nutrient broth was added to all wells. Then 100 μl of disinfectant was added to well one, after serial dilution, 100 μl of bacterial inoculum (106 cfu/ml) was added to all wells. Micro plates were then incubated at 37 °C for 24 h [31, 73, 74]. The dilutions included 1/2, 1/4, 1/8, 1/16, 1/32, 1/64, 1/128, 1/256, and 1/512, Active ingredient of disinfectants are available in Table 6. MICs and MBCs were calculated. wells 11 and 12 are positive (TSB + inoculation) and negative (TSB + antimicrobial) controls [75,76,77]. The lowest concentration of biocide that inhibits bacterial growth and does not show turbidity is reported as the minimum inhibitory concentration (MIC). Subsequently, 100 μl of four final clear diluted wells of each disinfectant were cultured on Muller Hinton agar medium and if after 48 h at 37° C 99.9% of the bacteria did not grow (i.e. no growth or growth Less than 15 colonies) That dilution is considered the minimum bactericidal concentration (MBC) [78, 79]. Rideal-Walker Phenol Coefficient Test was used to determine the efficacy of the disinfectants (Table 7) [80].
Efficacy of Ethylene-diamine-tetra acetic acid (EDTA) on selected biocides
For this purpose, the selected disinfectants were mixed equally with the EDTA 17% and placed at room temperature for 15 min. Then, for all isolates MIC and MBC with a new mixture were calculated. The obtained results were compared with the previous results and its synergistic effect was investigated [39, 43].
Assessment of biofilm formation capacity
The biofilm-forming ability was determined using the crystal violet staining method in triplicates and repeated three times for each strain, as previously described [81, 82]. P. aeruginosa ATCC 27,853 was used as a positive control, and LB medium was used as a negative control [81]. The bacterial isolates were inoculated with turbidity equal to 0.5 McFarland (1.5 × 108 CFU mL− 1). A 200-µL suspension was incubated in each well at 35 °C. After 48 h, the wells were washed three times with phosphate buffer. Following incubation with 1% crystal violet dye (200 µL/well) at 25˚C for 20 min, the wells were washed three times with phosphate buffer and dried. Finally, Ethanol 95% (200µL/well) was added, and optical absorbance was measured at 550 nm (Thermo Scientific GmbH, Driesch, Germany). Biofilm formation was classified into four different groups using the following formulas: If OD < ODc, the biofilm was not formed (negative), If ODc < OD < 2xODc, the biofilm was weak, if 2xODc < OD < 4xODc, the biofilm was moderate. If 4xODc < OD, the biofilm was strong (Table 8) [81].
Molecular method for detection of antiseptic resistance gene
Detection of antiseptic resistance gene was performed by polymerase chain reaction (PCR) method; primer sequences used are available in Table 9. Genomic DNA was extracted using the boiling method [83]. Briefly, the isolates were cultured on trypticase soy agar (TSA) and incubated for 20 h at 37 °C. Three colonies were selected and inoculated into 400 μL of Tris–EDTA (TE) buffer, in the next stage heated at boiling temperature (100 °C) for 10 min, and then cooled down on the ice for 15 min. Next, the tube was centrifuged at 11,000 rpm, and the supernatant was used as genomic DNA for the PCR assay. For PCR amplification, each reaction was performed in a final volume of 25 μL containing 12.5 μL of 2 × Taq PCR Master Mix (SinaClon Bioscience Co, Tehran, Iran), 0.5 μL 10 pM of each forward and reverse primer, 3 μL of DNA template, and 8.5 μL Sterile distilled water [83]. The PCR conditions were as follows: pre-denaturation at 94 °C for 5 min, 30 cycles of DNA denaturation for 1 min at 94 °C, annealing based on Table 10, extension for the 50 s at 72 °C, and a final extension at 72 °C for 10 min. The PCR products were electrophoresed on 1% agarose gel with 100 V for 50 min, and stained with DNA safe stain. All primers included in this study designed by AlleleID6. In the next stage all primers were tested in the BLAST program at the NCBI Gene Bank and verified.
Statistical analysis
Descriptive statistics were used to measure the characteristics of the study. Pearson chi-square or Fisher's exact test and McNemar’s test was used to determine significant differences between proportion. P values of < 0.05 were considered significant. Statistical analysis was performed by using SPSS version 16.0 statistical software (SPSS Inc., Chicago, IL, USA).
Availability of data and materials
All data and material are available upon request to correspondence author.
Abbreviations
- MDR:
-
Multidrug-resistant
- XDR:
-
Extensively drug-resistant
- MIC:
-
Minimum inhibitory concentration
- MBC:
-
Minimum bactericidal concentration
- WHO:
-
World Health Organization
- EDTA:
-
Ethylene-diamine-tetra acetic acid
References
Buhl M, Peter S, Willmann M. Prevalence and risk factors associated with colonization and infection of extensively drug-resistant pseudomonas aeruginosa: a systematic review. Expert Rev Anti Infect Ther. 2015;13(9):1159–70.
Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas M, Giske C, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81.
Medina E, Pieper DH. Tackling threats and future problems of multidrug-resistant bacteria. How to overcome the antibiotic crisis. Current Topics in Microbiology and Immunology. 2016;398:3–33.
Elbadawi HS, Elhag KM, Mahgoub E, Altayb HN, Ntoumi F, Elton L, et al. Detection and characterization of carbapenem resistant Gram-negative bacilli isolates recovered from hospitalized patients at Soba University Hospital Sudan. BMC Microbiol. 2021;21(1):1–9.
de Abreu PM, Farias PG, Paiva GS, Almeida AM, Morais PV. Persistence of microbial communities including Pseudomonas aeruginosa in a hospital environment: a potential health hazard. BMC Microbiol. 2014;14(1):1–10.
Firesbhat A, Tigabu A, Tegene B, Gelaw B. Bacterial profile of high-touch surfaces, leftover drugs and antiseptics together with their antimicrobial susceptibility patterns at University of Gondar Comprehensive Specialized Hospital Northwest Ethiopia. BMC Microbiol. 2021;21(1):1–13.
Rutala WA, Weber DJ. Guideline for disinfection and sterilization in healthcare facilities, 2008. 2008. Update: May 2019.
Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128(6):1037–50.
WHO. Worldwide country situation analysis: response to antimicrobial resistance. Geneva: World Health Organization; 2015.
Tezel U, Pavlostathis SG. Quaternary ammonium disinfectants: microbial adaptation, degradation and ecology. Curr Opin Biotechnol. 2015;33:296–304.
Gadea R, Fuentes MÁF, Pulido RP, Gálvez A, Ortega E. Effects of exposure to quaternary-ammonium-based biocides on antimicrobial susceptibility and tolerance to physical stresses in bacteria from organic foods. Food Microbiol. 2017;63:58–71.
Molina-González D, Alonso-Calleja C, Alonso-Hernando A, Capita R. Effect of sub-lethal concentrations of biocides on the susceptibility to antibiotics of multi-drug resistant Salmonella enterica strains. Food Control. 2014;40:329–34.
Capita R, Riesco-Peláez F, Alonso-Hernando A, Alonso-Calleja C. Exposure of escherichia coli ATCC 12806 to sublethal concentrations of food-grade biocides influences its ability to form biofilm, resistance to antimicrobials, and ultrastructure. Appl Environ Microbiol. 2014;80(4):1268–80.
Webber MA, Whitehead RN, Mount M, Loman NJ, Pallen MJ, Piddock LJ. Parallel evolutionary pathways to antibiotic resistance selected by biocide exposure. J Antimicrob Chemother. 2015;70(8):2241–8.
Gerba CP. Quaternary ammonium biocides: efficacy in application. Appl Environ Microbiol. 2015;81(2):464–9.
Davis GS, Waits K, Nordstrom L, Grande H, Weaver B, Papp K, et al. Antibiotic-resistant Escherichia coli from retail poultry meat with different antibiotic use claims. BMC Microbiol. 2018;18(1):1–7.
Mahzounieh M, Khoshnood S, Ebrahimi A, Habibian S, Yaghoubian M. Detection of antiseptic-resistance genes in Pseudomonas and Acinetobacter spp. isolated from burn patients. Jundishapur J Nat Pharm Prod. 2014;9(2):e15402.
Bay DC, Rommens KL, Turner RJ. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim Biophys Acta. 2008;1778(9):1814–38.
Jaglic Z, Cervinkova D. Genetic basis of resistance to quaternary ammonium compounds--the qac genes and their role: a review. Vet Med. 2012;57(6):275-281.
Kampf G. Adaptive bacterial response to low level chlorhexidine exposure and its implications for hand hygiene. Microbial Cell. 2019;6(7):307.
Xavier DE, Picão RC, Girardello R, Fehlberg LC, Gales AC. Efflux pumps expression and its association with porin down-regulation and β-lactamase production among Pseudomonas aeruginosa causing bloodstream infections in Brazil. BMC Microbiol. 2010;10(1):1–7.
Gomaa FAM, Helal ZH, Khan MI. High prevalence of blaNDM-1, blaVIM, qacE, and qacEΔ1 genes and their association with decreased susceptibility to antibiotics and common hospital biocides in clinical isolates of Acinetobacter baumannii. Microorganisms. 2017;5(2):18.
Zhao W-H, Chen G, Ito R, Kimura S, Hu Z-Q. Identification of a plasmid-borne blaIMP-11 gene in clinical isolates of Escherichia coli and Klebsiella pneumoniae. J Med Microbiol. 2012;61(2):246–51.
Hadadi-Fishani M, Khaledi A, Fatemi-Nasab ZS. Correlation between biofilm formation and antibiotic resistance in Pseudomonas aeruginosa: a meta-analysis. Infezioni in Medicina. 2020;28(1):47–54.
Lila G, Mulliqi-Osmani G, Bajrami R, Kurti A, Azizi E, Raka L. The prevalence and resistance patterns of Pseudomonas aeruginosa in a tertiary care hospital in Kosovo. Infez Med. 2017;25(1):21–6.
Pournajaf A, Razavi S, Irajian G, Ardebili A, Erfani Y, Solgi S, et al. Integron types, antimicrobial resistance genes, virulence gene profile, alginate production and biofilm formation in Iranian cystic fibrosis Pseudomonas aeruginosa isolates. Infez Med. 2018;26(3):226–36.
Gholami A, Barati M, Vahdani M, Vahdani H, Karimi M. Pattern of empirical antibiotic administration in emergency department of an educational hospital in Tehran. Razi J Med Sci. 2011;18(82):17–23.
Tripathi A, Shukla S, Singh A, Prasad K. Prevalence, outcome and risk factor associated with vancomycin-resistant Enterococcus faecalis and Enterococcus faecium at a Tertiary Care Hospital in Northern India. Indian J Med Microbiol. 2016;34(1):38–45.
Suleyman G, Zervos MJ. Safety and efficacy of commonly used antimicrobial agents in the treatment of enterococcal infections: a review. Expert Opin Drug Saf. 2016;15(2):153–67.
Di Muzio M, Cammilletti V, Petrelli E, Di Simone E. Hand hygiene in preventing nosocomial infections: a nursing research. Ann Ig. 2015;27(2):485–91.
Mitiku M, Ali S, Kibru G. Antimicrobial drug resistance and disinfectants susceptibility of Pseudomonas aeruginosa isolates from clinical and environmental samples in Jimma University specialized hospital, Southwest Ethiopia. Ame J Biom Life Sci. 2014;2(2):40–5.
Bouzada MLM, Silva VL, Moreira FAS, Silva GA, Diniz CG. Antimicrobial resistance and disinfectants susceptibility of persistent bacteria in a tertiary care hospital. J Microbiol Antimicrob. 2010;2(8):105–12.
Nasr AM, Mostafa MS, Arnaout HH, Elshimy AAA. The effect of exposure to sub-inhibitory concentrations of hypochlorite and quaternary ammonium compounds on antimicrobial susceptibility of Pseudomonas aeruginosa. Am J Infect Control. 2018;46(7):e57–63.
Deshaies F, Ahmad D, Massicotte R, Pichette G, Belhumeur P, Mafu AA. Comparison of efficacy profiles for minimum lethal concentrations (MLCs) of some commonly used commercial hospital microbicidal detergent-disinfectant products for disinfectants and sporicidal activity. Int J Infect Control. 2012;v8i2.013.12.
Röhner E, Jacob B, Böhle S, Rohe S, Löffler B, Matziolis G, et al. Sodium hypochlorite is more effective than chlorhexidine for eradication of bacterial biofilm of staphylococci and Pseudomonas aeruginosa. Knee Surg Sports Traumatol Arthrosc. 2020:28(12):3912-8.
Brost A. Chelation-based enhancement of novel and commercially available antimicrobials against foodborne pathogens: Iowa State University; 2020.
Liu F, Hansra S, Crockford G, Köster W, Allan BJ, Blondeau JM, et al. Tetrasodium EDTA is effective at eradicating biofilms formed by clinically relevant microorganisms from patients’ central venous catheters. Msphere. 2018;3(6):e00525.
Dall G, Tsang SJ, Gwynne P, MacKenzie S, Simpson A, Breusch S, et al. Unexpected synergistic and antagonistic antibiotic activity against Staphylococcus biofilms. J Antimicrob Chemother. 2018;73(7):1830–40.
Raad I, Hanna H, Dvorak T, Chaiban G, Hachem R. Optimal antimicrobial catheter lock solution, using different combinations of minocycline, EDTA, and 25-percent ethanol, rapidly eradicates organisms embedded in biofilm. Antimicrob Agents Chemother. 2007;51(1):78–83.
Raad II, Hachem RY, Hanna HA, Fang X, Jiang Y, Dvorak T, et al. Role of ethylene diamine tetra-acetic acid (EDTA) in catheter lock solutions: EDTA enhances the antifungal activity of amphotericin B lipid complex against Candida embedded in biofilm. Int J Antimicrob Agents. 2008;32(6):515–8.
Percival SL, Kite P, Eastwood K, Murga R, Carr J, Arduino MJ, et al. Tetrasodium EDTA as a novel central venous catheter lock solution against biofilm. Infect Control Hosp Epidemiol. 2005;26(6):515–9.
Chauhan A, Lebeaux D, Ghigo J-M, Beloin C. Full and broad-spectrum in vivo eradication of catheter-associated biofilms using gentamicin-EDTA antibiotic lock therapy. Antimicrob Agents Chemother. 2012;56(12):6310–8.
Lebeaux D, Leflon-Guibout V, Ghigo J-M, Beloin C. In vitro activity of gentamicin, vancomycin or amikacin combined with EDTA or l-arginine as lock therapy against a wide spectrum of biofilm-forming clinical strains isolated from catheter-related infections. J Antimicrob Chemother. 2015;70(6):1704–12.
Guardabassi L, Ghibaudo G, Damborg P. In vitro antimicrobial activity of a commercial ear antiseptic containing chlorhexidine and Tris–EDTA. Vet Dermatol. 2010;21(3):282–6.
Henshaw DL, O’Carroll MJ. Scientific Committee on Emerging and Newly Identified Health Risks (SCENIHR). Brussels: European Commission; 2009.
Leitgeb N, Auvinen A, Danker-hopfe H, Mild K. SCENIHR (Scientific Committee on Emerging and Newly Identified Health Risks). Potential health effects of exposure to electromagnetic fields (EMF), Scientific Committee on Emerging and Newly Identified Health Risks SCENIHR Opinion on Potential health. 2016;10:75635.
Christensen EG, Gram L, Kastbjerg VG. Sublethal triclosan exposure decreases susceptibility to gentamicin and other aminoglycosides in Listeria monocytogenes. Antimicrob Agents Chemother. 2011;55(9):4064–71.
Pal C. Effects of biocides and metals on antibiotic resistance: a genomic and metagenomic perspective. 2017.
Buffet-Bataillon S, Le Jeune A, Le Gall-David S, Bonnaure-Mallet M, Jolivet-Gougeon A. Molecular mechanisms of higher MICs of antibiotics and quaternary ammonium compounds for Escherichia coli isolated from bacteraemia. J Antimicrob Chemother. 2012;67(12):2837–42.
Meyer B, Cookson B. Does microbial resistance or adaptation to biocides create a hazard in infection prevention and control? J Hosp Infect. 2010;76(3):200–5.
Kastbjerg VG, Gram L. Industrial disinfectants do not select for resistance in Listeria monocytogenes following long term exposure. Int J Food Microbiol. 2012;160(1):11–5.
Harbarth S, Soh ST, Horner C, Wilcox M. Is reduced susceptibility to disinfectants and antiseptics a risk in healthcare settings? A point/counterpoint review. J Hosp Infect. 2014;87(4):194–202.
Wales AD, Davies RH. Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics. 2015;4(4):567–604.
El-Ganiny AM, Shaker GH, Aboelazm AA, El-Dash HA. Prevention of bacterial biofilm formation on soft contact lenses using natural compounds. J Ophthalmic Inflamm Infect. 2017;7(1):1–7.
Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Brazilian J infect Dis. 2011;15(4):305–11.
Salman M, Rizwana R, Khan H, Munir I, Hamayun M, Iqbal A, et al. Synergistic effect of silver nanoparticles and polymyxin B against biofilm produced by Pseudomonas aeruginosa isolates of pus samples in vitro. Artif Cells Nanomed Biotechnol. 2019;47(1):2465–72.
Shi H, Trinh Q, Xu W, Zhai B, Luo Y, Huang K. A universal primer multiplex PCR method for typing of toxinogenic Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2012;95(6):1579–87.
Gurung J, Khyriem AB, Banik A, Lyngdoh WV, Choudhury B, Bhattacharyya P. Association of biofilm production with multidrug resistance among clinical isolates of Acinetobacter baumannii and Pseudomonas aeruginosa from intensive care unit. Indian J Crit Care Med. 2013;17(4):214.
de Almeida KdCF, Calomino MA, Deutsch G, de Castilho SR, de Paula GR, Esper LMR, et al. Molecular characterization of multidrug-resistant (MDR) Pseudomonas aeruginosa isolated in a burn center. Burns. 2017;43(1):137–43.
Karami P, Mohajeri P, Mashouf RY, Karami M, Yaghoobi MH, Dastan D, et al. Molecular characterization of clinical and environmental Pseudomonas aeruginosa isolated in a burn center. Saudi J Biol Sci. 2019;26(7):1731–6.
Vasiljević Z, Jovčić B, Ćirković I, Đukić S. An examination of potential differences in biofilm production among different genotypes of Pseudomonas aeruginosa. Arch Biol Sci. 2014;66(1):117–21.
Senturk S, Ulusoy S, Bosgelmez-Tinaz G, Yagci A. Quorum sensing and virulence of Pseudomonas aeruginosa during urinary tract infections. J Infect Dev Ctries. 2012;6(06):501–7.
Karami P, Khaledi A, Mashoof RY, Yaghoobi MH, Karami M, Dastan D, et al. The correlation between biofilm formation capability and antibiotic resistance pattern in Pseudomonas aeruginosa. Gene Rep. 2020;18:100561.
Kádár B, Szász M, Kristóf K, Pesti N, Krizsan G, Szentandrássy J, et al. In vitro activity of clarithromycin in combination with other antimicrobial agents against biofilm-forming Pseudomonas aeruginosa strains. Acta Microbiol Immunol Hung. 2010;57(3):235–45.
Hou W, Sun X, Wang Z, Zhang Y. Biofilm-forming capacity of Staphylococcus epidermidis, Staphylococcus aureus, and Pseudomonas aeruginosa from ocular infections. Invest Ophthalmol Vis Sci. 2012;53(9):5624–31.
Chen J, Su Z, Liu Y, Wang S, Dai X, Li Y, et al. Identification and characterization of class 1 integrons among Pseudomonas aeruginosa isolates from patients in Zhenjiang China. Int J Infect Diseases. 2009;13(6):717–21.
Wang C, Zhan Q, Mi Z, Huang Z, Chen G. Distribution of the antiseptic-resistance gene qacEΔ1 in 283 clinical isolates of Gram-negative bacteria in China. J Hosp Infect. 2008;69(4):394–6.
Helal ZH, Khan MI. QacE and QacEΔ1 Genes and Their Correlation to Antibiotics and Biocides Resistance Pseudomonas aeruginosa. Am J Biomed Sci. 2015;7(2):52-62.
Subedi D, Vijay AK, Willcox M. Study of disinfectant resistance genes in ocular isolates of Pseudomonas aeruginosa. Antibiotics. 2018;7(4):88.
Krieg NR, Manual H. Systematic bacteriology. Baltimore: Williams; 1984.
Clinical and Laboratory Standard Institute (CLSI). Performance standards for antimicrobial susceptibility testing. M100-S30th. Clinical and laboratory standards institute. 2020.
Montrucchio G, Corcione S, Sales G, Curtoni A, De Rosa F, Brazzi L. Carbapenem-resistant Klebsiella pneumoniae in ICU-admitted COVID-19 patients: keep an eye on the ball. J Global Antimicrob Resist. 2020;23:398–400.
Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: a review. J Pharm Anal. 2016;6(2):71–9.
Wu X, Fan W, Fan B. Synergistic effects of silver ions and metformin against enterococcus faecalis under high-glucose conditions in vitro. BMC Microbiol. 2021;21(1):1–9.
Mazzola PG, Jozala AF, Novaes LCdL, Moriel P, Penna TCV. Minimal inhibitory concentration (MIC) determination of disinfectant and/or sterilizing agents. Braz J Pharm Sci. 2009;45(2):241–8.
Babaei MR, Sulong A, Hamat RA, Nordin SA, Neela VK. Extremely high prevalence of antiseptic resistant quaternary ammonium compound E gene among clinical isolates of multiple drug resistant Acinetobacter baumannii in Malaysia. Ann Clin Microbiol Antimicrob. 2015;14(1):1–5.
Maertens H, De Reu K, Meyer E, Van Coillie E, Dewulf J. Limited association between disinfectant use and either antibiotic or disinfectant susceptibility of Escherichia coli in both poultry and pig husbandry. BMC Vet Res. 2019;15(1):1–12.
Kawamura-Sato K, Wachino J-i, Kondo T, Ito H, Arakawa Y. Reduction of disinfectant bactericidal activities in clinically isolated Acinetobacter species in the presence of organic material. J Antimicrob Chemother. 2008;61(3):568–76.
Zareniya M, Hallaj-Nezhadi S, Dinmohamadi F, Haghi F, Hassan M. Study the efficacy of antimicrobial activities of eight clinically applied disinfectants against clinical isolated of Enterococci and Pseudomonas aeruginosa. Pharm Sci. 2017;23(2):159–65.
Dapgh AN, Hakim AS, Shawky H, Ibrahim ES. Study of some disinfectants efficacy on Aeromonas hydrophila recovered from local animal and water sources. IOSR J Agric Vet Sci. 2019;12(2):41–7.
Rahimi S, Farshadzadeh Z, Taheri B, Mohammadi M, Haghighi M-A, Bahador A. The relationship between antibiotic resistance phenotypes and biofilm formation capacity in clinical isolates of Acinetobacter baumannii. Jundishapur J Microbiol. 2018;11(8):e74315.
Banar M, Emaneini M, Beigverdi R, Pirlar RF, Farahani NN, van Leeuwen WB, et al. The efficacy of lyticase and β-glucosidase enzymes on biofilm degradation of Pseudomonas aeruginosa strains with different gene profiles. BMC Microbiol. 2019;19(1):1–10.
Robatjazi S, Nikkhahi F, Niazadeh M, Marashi SMA, Peymani A, Javadi A, et al. Phenotypic identification and genotypic characterization of plasmid-mediated AmpC β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates in Iran. Curr Microbiol. 2021;78(6):2317–23.
Acknowledgements
We gratefully thank the four educational Hospitals in Qazvin, Iran (boali, Ghods, Kosar, and velayat). This work would not have been possible without their support of them.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Farhad Nikkhahi designed the study, Mehdi Bakht, Sara Rahimi, Raana Kazemzadeh Anari, Mohammad Rostamani performed the study. Mehdi Bakht and Sara Rahimi wrote the manuscript. Amir Javadi Performed statistical work on the article. Amir peymani revise the manuscript. Safar Ali Alizadeh, Amir peymani and Mahmood Amin Marashi were the project consultants. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The current study was performed by approval of the Ethics Committee of Qazvin Medical University with approval number IR.QUMS.REC.1398.156. In addition, the committee approved the utilization of human samples within this study. The clinical samples were taken as part of standard patient care and therefore no informed consent was applied from participants. Hospitals provided the clinical samples. Also, it should be noted that biological samples are handled by the authors in the present study. The adopted methods for handling human samples were carried out in accordance with relevant guidelines and regulations provided in the Declaration of Helsinki. The research protocol was approved by the Research Ethics Committee at the Qazvin Medical University, Iran.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bakht, M., Alizadeh, S.A., Rahimi, S. et al. Phenotype and genetic determination of resistance to common disinfectants among biofilm-producing and non-producing Pseudomonas aeruginosa strains from clinical specimens in Iran. BMC Microbiol 22, 124 (2022). https://doi.org/10.1186/s12866-022-02524-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-022-02524-y