Abstract
Background
T-helper cell type 1 (Th1) polarization in chronic immune thrombocytopenia (cITP) has been reported at the protein and mRNA levels. We evaluated the impact of Th1/Th2 cytokine and cytokine receptor functional polymorphisms on both susceptibility to, and severity of, cITP. We analysed IFN-γ + 874 T/A, IFN-γR -611G/A, IL-4 -590C/T, and IL-4Rα Q576R polymorphisms in 126 cITP patients (male/female: 34/92; median age: 47.7 years) and 202 healthy control donors. Genotyping was determined by PCR and direct sequencing. The Th1/Th2 ratio was detected in peripheral blood mononuclear cells via flow cytometry.
Results
cITP patients had a higher frequency of the IL-4Rα 576 non-QQ genotype compared to healthy subjects (P = 0.04). cITP patients with the IFN-γ +874 non-AA genotype (high expression type) showed more severe thrombocytopenia than those with the AA genotype (P < 0.05). cITP patients had a significantly higher Th1/Th2 ratio than control patients (P < 0.01); this ratio was inversely correlated with platelet counts. Furthermore, patients with both IFN-γ +874 non-AA genotype (high expression type) and IFN-γR −611 non-AA genotype (high-function type) had a significantly higher Th1/Th2 ratio (P < 0.05).
Conclusions
The cytokine polymorphisms affecting Th1/Th2 increase the susceptibility to, and severity of, chronic ITP.
Similar content being viewed by others
Background
Chronic immune thrombocytopenia (cITP) is an acquired immune-mediated disorder characterized by isolated thrombocytopenia. Several immune mechanisms contribute to the pathogenesis of cITP, including increased platelet destruction in the reticuloendothelial system and impaired platelet production in the bone marrow [1]. Recent studies have revealed that T helper (Th) type 1 (Th1) cytokine polarization occurs in cITP patients [2,3,4,5]; several investigators have also reported that the Th1/Th2 ratio is inversely correlated with disease progression [3].
Naive CD4+ T cells differentiate into several different Th cells, including Th1, Th2, Th17, and regulatory T (Treg) cells. Th cell functions are determined by their cytokine secretion patterns; Th1 and Th2 cells are defined by their ability to produce interferon-γ (IFN-γ) but not interleukin (IL)-4, and IL-4 but not IFN-γ, respectively [6,7,8]. Many investigators have shown IFN-γ to be upregulated at both the mRNA and protein levels in cITP patients [3, 9]. It was also reported that serum levels of IL-4 were decreased in cITP patients [9, 10]. Such findings demonstrate that Th1 polarization is consistent with characteristics of cITP. However, it remains unclear whether the influences of Th1/Th2 cytokines on cITP are due to genetic factors.
We investigated the existence of a role for Th1/Th2 cytokine and cytokine receptor functional polymorphisms, including IFN-γ +874 T/A, IFN-γR -611G/A, IL-4 -590C/T, and IL-4Rα Q576R, on susceptibility to cITP, as well as on its clinical features. Furthermore, we explored the association between the Th1/Th2 ratio and these polymorphisms in both healthy donors and cITP patients.
Methods
Patients and control subjects
In this study, 126 Japanese cITP patients (92 females and 34 males with a median age of 47.7 [range: 2.4–82.3] years), as well as 202 race- and sex-matched healthy subjects were examined. cITP was defined as isolated thrombocytopenia (platelet count <100 × 109/L) in the absence of other causes or disorders that may be associated with thrombocytopenia according to the criteria of the ITP International Working Group (IWG) [11]. cITP was also diagnosed if the disease lasted longer than 12 months [11]. “Very severe thrombocytopenia” was defined as a platelet count <10 × 109/L at presentation of cITP. “Severe bleeding tendency” was defined as gastrointestinal bleeding and cerebral haemorrhage [11]. The responses were assessed according to the criteria of the ITP IWG [11]. In these guidelines, a complete response was defined as a platelet count of at least 100 × 109/L, and a response was defined as a platelet count between 30 and 100 × 109/L on condition that it was at least double the baseline count. “Loss of R” was defined as a platelet count <30 × 109/L, a less than 2-fold increase in platelet count from baseline, or the presence of bleeding. “Corticosteroid-dependence” was defined as the ongoing need for continuous corticosteroid administration or frequent courses of corticosteroids to maintain a platelet count at or above 30 × 109/L and/or to avoid bleeding [11]. “Severe cITP” was defined by the presence of bleeding symptoms at presentation of severity sufficient to mandate treatment, or by the occurrence of new bleeding symptoms requiring additional therapeutic intervention via increasing the dose of the platelet-enhancing agent or replacing the agent [11]. “Refractory ITP” was defined as failure to achieve at least R or loss of R after splenectomy [11]. “Second-line treatment” was defined as additional therapy beyond glucocorticoids.
Genotyping
The IL-4 -590C/T (rs2243250), IL-4Rα Q576R (rs1801275), and IFN-γR -611G/A (rs1327474) genotypes were determined using the polymerase chain reaction restriction fragment length polymorphism method. Genomic DNA was extracted from whole blood using a DNA Isolation kit (Qiagen GmbH, Hilden, Germany). The reaction volume was 20 μL, comprising 1 μL of genomic DNA, 0.2 mM of dNTPs, 0.5 U of TaKaRa Ex Taq HS DNA polymerase (TaKaRa Bio, Japan), and 0.5 μM of each of 2 primers. We used the primers 5'- ACTAGGCCTCACCTGATACG -3' (forward) and 5'- GTTGTAATGCAGTCCTCCTG -3' (reverse) [12] for analysis of IL-4 -590C/T, the primers 5'- GCCCCCACCAGTGGCTACC -3' (forward) and 5'- GAGGTCTTGGAAAGGCTTATAC -3' (reverse) [13] for analysis of IL-4Rα Q576R, the primers 5'- CTCTTCATGAGAGGCTGTCT -3' (forward) and 5'- TAACTCTTGGAGTTCACCTGG -3' (reverse) [14] for analysis of IFN-γR -611G/A. Identification of the alleles at each polymorphic site was performed by incubating the PCR product with the restriction enzyme BsmFI (IL-4), MspI (IL-4Rα), and Hpy188I (IFN-γR) (New England Biolabs, Ipswich, MA, USA) followed by electrophoresis through a 2.0% agarose gel (for IL-4) or a 6% polyacrylamide gel (for IL-4Rα and IFN-γR).
Genotyping by allele-specific PCR
The IFN-γ +874 T/A (rs2430561) genotype was determined using the allele-specific PCR method. Genomic DNA was extracted from whole blood using a DNA Isolation kit (Qiagen). The reaction volume was 20 μL, comprising 1 μL of genomic DNA, 0.2 mM of dNTPs, 0.5 U of TaKaRa Ex Taq HS DNA polymerase (TaKaRa Bio, Japan), and 0.5 μM of each of 3 primers: 5'-TCA ACA AAG CTG ATA CTC CA-3' (common reverse), 5'-TTC TTA CAA CAC AAA ATC AAA TCT -3' (T allele specific forward), and 5'-TTC TTA CAA CAC AAA ATC AAA TCA-3' (A allele specific forward) [15]. The amplified product was analysed by electrophoresis on a 2% agarose gel.
Genotyping by sequencing
To confirm the accuracy of our assays, several PCR products were sequenced and analysed using an ABI Prism Genetic Analyzer (Applied Biosystems, CA, USA).
Flow cytometry for analysis of the Th1/Th2 ratio
We determined the Th1/Th2 ratio using flow cytometry as previously described by Ogawara et al. [2]. Whole heparinized blood was added to an equal volume of RPMI 1640 medium (Gibco, Grand Island, NY, USA) in 15 ml centrifuge tubes. Twenty-five ng/mL of phorbol 12-myristate 13-acetate and 1 μg/mL of ionomycin (Sigma-Aldrich, St. Louis, MO, USA) were added to all tubes except the resting controls; all tubes were supplemented with 10 μg/mL Brefeldin A (Sigma-Aldrich) except the activation controls. Tubes were incubated at 37 °C in 7% CO2 for four hours. After incubation with FACS lysing solution and FACS permeabilizing solution, cells were stained at 4 °C for 30 min with antihuman CD4-PE-Cy5 (BD Biosciences, CA, USA), FastImmune™ IFN-γ FITC/IL-4 PE (BD Biosciences) for intracellular cytokine staining and CD69 PE (BD Biosciences) for activation markers. FastImmune™ IgG2a FITC/IgG1 PE isotype control (BD Biosciences) and mouse IgG1 PE control (BD Biosciences) were used as negative controls. Three-color flow cytometric analysis was performed on a FACS Canto flow cytometer (BD Biosciences) using the FACS Diva software (BD Biosciences). Cells were logically gated on CD4 vs. side-scattered light (SSC) and forward-scattered light vs. SSC. Data were analysed using the FACS Diva software and displayed as dot plots of IFN-γ FITC vs. IL-4 PE. IFN-γ +and IL-4 − cells were defined as Th1 cells, while IFN-γ − and IL-4 + cells were deemed Th2 cells. Analysis of the stimulation effect was based on the fraction of CD69-positive cells after activation, as evaluated using histograms. In all analyses, CD69 positivity exceeded 95%.
Statistical analysis
The measured continuous data were expressed as mean ± standard deviation. Allele and genotype frequencies were analysed using the chi-square test, and clinical characteristics and treatment response were analysed using the chi-square test and student’s T test. The Th1/Th2 ratio and age were determined using the non-parametric Mann–Whitney U test. P-values were two-tailed, and P-values < 0.05 were considered statistically significant. We also compared the genotype frequencies with those calculated using the Hardy-Weinberg equilibrium theory (p2 + q2 + 2pq = 1, where q is the variant allele frequency).
Results
Patients’ characteristics
Of 126 patients, 92 were female (73.0%) and 34 were male (27.0%). Their median age at diagnosis was 47.7 years (range, 2.4–82.3 years). The platelet count ranged from 1.0 × 109/L to 96.0 × 109/L with a median count of 20.0 × 109/L at the initial diagnosis. The patients’ characteristics are shown in Table 1.
Genotype and allele frequencies of IFN-γ, IFN-γR, IL-4, and IL-4Rα polymorphism in patients with cITP and healthy controls
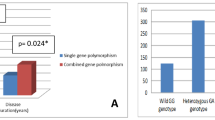
The genotype and allele frequencies of IFN-γ +874 T/A, IFN-γR -611G/A, IL-4 -590C/T, and IL-4Rα Q576R are shown in Table 2. The genotype distributions of these four polymorphisms in healthy subjects were similar to those observed in previous studies of Japanese populations [16,17,18,19,20,21]. Patients with cITP had a significantly lower frequency of the IL-4Rα 576 QQ genotype compared to healthy controls using a dominant model (69.8% vs. 79.7% respectively, odds ratio [OR] = 0.59, 95% confidence interval [CI] = 0.35–0.98, P = 0.04). However, no significant differences in the genotype frequencies of IFN-γ, IFN-γR, and IL-4 were observed between cITP patients and healthy controls using a dominant model and recessive model. Genotype frequencies of the four polymorphisms were in Hardy-Weinberg equilibrium in both cITP patients and healthy controls. These data show that IL-4Rα Q576R is associated with susceptibility to cITP.
The clinical characteristics of the patients with cITP and the treatment response according to IFN-γ, IFN-γR, IL-4, and IL-4Rα polymorphisms
We examined the association between the polymorphisms and the clinical characteristics of cITP patients (Tables 3 and 4). cITP patients with IFN-γ +874 non-AA genotype (high expression type) showed a lower minimum platelet count than those with an AA genotype (12.9 × 109/L ± 10.7 × 109/L vs 19.4 × 109/L ± 18.9 × 109/L, P = 0.045). However, there was no significant association between the other three genotype distributions and their various clinical features. We also explored the association between the four polymorphisms and treatment response (Tables 5 and 6). cITP patients with the IL-4 -590 cm3 genotype (low expression type) had a higher incidence of second-line treatment than those with non-CC genotypes (52.9% vs 25.7% respectively, OR = 3.25, 95% CI = 1.15–9.25, P = 0.04). No significant difference in treatment response was observed in other genotype distributions. These data suggest that Th1 polarization due to Th1/Th2 gene polymorphisms plays a role in the clinical features of cITP was well as in the response to treatment.
Th1/Th2 ratio in patients with cITP and healthy controls
The median Th1/Th2 ratio in patients with cITP was significantly higher than that of healthy controls (31.4, range 0.6–98.8 vs. 17.8, range, 2.2–52.5 respectively; P = 0.002) (Fig. 1a). As the median Th1/Th2 ratio was approximately 20 in the control group, we divided cITP patients into 2 groups; high Th1/Th2 (Th1/Th2 ratio ≥20) and low Th1/Th2 (Th1/Th2 ratio <20). The high Th1/Th2 group had a significantly lower platelet count at diagnosis than the low Th1/Th2 group (22.5 × 109⁄L, range 4.0–88.0 × 109⁄L vs. 53.0 × 109⁄L, range 2.0–86.0 × 109⁄L, respectively; P = 0.02) (Fig. 1b). The minimum platelet count during the clinical course in the high Th1/Th2 group was significantly lower than in the low Th1/Th2 group (median 13.0 × 109⁄L vs. 28.0 × 109⁄L respectively, P = 0.04) (Fig. 1c). These data suggest a role for the Th1/Th2 ratio in the pathogenesis of cITP.
Th1/Th2 ratio of patients with chronic ITP and healthy controls. a Th1/Th2 ratio of patients with chronic ITP was significantly higher than healthy controls (median, 31.4 vs. 17.8, P = 0.002). b Platelet count (×109⁄L) at diagnosis of chronic ITP with Th1/Th2 ratio ≧ 20 was significantly lower than that with Th1/Th2 ratio < 20 (median, 22.5 vs. 53.0, P = 0.02). c Minimum platelet count (×109⁄L) of chronic ITP with Th1/Th2 ratio ≧ 20 was significantly lower than that with Th1/Th2 ratio < 20 (median, 13.0 vs. 28.0, P = 0.04). d Th1/Th2 ratio of patients with the IFN-γ +874 non-AA genotype was significantly higher than that of the AA genotype (median, 71.5 vs. 27.5, P = 0.04). Th1/Th2 ratio of controls was similar in IFN-γ +874 polymorphism. e Th1/Th2 ratio of patients with the IFN-γR −611 non-AA genotype was significantly higher than that of the AA genotype (median, 78.5 vs. 28.4, P = 0.01). Th1/Th2 ratio of controls was similar in IFN-γR −611 polymorphism. f Th1/Th2 ratio of both patients and controls with the IL-4–590 non-CC genotype was not significantly different from that of the CC genotype. g Th1/Th2 ratio of both patients and controls with the IL-4Rα Q576R non-QQ genotype was not significantly different from that of the QQ genotype
Th1/Th2 ratio of both patients and healthy controls with IFN-γ, IFN-γR, IL-4, and IL-4Rα polymorphisms
Patients with the IFN-γ +874 non-AA genotype (high expression type) had a significantly higher Th1/Th2 ratio compared to those with the IFN-γ +874 AA genotype (low expression type) (71.5 [range 29.4–92.8] vs. 27.5 [range 0.6–98.8] respectively; P = 0.04) (Fig. 1d). Furthermore, patients with the IFN-γR −611 non-AA genotype (high-function type) had a significantly higher Th1/Th2 ratio compared to those with the IFN-γR −611 AA genotype (low function type) (medians, 78.5 vs. 28.4 respectively, P = 0.01) (Fig. 1e). However, there was no significant association between the Th1/Th2 ratio and IL-4/IL-4Rα polymorphisms (Fig. 1f, g). In contrast to cITP, there was no significant association between the Th1/Th2 ratio and all these four polymorphisms in the control group (Fig. 1d, e, f, g). These data confirm that Th1 polarization due to IFN-γ and IFN-γR polymorphisms is associated with the Th1/Th2 ratio in cITP.
Discussion
Although the immune mechanism that initiates cITP has not been identified, several processes of immune dysregulation have been reported [22]. It is well known that autoantibodies to GpIb/IX and/or GPIIb/IIIa induce the destruction of platelets in peripheral blood, as well as the production of platelets in bone marrow. T cell abnormalities have also been reported in cITP, including a high Th1/Th2 ratio, a high cytotoxic T cell type 1/cytotoxic T cell type 2 lymphocyte ratio, high Th17 cell levels, and decreased Treg cells. Furthermore, genetic factors such as polymorphisms of the cytokine genes FcgR and HLA were reported to contribute to the pathogenesis of cITP. Thus, cITP is considered to be a consequence of complex immune dysregulation events in conjunction with the presence of genetic risk factors.
Recent studies using flow cytometry and real-time PCR have revealed a clear Th1-polarized cytokine profile both at the protein and mRNA levels in cITP [2, 3, 5, 10]. However, it is unclear whether fluxes in the Th1/Th2 ratio that lead to cITP pathogenesis involve genetic factors.
Numerous studies have shown that patients with autoimmune disease have polarized Th1 or Th2 responses [23]. IFN-γ is one of the main cytokines used to distinguish Th1 from other CD4+ subsets. IFN-γ, which is secreted mainly by Th1 and natural killer cells, promotes inflammation, responses to intracellular pathogens, and switching to the IgG2a, IgG2b, and IgG3 subclasses [24, 25]. IFN-γ exerts its biological effect by binding to the IFN-γ receptor (IFNGR), which consists of two chains: a receptor α chain (IFNGR1) and a receptor β chain (IFNGR2). Dysregulated IFN-γ production has been reported in many autoimmune diseases, including Hashimoto’s disease, type I diabetes, and multiple sclerosis. Panitsas et al. showed that serum levels and leukocyte gene expression of IFN-γ are markedly elevated in patients with cITP [3]. A high Th1/Th2 ratio showing a Th1-polarized response was also reported in cITP patients; this was reversed by treatment with dexamethasone [26, 27]. Thus, Th1 polarization may comprise a pivotal event in the pathogenesis of cITP [10].
Furthermore, recent studies have shown the association between IFN-γ +874 T/A polymorphism and various diseases such as cancer and autoimmune disorders. IFN-γ +874 T/A polymorphism has been reported to affect the production of IFN-γ; the TT genotype has been linked to higher production of IFN-γ compared to the A/A genotype [15, 28]. Although there are few studies on IFN-γ polymorphisms in cITP patients, they produced inconsistent findings. Pehlivan et al. [29] reported that cITP patients had a significantly higher frequency of the IFN-γ +874 TT genotype (high expression type) compared to healthy controls, while Chen et al. reported no significant association between IFN-γ +874 T/A polymorphism and infant ITP in Chinese patients [30]. We also found no significant association between IFN-γ +874 T/A polymorphism and Japanese patients with cITP. The frequency of IFN-γ +874 T/A polymorphism has been reported to differ by race [19, 20], which may explain the inconsistencies in various reports; in our control group, the rate was similar to that previously reported in healthy Asian control subjects [20]. Although Chen et al’s results were in accordance with our findings, they did not show the clinical characteristics of cITP according to IFN-γ +874 T/A polymorphism.
In contrast to cITP susceptibility, our data showed that cITP patients with IFN-γ +874 non-AA genotype (high expression type) had a lower minimum platelet count than those with the AA genotype. Panitsas et al. showed that lower peripheral platelet counts correlated with higher IFN-γ mRNA levels [3]. Thus, the patients with IFN-γ +874 non-AA genotype may be susceptible to severe cITP. Further studies may be needed to confirm the involvement of the IFN-γ gene in the pathogenesis of cITP.
IL-4 is the Th2 cytokine that is pivotal for the pathogenesis of many autoimmune diseases; it induces the differentiation of Th0 cells to Th2 cells [31]. Th2 cells subsequently produce additional IL-4 in a positive feedback mechanism upon activation by IL-4 [31]. There are some IL-4 polymorphisms which affect the expression level of IL-4, including IL-4 VNTR intron 3 and IL-4-590C/T. Rosenwasser et al. [32] analysed the association between IL-4 production and the IL-4 -590C/T polymorphism, and reported the TT genotype was linked to higher IL-4 levels compared to the C/C genotype. Several IL-4 -590C/T polymorphism studies were reported in various autoimmune diseases, including asthma, rheumatoid arthritis, and multiple sclerosis [33,34,35]. We found no association between IL-4 -590C/T polymorphism and susceptibility to cITP. A similar finding has been reported by Foster et al. [36]. In contrast to IL-4 -590C/T polymorphism, they produced inconsistent findings of the association between IL-4 VNTR intron 3 and ITP susceptibility. Makhlouf et al. [37] reported that Egyptian cITP patients had a significantly association with IL-4 VNTR, however Chen et al. reported showed no significant association between IL-4 VNTR intron 3 and Chinese ITP patients [30, 37].
cITP patients with the IL-4 -590 CC genotype (low expression type) had a higher number of treatment regimens than those with the non-CC genotype. Thus, the IL-4 -590 CC genotype, which predominantly induced Th1, appears to contribute to poor response to treatment.
IL-4 exerts its biological effects via signalling through its receptor, IL-4R. There are two types of IL-4 receptor complexes; the type I receptor consisting of IL-4Rα and the common gamma chain, and the type II receptor consisting of IL-4Rα and IL-13Rα [38]. IL-4Rα Q576R, which can affect the binding of IL-4 and phosphorylation of intracellular substrates including signal transducer and activator of transcription 6 (STAT6), has been linked to many autoimmune disorders such as asthma, atopy, and allergy [39]. Our study demonstrated that cITP patients had significantly higher frequencies of the IL-4Rα 576 non-QQ genotype. To our knowledge, ours is the first report demonstrating that IL-4Rα Q576R influences susceptibility to cITP. Recently Massoud et al. have shown that IL-4Rα Q576R promotes conversion of induced Treg cells toward a Th17 cell fate using Asthma mouse model [40]. Many investigators have shown that ITP patients have higher Th17/Treg compared to normal controls [4]. Th17 polarization by IL-4Rα Q576R polymorphism may affect the susceptibility to cITP in our study.
In this study, cITP patients had a higher Th1/Th2 ratio compared to healthy subjects. Among cITP patients, those with a higher Th1/Th2 ratio had a significantly lower platelet count than those with a lower ratio; these results are consistent with previous studies [2, 3]. Our study showed that the patients with IFN-γ non-AA and IFN-γR non-AA genotypes, which are related to higher IFN-γ expression and higher IFN-γR function, respectively, had a significantly higher Th1/Th2 ratio (P < 0.05). Thus, such genotypes of the IFN-γ pathway components might contribute to a higher Th1/Th2 ratio in cITP patients, and may thus increase the severity of cITP. We also explored the association between these four polymorphisms and Th1/Th2 ratios in healthy control subjects; however, we found no significant association between these polymorphisms and Th1/Th2 ratios.
Conclusion
In conclusion, our study revealed that the IL-4Rα polymorphism is associated with susceptibility to cITP. Moreover, the IFN-γ +874 non-AA genotype is associated with more severe thrombocytopenia and a higher Th1/Th2 ratio in cITP, indicating that the cytokine polymorphisms affecting Th1/Th2 increase the susceptibility to, and severity of, chronic ITP.
Abbreviations
- CI:
-
Confidence interval
- cITP:
-
Chronic immune thrombocytopenia
- IFN:
-
Interferon
- IFNGR:
-
Interferon-γ receptor
- IL:
-
Interleukin
- OR:
-
Odds ratio
- SD:
-
Standard deviation
- STAT6:
-
Signal transducer and activator of transcription 6
- Th1:
-
T-helper cell type 1
- Th2:
-
T-helper cell type 2
- Treg:
-
Regulatory T
References
Chang M, Nakagawa PA, Williams SA, Schwartz MR, Imfeld KL, Buzby JS, Nugent DJ. Immune thrombocytopenic purpura (ITP) plasma and purified ITP monoclonal autoantibodies inhibit megakaryocytopoiesis in vitro. Blood. 2003;102:887–95.
Ogawara H, Handa H, Morita K, Hayakawa M, Kojima J, Amagai H, Tsumita Y, Kaneko Y, Tsukamoto N, Nojima Y, Murakami H. High Th1/Th2 ratio in patients with chronic idiopathic thrombocytopenic purpura. Eur J Haematol. 2003;71:283–8.
Panitsas FP, Theodoropoulou M, Kouraklis A, Karakantza M, Theodorou GL, Zoumbos NC, Maniatis A, Mouzaki A. Adult chronic idiopathic thrombocytopenic purpura (ITP) is the manifestation of a type-1 polarized immune response. Blood. 2004;103:2645–7.
Semple JW, Provan D. The immunopathogenesis of immune thrombocytopenia: T cells still take center-stage. Curr Opin Hematol. 2012;19:357–62.
Semple JW, Milev Y, Cosgrave D, Mody M, Hornstein A, Blanchette V, Freedman J. Differences in serum cytokine levels in acute and chronic autoimmune thrombocytopenic purpura: relationship to platelet phenotype and antiplatelet T cell reactivity. Blood. 1996;87:4245–54.
Mosmann TR, Coffman RL. Th1 and Th2 cells: differentpatterns of lymphokine secretion lead to different func-tional properties. Annu Rev Immunol. 1989;7:145–73.
Romagnani S. Lymphokine production by human T cellsin disease states. Annu Rev Immunol. 1994;12:227–57.
Mosmann TR, Sad S. The expanding universe of T-cellsubsets: Th1, Th2 and more. Immunol Today. 1996;17:138–46.
Zhao H, Du W, Wang D, Gu D, Xue F, Ge J, Sui T, Yang R. The expression of IFN-gamma, IL-4, Foxp3 and perforin genes are not correlated with DNA methylation status in patients with immune thrombocytopenic purpura. Platelets. 2010;21:137–43.
Andersson J. Cytokines in idiopathic thrombocytopenic purpura (ITP). Acta Paediatr Suppl. 1998;424:61–4.
Rodeghiero F, Stasi R, Gernsheimer T, Michel M, Provan D, Arnold DM, Bussel JB, Cines DB, Chong BH, Cooper N, Godeau B, Lechner K, Mazzucconi MG, McMillan R, Sanz MA, Imbach P, Blanchette V, Kühne T, Ruggeri M, George JN. Standardization of terminology, definitions and outcome criteria in immune thrombocytopenic purpura of adults and children: report from an international working group. Blood. 2009;113:2386–93.
Li W, Qian X, Teng H, Ding Y, Zhang L. Association of interleukin-4 genetic polymorphisms with sporadic Alzheimer’s disease in Chinese Han population. Neurosci Lett. 2014;563:17–21.
Huang CZ, Yang J, Qiao HL, Jia LJ. Polymorphisms and haplotype analysis of IL-4Ralpha Q576R and I75V in patients with penicillin allergy. Eur J Clin Pharmacol. 2009;65:895–902.
Khanizadeh S, Ravanshad M, Mohebbi SR, Naghoosi H, Abrahim Tahaei M, Mousavi Nasab SD, Romani S, Azimzadeh P, Sanati A, Zali MR. Polymorphisms within the promoter region of the gamma interferon (IFN-γ) Receptor1 gene are associated with the susceptibility to chronic HBV infection in an Iranian population. Hepat Mon. 2012;12:e7283.
Pravica V, Perrey C, Stevens A, Lee JH, Hutchinson IV. A single nucleotide polymorphism in the first intron of the human IFN-gamma gene: absolute correlation with a polymorphic CA microsatellite marker of high IFN-gamma production. Hum Immunol. 2000;61:863–6.
Noguchi E, Shibasaki M, Arinami T, Takeda K, Yokouchi Y, Kawashima T, Yanagi H, Matsui A, Hamaguchi H. Association of asthma and the interleukin-4 promoter gene in Japanese. Clin Exp Allergy. 1998;28:449–53.
Nakamura H, Miyagawa K, Ogino K, Endo T, Imai T, Ozasa K, Motohashi Y, Matsuzaki I, Sasahara S, Hatta K, Eboshida A. High contribution contrast between the genes of eosinophil peroxidase and IL-4 receptor alpha-chain in Japanese cedar pollinosis. J Allergy Clin Immunol. 2003;112:1127–31.
Horie Y, Kitaichi N, Takemoto Y, Namba K, Yoshida K, Hirose S, Hasumi Y, Ota M, Inoko H, Mizuki N, Ohno S. Polymorphism of IFN-gamma gene and Vogt-koyanagi-Harada disease. Mol Vis. 2007;13:2334–48.
Popadic D, Savic E, Spuran Z, Markovic M, Mostarica Stojkovic M, Ramic Z, Pravica V. Distinctive frequencies of +874 T/a IFN-γ gene polymorphism in a healthy Serbian population. Clin Transl Sci. 2012;5:461–3.
Tian C, Zhang Y, Zhang J, Deng Y, Li X, Xu D, Huang H, Huang J, Fan H. The +874 T/a polymorphism in the interferon-γ gene and tuberculosis risk: an update by meta-analysis. Hum Immunol. 2011;72:1137–42.
Huang HR, Zhong YQ, Wu JF. The association between IFN-γ and IL-4 genetic polymorphisms and childhood susceptibility to bronchial asthma. Gene. 2012;494:96–101.
Liu YX, Zhang F, Yao QM, Yuan T, Xu J, Zhu XJ. Expression of CD11a in lymphocyte subpopulation in immune thrombocytopenia. Int J Clin Exp Pathol. 2015;8:15642–51.
Kidd P. Th1/Th2 balance: the hypothesis, its limitations, and implications for health and disease. Altern Med Rev. 2003;8:223–46.
Baudino L, Azeredo Da Silveira S, Nakata M, Izui S. Molecular and cellular basis for pathogenicity of autoantibodies: lessons from murine monoclonal autoantibodies. Springer Semin Immunopathol. 2006;28:175–84.
Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular Mechanisms Regulating Th1 Immune Responses. Annu Rev Immunol. 2003;21:713–58.
Lingjia Y, Chunmei Z, Liping Z, Yongyu S, Xuebin J. Biomarkers for immune thrombocytopenia. Biomark Res. 2015;3:19.
Ma D, Zhu X, Zhao P, Zhao C, Li X, Zhu Y, Li L, Sun J, Peng J, Ji C, Hou M. Profile of Th17 cytokines (IL-17, TGF-beta, IL-6) and Th1 cytokine (IFN-gamma) in patients with immune thrombocytopenic purpura. Ann Hematol. 2008;87:899–904.
Pravica V, Asderakis A, Perrey C, Hajeer A, Sinnott PJ, Hutchinson IV. In vitro production of IFN-gamma correlates with CA repeat polymorphism in the human IFN-gamma gene. Eur J Immunogenet. 1999;26:1–3.
Pehlivan M, Okan V, Sever T, Balci SO, Yilmaz M, Babacan T, Pehlıvan S. Investigation of TNF-alpha, TGF-beta 1, IL-10, IL-6, IFN-gamma, MBL, GPIA, and IL1A gene polymorphisms in patients with idiopathic thrombocytopenic purpura. Platelets. 2011;22:588–95.
Chen X, Xu J, Chen Z, Zhou Z, Feng X, Zhou Y, Ren Q, Yang R, Han ZC. Interferon-gamma +874A/T and interleukin-4 intron3 VNTR gene polymorphisms in Chinese patients with idiopathic thrombocytopenic purpura. Eur J Haematol. 2007;79:191–7.
Gadani SP, Cronk JC, Norris GT, Kipnis J. IL-4 in the brain: a cytokine to remember. J Immunol. 2012;189:4213–9.
Rosenwasser LJ, Klemm DJ, Dresback JK, Inamura H, Mascali JJ, Klinnert M, Borish L. Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy. 1995;25 Suppl 2:74–8.
Anabela Gonçalves B, Ana Teresa F, Susana O, Mariana R, Pedro O, Diogo R, Tânia S, Alexandra R, Rita C. Genetic polymorphisms and asthma: findings from a case–control study in the Madeira island population. Biol Res. 2014;47:40.
Li X, Chai W, Ni M, Xu M, Lian Z, Shi L, Bai Y, Wang Y. The effects of gene polymorphisms in interleukin-4 and interleukin-6 on the susceptibility of rheumatoid arthritis in a Chinese population. Biomed Res Int. 2014;2014:265435.
Arababadi MK, Mosavi R, Ravari A, Teimori H, Hassanshahi G. Association of interleukin-4 polymorphisms with multiple sclerosis in southeastern Iranian patients. Ann Saudi Med. 2012;32:127–30.
Foster CB, Zhu S, Erichsen HC, Lehrnbecher T, Hart ES, Choi E, Stein S, Smith MW, Steinberg SM, Imbach P, Kühne T, Chanock SJ. Early Chronic ITP Study Group. Polymorphisms in inflammatory cytokines and Fcgamma receptors in childhood chronic immune thrombocytopenic purpura: a pilot study. Br J Haematol. 2001;113:596–9.
Makhlouf MM, Abd Elhamid SM. Expression of IL4 (VNTR intron 3) and IL10 (−627) genes polymorphisms in childhood immune thrombocytopenic purpura. Lab Med. 2014;45:211–9.
Andrews AL, Holloway JW, Holgate ST, Davies DE. IL-4 receptor alpha is an important modulator of IL-4 and IL-13 receptor binding: implications for the development of therapeutic targets. J Immunol. 2006;176:7456–61.
Kruse S, Japha T, Tedner M, Sparholt SH, Forster J, Kuehr J, Deichmann KA. The polymorphisms S503P and Q576R in the interleukin-4 receptor alpha gene are associated with atopy and influence the signal transduction. Immunology. 1999;96:365–71.
Massoud AH, Charbonnier LM, Lopez D, Pellegrini M, Phipatanakul W, Chatila TA. An asthma-associated IL4R variant exacerbates airway inflammation by promoting conversion of regulatory T cells to TH17-like cells. Nat Med. 2016;22:1013–22.
Acknowledgments
The authors thank Ms. Rumi Ino and Mr. Yuya Kitamura for providing technical assistance. The authors were supported in part by Gunma University Polymorphism Study Program.
Availability of data and materials
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Authors’ contributions
TS, NT, YM, and HM designed the experiments, NT, YN, NG, TK, LA and TS performed the experiment and analyzed the data, TS, NT, AK, and HH supplied materials, NT and TS wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was approved by the Institutional Research Board of Gunma University Hospital (Approval #770). Patients and control donors gave written informed consents about this study.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Takahashi, N., Saitoh, T., Gotoh, N. et al. The cytokine polymorphisms affecting Th1/Th2 increase the susceptibility to, and severity of, chronic ITP. BMC Immunol 18, 26 (2017). https://doi.org/10.1186/s12865-017-0210-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12865-017-0210-3