Abstract
Background
Immune thrombocytopenia (ITP) is characterized by immune response dysregulations. Cytotoxic T lymphocyte‐associated antigen‐4 (CTLA‐4) plays a central role in immune checkpoint pathways and preventing autoimmune diseases by regulating immune tolerance. We aimed to explore the potential association between CTLA-4 gene polymorphisms and ITP as well as study their impact on the response to therapy.
Methods
We investigated two CTLA-4 single‐nucleotide polymorphisms (SNPs; rs: 231775 and rs: 3087243) using real-time PCR as well as the plasma levels of CTLA-4 by ELISA in 88 patients with ITP and 44 healthy participants (HC).
Results
CTLA-4 (rs: 3087243) A > G polymorphism analysis showed most HC had the homozygous AA genotype, which was statistically significant compared to patients with ITP. Plasma levels of CTLA4 were statistically lower in patients with acute ITP. There was no correlation between CTLA-4 (rs: 231775 and rs: 3087243) A/G SNPs were not correlated to the response to all lines of therapy assessed (corticosteroids, thrombopoietin receptor agonists, splenectomy, and rituximab).
Conclusion
CTLA-4 CT 60 A/G may affect the susceptibility of ITP, but both CTLA-4 + 49 A/G and CT60 A/G did not impact the response of patients with ITP to different lines of therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Immune thrombocytopenia (ITP), one of the most common acquired bleeding disorders, is characterized by reduced platelet count and increased bleeding risk [1].
Several immune mechanisms, including increased platelet destruction in the reticuloendothelial system and disturbed platelet production in the bone marrow, contribute to ITP pathogenesis. However, the exact pathogenesis of ITP is not clear yet [2].
Although antibody-mediated platelet destruction is a well-known primary immunologic defect, ITP pathogenesis also involves a hyper-activated T-cell response, which is important for cell-mediated platelet destruction [3]. Therefore, investigating T-cell abnormalities in patients with ITP may resolve the mechanism of pathogenesis and behavior of ITP as well as the diverse response of therapies.
The immune checkpoint pathways are critical modulators of the immune system, allowing immune response initiation and preventing autoimmunity onset. Immune checkpoints, including co-stimulation and co-inhibition signal pathways, are among the central mechanisms that regulate T-cell-mediated immune responses [4].
A "co-stimulation" signal, which is provided by CD28 on T-cells engaged with B7 family members on antigen-presenting cells (APCs), is necessary for efficient T-cell activation [5].
Cytotoxic T lymphocyte‐associated antigen‐4 (CTLA‐4) is a CD28 structural homolog, a co-inhibitory molecule expressed by activated cytotoxic T-cells, and a specific surface marker of T-reg, which plays a central role in immune checkpoint pathways. CTLA‐4 has a higher affinity for B7 than CD28 and is responsible for T-cell inactivation. Therefore, CTLA‐4 has a role in preventing autoimmune diseases by regulating immune tolerance [6].
CTLA‐4 production is strongly influenced by genetic factors. Single‐nucleotide polymorphisms (SNPs) of the CTLA-4-encoding genes are involved in the pathogenesis of many autoimmune diseases such as systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA) [7]. To explore the potential role of CTLA‐4 in the pathogenesis of primary ITP, we aimed to explore the potential association between single gene polymorphisms of CTLA-4 and primary ITP as well as study their impact on the response to therapy.
Patients and methods
Patients
We enrolled 88 Egyptian patients with primary ITP as well as 44 age- and sex-matched unrelated Egyptian individuals living in the same geographical region as healthy participants (HC) from the clinical hematology unit, Internal Medicine Department of the Kasr Al-Ainy teaching hospital. They were diagnosed according to the criteria of the ITP International Working Group (IWG) and followed up prospectively between September 2021 and March 2023 [8]. Patients with disorders possibly associated with secondary thrombocytopenia were excluded from the study. We conducted a single-institution case–control study. Informed consent was obtained from all patients before study initiation and patient recruitment, the study was approved by the local ethical committee (MD-225–2021). The study complied with good clinical practice protocols and with the ethical rules stated in the Declaration of Helsinki (as revised in Tokyo 2004).
Methods
All participants with ITP as well as HCs were subjected to full history taking (particularly bleeding phenotypes, drug intake, family history, recent immunizations as well as Obstetrics history) thorough clinical examination and laboratory investigations, including complete blood count (CBC) & film, reticulocyte count, and erythrocyte sedimentation rate (ESR). Patients with thyroid diseases or secondary ITP due to viral infections (hepatitis B virus, hepatitis C virus, human immunodeficiency virus), drugs, Helicobacter pylori infection, and autoimmune diseases such as SLE were excluded.
ITP was defined as isolated thrombocytopenia (platelet count < 100 × 103/µL) with no evidence of another underlying disorder.
All patients when indicated for therapy received corticosteroids (oral prednisolone 1 mg/kg/day) as the first line of therapy for maximum 21 days and gradually withdrawn. The patients who failed the first line of therapy received second line therapies such as thrombopoietin receptor agonists (TPO-RAs; eltrombopag), monoclonal anti (CD20) rituximab, or splenectomy. Some patients may fail one line and shift to another line (receive more than one line of therapy). The choice of the second line of therapy was according to patient preference, local availability, and physician choice.
Response to therapy was assessed after three months of therapies as follows:
-
Complete response (CR): Platelet count ≥ 100 × 103/µL and absence of bleeding.
-
Response (R): Platelet count ≥ 30 × 103/µL and at least a twofold increase in the baseline count and absence of bleeding.
-
No response (NR): Platelet count < 30 × 103/µL or less than twofold increase of baseline platelet count or bleeding [9].
Genotyping
Genomic DNA was extracted from whole blood collected on EDTA using a DNA extraction kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocol. Real-Time PCR with sequence-specific primers was used to assess the CTLA-4 genotype (+ 49 A/G rs: 231,775 and CT60 A/G rs: 3,087,243). Variants were determined using real-time PCR and high-resolution melt analysis on the CFX95 real-time system C 1000 thermal cycle (Biorad). The context sequence for CTLA-4 (+ 49 A/G rs: 231,775) was: forward primer: (5-GCTCTACTTCCTGAAGACCT-3 and reverse primer: (5-AGTCTCACTCACCTTTGCAG-3), The context sequence for CTLA-4 (CT60 A/G rs: 3,087,243) was: forward primer: (5′-ATAATGCTTCATGAGTCAGCTT-3′) and a reverse primer: (5′-GAGGTGAAGAACCTGTGTTAAA-3′). All reactions were performed in 20 μL reaction mix containing 10 μL master mix, 0.5 μL SNP-readymade assay, 1–5 μL purified DNA solution according to DNA concentration, completed to 20 μL with nuclease-free water.
Steps of performing PCR and the thermal cycling conditions.
PCR (40 Cycles) | AmpliTaq Gold Enzyme Activation | |
|---|---|---|
Anneal/Extend | Denature | HOLD |
1 min at 60 °C | 15 s at 95 °C | 10 min at 95 °C |
Specify the reaction volume (20 μL/well) in a 48-well plate | ||
Load the reaction plate into the thermal cycler, and then start the run | ||
Relation between fluorescence signals and sequences in a sample.
Indicates | A substantial increase in |
|---|---|
Homozygosity for Allele 2 | FAM-dye fluorescence only |
Homozygosity for Allele 1 | VIC-dye fluorescence only |
Allele 1- Allele 2 heterozygosity | Both VIC- and FAM-dye fluorescence |
After PCR amplification, an endpoint plate read was performed using a DNA-Technology Real-Time PCR System. The DT master Software used the fluorescence measurements made during the plate read to plot fluorescence values based on the signals from each well. The plotted fluorescence signals indicated which alleles were in each sample. The plate-read document was analyzed. Automatic allele calls were made. Allele calls were converted to genotypes.
Statistical analysis
Data was collected, tabulated, and statistically analyzed using an IBM-compatible personal computer with Statistical Package for the Social Sciences (SPSS) version 26. The quantitative data are presented in the form of mean, standard deviation (SD), median, and range, and qualitative data were presented in the form of numbers (N) and percentages (%). Chi-square test (χ2) or Fisher’s Exact test were used to study the association between two qualitative variables, Student’s t-test (t) was used to compare the quantitative variables between two groups of normally distributed data, Mann–Whitney U test was used to compare the quantitative variables between two groups of non-normally distributed data.
Results were considered statistically significant at P < 0.05.
Results
Clinical and laboratory features
In this case–control study, the median age of the patients with ITP and HCs was 32.5 (15–50) and 30.5 (15–46) years, respectively. Moreover, most of the patients with ITP were females (73.9%). The median disease duration was 5.4 (0–11) years. Regarding the clinical presentation, no patients had a history of NSAID use or herbal supplement intake and all patients presented with mucocutaneous bleeding in the form of epistaxis, purpura, or vaginal bleeding. Only one patient (1.1%) presented with severe bleeding (internal bleeding) and another patient (1.1%) had a history of previous repeated abortions, but no history of thrombotic events. None of the patients had lymphadenopathy or splenomegaly. The mean platelet count at diagnosis was 17.2 × 103/µL ± 12.5 (2–60 × 103/µL). Since all studied patients had primary ITP, the virological screening was negative for all patients. Autoimmune screening, including anti-nuclear antibody (ANA) and anti-dsDNA, of the patients was negative. Screening for antiphospholipids was also negative and only one patient had subclinical hypothyroid. The clinical and demographic characteristics as well as response of patients with ITP to therapies are summarized in Table 1.
Response to therapies
Briefly, 88 (100%) patients received corticosteroids as the first line of therapy. Among them, 8 (9.1%) patients had a complete response (CR). The patients received second line therapies when they failed to respond to or were dependent on the first line of therapy to maintain platelet response. Some patients may fail one line and be shifted to another line (received more than one line of therapy). A total of 67 and 38 patients received eltrombopag and anti (CD20) rituximab, respectively, of whom 18 (26.9%) and 10 (26.3%), respectively, had a CR. Additionally, 11 patients underwent splenectomy and 4 (36.4%) of them had CR. Unfortunately, the duration of maintaining CR in each line was not documented.
Genotyping of CTLA-4
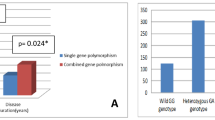
The two CTLA-4 gene SNPs (+ 49A/G rs231775 and CT60 A/G rs3087243) were assessed. The CT 60 A/G rs:3087243 (P = 0.001) was significantly different between patients with ITP and HCs, while SNP + 49A/G rs:231,775 was not significantly different between patients with ITP and HCs (Table 2).
We further investigated the correlation between CTLA-4 genotypes (+ 49A/G rs; 231,775 and CT60 A/G rs: 3,087,243) and the response to different lines of therapy; however, it was not statistically significant in our studied patients with ITP (Table 3).
Discussion
Several immune mechanisms contribute to ITP pathogenesis and interact to affect the response to therapy. The involvement of all T-cell subtypes in ITP pathogenesis has been explored for many years [10,11,12]. CTLA-4 is a competitive antagonist for B7 on the surface of APCs and responsible for T-cell inactivation and immune tolerance owing to its involvement in the immune checkpoint pathways [13]. The SNPs of CTLA-4-encoding genes are involved in the pathogenesis of many autoimmune diseases [7]. However, the association between the immune checkpoint pathway and ITP pathogenesis is still unclear.
We investigated the impact of CTLA-4 SNPs [+ 49A/G (rs231775) and CT60 A/G (rs3087243)] on the susceptibility and response to therapy in primary ITP for finding new clues in the pathogenesis of primary ITP.
A total of 88 patients with ITP (male:female = 21%:79%) and 44 HCs were enrolled. Although there were conflicting reports regarding sex predominance, our results come in agreement with those of many studies reporting that ITP is common in young and middle-aged females [14,15,16], while few reports reporting that it is common in older men [17, 18].
To the best of our knowledge, few studies have addressed the influence of SNPs of CTLA-4 in ITP (Table 4). However, this is the first study to find a significant difference in CT60 A/G (rs3087243) genotype between patients with ITP and HCs as most HCs had AA genotypes and lack heterozygous GA genotypes, which may be correlated to the susceptibility of ITP. Moreover, we did not find a difference in + 49 A/G (rs231775) genotype between patients with ITP and HC. Interestingly, Kasamatsu et al. did not find a difference in + 49 A/G and CT60 A/G genotype between patients with ITP and HCs, but reported a correlation between CTLA4 CT60 GG genotype (low expression type) and severe clinical presentation at diagnosis while studying four CTLA-4 SNPs in 119 patients with chronic ITP [19]. Moreover, Chen et al., studied nine SNPs of the promoter region of CTLA-4 in 32 patients with ITP and showed no difference in + 49 A/G and CT60 A/G genotype between patients with ITP and HC, while a different SNP (rs11571315) was a susceptible SNP for primary ITP risk in the Taiwanese population [20]. Yao et al. studied CT60 A/G (rs3087243) genotype in 102 Chinese Han pediatric patients with ITP and showed that the genotypes were not significantly different between the pediatric patients with ITP and healthy participants. However, they also found decreased CTLA4 expression in patients with ITP, supporting that CTLA4 gene expression instead of gene mutation is one of the factors for ITP [21].
The conflicting results may be due to different studied ethnic populations, disease stages, or the potential role of epigenetic modifiers that may need more exploration in future studies.
Although we did not find a correlation regarding CTLA-4 + 49 A/G (rs231775) between patients with ITP and HCs, a large metanalysis by Wang et al., who searched the same SNP in different autoimmune diseases like RA and type 1 DM including a considerable number of patients (N, 4732), found that CTLA-4 + 49 G/A (rs231775) is associated with the susceptibility of autoimmune disease in Asian and Caucasian populations [26].
Pavkovic et al. studied CTLA-4 + 49 A/G in different hematological diseases (AIHA, ITP, and CLL) and found no difference between patients with ITP (N, 60) and healthy participants, but the G allele of CTLA-4 predisposes to AIHA development, particularly among patients with CLL [27].
Wang et al. studied different immune checkpoint-related gene polymorphisms including CTLA-4 (rs231779) in 307 patients with ITP and stated that immune checkpoint-related SNPs, particularly CD28 rs1980422, may be genetic factors associated with ITP development and treatment. However, neither allelic nor genotypic frequencies of CTLA4 rs231779 were significantly associated with the susceptibility to or severity of ITP [22].
In this study, we did not find any correlation between the two CTLA4 SNPs and the response to all therapies including corticosteroids, TPO-RAs, splenectomy, and rituximab in our studied patients. To the best of our knowledge, very few studies have assessed the correlation between CTLA-4 SNPs and the response to therapy in ITP, particularly the response to corticosteroids. This is the first study to assess this correlation with different lines of therapy but a bigger sample size of patients in each group of therapy may be needed. Kasamatsu et al., found no correlation between CTLA4 polymorphisms and the response to prednisolone therapy and splenectomy in 52 patients with ITP [19]. Moreover, Wang et al., found that the allelic or genotypic frequencies of CTLA4 (rs231779) were associated with corticosteroid sensitivity of ITP [22]. Zhu et al. monitored plasma levels of CTLA-4 in 37 patients with ITP who received 40 mg/day dexamethasone for four consecutive days and found that CTLA-4 levels were significantly elevated in not only patients with acute ITP, but also responders with acute ITP, suggesting that CTLA-4 might be associated with the pathogenesis of acute ITP and reflect treatment efficacy [28]. Moreover, Guo et al., observed dynamic changes in CTLA-4 and CD28 expression after high-dose dexamethasone therapy in 28 patients with ITP, suggesting that a disturbed CD28/CTLA-4 balance may contribute to ITP immunopathogenesis [29].
Conclusion
CTLA-4 CT 60 A/G may affect the susceptibility of ITP, but both CTLA-4 + 49 A/G and CT60 A/G had no impact on the response to different lines of therapy in patients with ITP. Thus, further studies should include more patients with ITP in each group of therapy as well as focus on more immune checkpoint-related gene polymorphisms.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- ANA:
-
Anti-nuclear antibody
- APCs:
-
Antigen-presenting cells
- CBC:
-
Complete blood count
- cITP:
-
Chronic immune thrombocytopenia
- CR:
-
Complete response
- CTLA-4:
-
Cytotoxic T lymphocyte‐associated antigen‐4
- DM:
-
Diabetes mellitus
- DNA:
-
Deoxyribonucleic acid
- EDTA:
-
Ethylenediaminetetraacetic acid
- ESR:
-
Erythrocyte sedimentation rate
- HC:
-
Healthy participants
- ITP:
-
Immune thrombocytopenia
- IWG:
-
International Working Group
- N:
-
Number
- NR:
-
No response
- PCR:
-
Polymerase chain reaction
- R:
-
Response
- RA:
-
Rheumatoid arthritis
- SLE:
-
Systemic lupus erythematosus
- SNPs:
-
Single‐nucleotide polymorphisms
- SPSS:
-
Statistical Package for the Social Sciences
- TPO-RAs:
-
Thrombopoietin receptor agonists
References
Cooper N, Ghanima W. Immune thrombocytopenia. N Engl J Med. 2019;381:945–55.
Zufferey A, Kapur R, Semple JW. Pathogenesis and therapeutic mechanisms in immune thrombocytopenia (ITP). J Clin Med. 2017;6:16.
Audia S, Mahévas M, Nivet M, Ouandji S, Ciudad M, Bonnotte B. Immune thrombocytopenia: recent advances in pathogenesis and treatments. Hemasphere. 2021;5:e574.
Kim GR, Choi JM. Current understanding of cytotoxic T lymphocyte antigen-4 (CTLA-4) signaling in T-cell biology and disease therapy. Mol Cells. 2022;45:513–21.
Lenschow DJ, Walunas TL, Bluestone JA. CD28/B7 system of T cell costimulation. Annu Rev Immunol. 1996;14:233–58.
Linsley PS, Brady W, Urnes M, Grosmaire LS, Damle NK, Ledbetter JA. CTLA-4 is a second receptor for the B cell activation antigen B7. J Exp Med. 1991;174:561–9.
Zhou C, Gao S, Yuan X, et al. Association between CTLA-4 gene polymorphism and risk of rheumatoid arthritis: a meta-analysis. Aging (Albany NY). 2021;13:19397–414.
Provan D, Arnold DM, Bussel JB, et al. Updated international consensus report on the investigation and management of primary immune thrombocytopenia. Blood Adv. 2019;3:3780–817.
Rodeghiero F, Stasi R, Gernsheimer T, et al. Standardization of terminology, definitions and outcome criteria in immune thrombocytopenic purpura of adults and children: report from an international working group. Blood. 2009;113:2386–93.
Ogawara H, Handa H, Morita K, et al. High Th1/Th2 ratio in patients with chronic idiopathic thrombocytopenic purpura. Eur J Haematol. 2003;71:283–8.
Stasi R, Cooper N, Del Poeta G, et al. Analysis of regulatory T-cell changes in patients with idiopathic thrombocytopenic purpura receiving B cell-depleting therapy with rituximab. Blood. 2008;112:1147–50.
Bao W, Bussel JB, Heck S, et al. Improved regulatory T-cell activity in patients with chronic immune thrombocytopenia treated with thrombopoietic agents. Blood. 2010;116:4639–45.
Collins AV, Brodie DW, Gilbert RJ, et al. The interaction properties of costimulatory molecules revisited. Immunity. 2002;17:201–10.
Doobaree IU, Conway K, Miah H, et al. Incidence of adult primary immune thrombocytopenia in England-an update. Eur J Haematol. 2022;109:238–49.
Palau J, Sancho E, Herrera M, et al. Characteristics and management of primary and other immune thrombocytopenias: Spanish registry study. Hematology. 2017;22:484–92.
Lebowa W, Zdziarska J, Sacha T. Immune thrombocytopenia: characteristics of the population and treatment methods-one-center experience. Hamostaseologie. 2023;43:132–41.
Moulis G, Palmaro A, Montastruc JL, Godeau B, Lapeyre-Mestre M, Sailler L. Epidemiology of incident immune thrombocytopenia: a nationwide population-based study in France. Blood. 2014;124:3308–15.
Schoonen WM, Kucera G, Coalson J, et al. Epidemiology of immune thrombocytopenic purpura in the general practice research database. Br J Haematol. 2009;145:235–44.
Kasamatsu T, Ino R, Takahashi N, et al. PDCD1 and CTLA4 polymorphisms affect the susceptibility to, and clinical features of, chronic immune thrombocytopenia. Br J Haematol. 2018;180:705–14.
Chen DP, Lin WT, Wen YH, Wang WT. Investigation of the correlation between immune thrombocytopenia and T cell activity-regulated gene polymorphism using functional study. Sci Rep. 2022;12:6601.
Yao L, Liu B, Jiang L, Zhou L, Liu X. Association of cytotoxic T-lymphocyte antigen 4 gene with immune thrombocytopenia in Chinese Han children. Hematology. 2019;24:123–8.
Wang S, Zhang X, Leng S, et al. Immune checkpoint-related gene polymorphisms are associated with primary immune thrombocytopenia. Front Immunol. 2021;11:615941.
Aktürk F, Hançer VS, Küçükkaya R. Cytotoxic T lymphocyte antigen-4 (CTLA-4) A49G polymorphism and autoimmune blood diseases. Turk J Haematol. 2010;27:78–81.
Li H, Ge J, Zhao H, et al. Association of cytotoxic T-lymphocyte antigen 4 gene polymorphisms with idiopathic thrombocytopenic purpura in a Chinese population. Platelets. 2011;22:39–44.
Radwan ER, Goda RL. Lack of impact of cytotoxic T-lymphocyte antigen 4 gene exon 1 polymorphism on susceptibility to or clinical course of Egyptian childhood immune thrombocytopenic purpura. Clin Appl Thromb Hemost. 2015;21:378–82.
Wang K, Zhu Q, Lu Y, et al. CTLA-4 +49 g/a polymorphism confers autoimmune disease risk: an updated meta-analysis. Genet Test Mol Biomarkers. 2017;21:222–7.
Pavkovic M, Georgievski B, Cevreska L, Spiroski M, Efremov DG. CTLA-4 exon 1 polymorphism in patients with autoimmune blood disorders. Am J Hematol. 2003;72:147–9.
Zhu F, Qiao J, Cao J, et al. Decreased level of cytotoxic T lymphocyte antigen-4 (CTLA-4) in patients with acute immune thrombocytopenia (ITP). Thromb Res. 2015;136:797–802.
Guo X, Yasen H, Zhao F, et al. The effect of single course high dose dexamethasone on CD28/CTLA-4 balance in the treatment of patients with newly diagnosed primary immune thrombocytopenia. Hum Vaccin Immunother. 2016;12:97–103.
Acknowledgements
We deeply thank all patients who participated in this study.
Funding
Self-funding.
Author information
Authors and Affiliations
Contributions
Idea of the research done by Prof Dr DM & Dr MS, data collection done by all authors, Meticulous laboratory work done under the supervision of Dr. KG, Written by Dr. DM & Dr. MAM, and all work revised by all authors with an equal contribution.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Approved by the local ethical committee of internal medicine department, Faculty of medicine, Cairo University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
El Demerdash, D.M., Saber, M.M., Ayad, A. et al. Cytotoxic T lymphocyte‐associated antigen‐4 (CTLA-4) gene polymorphisms in a cohort of Egyptian patients with immune thrombocytopenia (ITP). Blood Res. 59, 8 (2024). https://doi.org/10.1007/s44313-024-00011-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44313-024-00011-z