Abstract
Background
ORP (Oxysterol-binding protein-related proteins) genes play a role in lipid metabolism, vesicular transferring and signaling, and non-vesicular sterol transport. However, no systematic identification and analysis of ORP genes have been reported in cotton.
Result
In this study, we identified 14, 14, 7, and 7 ORP genes in G. hirsutum, G. barbadense, G. arboreum, and G. raimondii, respectively. Phylogenetic analysis showed that all ORP genes could be classified into four groups. Gene structure and conserved motif analysis suggest that the function of this gene family was conserved. The Ka/Ks analysis showed that this gene family was exposed to purifying selection during evolution. Transcriptome data showed that four ORP genes, especially GhORP_A02, were induced by abiotic stress treatment. The cis-acting elements in the ORP promoters were responsive to phytohormones and various abiotic stresses. The silenced plants of GhORP_A02 were more sensitive to drought stress when compared to control.
Conclusion
The major finding of this study shed light on the potential role of ORP genes in abiotic stress and provided a fundamental resource for further analysis in cotton.
Similar content being viewed by others
Introduction
Cotton is the most important natural fiber crop and amounts to 35% of all fibers produced worldwide [49]. Cotton is a part of the Malvaceae family and belongs to the Gossypium genus with 45 diploids and five allotetraploid species found in Africa, America, Galapagos, India, Australia, Arabia, and Hawaii [16]. Eight diploid genomes (A-G and K) are assigned to these 50 species [1, 5]. Abiotic stresses significantly limit cotton growth, output, and development, resulting in a 50% decline in worldwide yield [7, 15]. Abiotic stresses such as drought, salinity, cold, and heat have negative effects on plant photosynthesis and respiration, attributed to disruption of various molecular pathways, such as Ca2+ signaling, abscisic acid (ABA) signaling, reactive oxygen species (ROS) metabolism, and sugar and lipid metabolism [6, 10, 18, 27]. Drought is considered as an important yield limiting factor [24, 40]. Cotton plants have evolved a range of sophisticated signaling networks, including metabolic, physiological, and morphological changes, to adapt to drought stress [24]. Drought tolerance mechanisms in cotton include drought avoidance, drought tolerance, drought recovery, and drought escape [14]. These tolerance mechanisms are aided by signal transduction and hormone regulation, such as jasmonic acid (JA), ABA, and ethylene synthesis [11, 13, 14, 20, 26, 32, 34, 52, 56].
ORP (oxysterol-binding protein-related proteins) genes play a key role in lipid metabolism, vesicular transferring and signaling, and non-vesicular sterol transport [41]. Previous studies about ORP genes in Arabidopsis, soybean, and petunia have also demonstrated its significant role in biotic stress, abiotic stress [31, 43, 45]. The Arabidopsis genome encodes 12 ORP genes, and the rice genome encodes six ORP genes [48]. Although the ORP gene in plants has been cloned, there are few studies focused on their functions. In Arabidopsis, ORP3a, located the endoplasmic reticulum, interacts with VAP33 family member PVA12 [43]. In Petunia inflata, PiORP1 participates in pollen growth and development by interacting with PRK1 receptor kinase on the plasma membrane of a hybrid pollen tube [45]. In soybean, the expression of GmOSBP was inhibited by salt stress but induced in aging leaves, indicating that GmOSBP may be involved in stress response and the cell aging process [31]. In this study, we performed genome-wide identification and investigated phylogenetic relationships, gene structure, conserved domains, gene duplication events and expression files of ORP genes. Our study may be useful for the future molecular and biological function of the ORP gene family in cotton.
Materials and methods
ORP gene identification in cotton species
The conserved domain PF15413 of ORP genes was obtained from PFAM (http://pfam.xfam.org) and used as a query sequence to retrieve the ORP genes in four cotton species by Hmmer 3.0 (http://hmmer.org), and the identity of the ORPs genes was analyzed by SMART (http://smart.embl-heidelberg.de/smart/). The physical and chemical characteristics of ORP proteins, including molecular weight, protein length, molecular charge, isoelectric point, and grand average of hydropathy, were obtained from CottonFGD (https://cottonfgd.net/).
Chromosomal mapping
We used the GFF3 files of the ORPs genes downloaded from CottonFGD to find the distribution on all chromosomes. TBtools software (version 1.098685) was then used to visualize the gene’s location on chromosomes [8].
Phylogenetic tree and collinearity analysis
The full-length protein sequences of ORP genes from Gossypium were downloaded and aligned using ClustalW with default settings. The phylogenetic tree was constructed using the neighbor-joining method in MEGA 6 with default parameters and 1000-bootstrap replicates (http://www.megasoftware.net/). The protein sequences of Gossypium hirsutum (G. hirsutum) have been searched along the protein databases of Gossypium arboreum (G. arboreum), Gossypium barbasense (G. barbasense) and Gossypium raimondii (G. raimondii) by BlastP to identify homologous genes and hits with E-values of 1.0E–5 and similarity of 90% were considered noteworthy. TBtools program was used to create the collinearity analysis using the GFF3 file, linked file, and gene IDs. Collinearity analysis was performed among three cotton species (G. hirsutum, G. arboreum, and G. raimondii) using Circle gene viewer in TBtools software to determine collinear gene pairs. Coding and protein sequences of all homolog genes were used to calculate the Ka/Ks (Non-synonymous substitution- rate/Synonymous substitution rate) value by TBtools [47].
Gene structure, conserved motif analysis and prediction of regulatory elements
The gene structures were analyzed using a gene structure displayer server (http://gsds.cbi.pku.edu.cn/). Conserved motifs of ORP genes were discovered with default settings of the MEME Suite (http://memesuite.org/index.html) [4]. The gene structure was visualized using TBtools (v1.098661) [8]. 1500 bp upstream sequences of ORPs genes from different cotton species were downloaded from CottonFGD and uploaded into PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to identify cis-regulatory elements [30].
Expression profile analysis of ORP gene family
FPKM values (fragments per kilobase of exon per million fragments mapped) of ORP genes were downloaded from CottonFGD. We analyzed the expression profiles of ORP genes under different stress treatments, which included PEG, salt, heat and cold treatments.
Virus-induced gene silencing (VIGS)
For virus-induced gene silencing, the cotton variety H117 was employed. H177 was developed by the Institute of Cotton Research Anyang of the Chinese Academy of Agricultural Sciences. This Variety was chosen because it is particularly susceptible to many environmental stresses, including drought. A 306-bp fragment of GH_A02G0809 was amplified from G. hirsutum acc. TM-1 with gene-specific primers. The PCR product was then digested with Spe I and Acs I and cloned into Spe I-Acs I -Cut pCLCrVA. The fusion vector was named pCLCrVA: GhORP_A02 and transformed into Agrobacterium tumefaciens strain LBA4404. The control vector pCLCrVA, pCLCrVA: GhORP_A02 and positive vector pCLCrVA: PDS were mixed with pCLCrVB at a 1:1 ratio [19]. The mixed Agrobacterium tumefaciens solutions were injected into the ten-day-old cotton cotyledons on the abaxial side with a needle-free syringe. The plants were placed at room temperature in the dark overnight and grew at 23 °C with a 16 h / 8 h light/dark cycle. Agrobacterium infection was carried out three times with 30 plants for each vector. The primers for VIGS vector construction are listed in Table S1. Wild type and the plants injected with pCLCrVA empty control and pCLCrVA: GhORP_A02 were subjected to drought treatment after four weeks. Drought treatments of the seedlings were irrigated with 15% PEG6000, while control plants were irrigated with 1/2 MS nutrient solution.
RNA extraction and quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from fresh leaves and roots using TRIzol® Plus RNA Purification Kit (Invitrogen, CA) based on the manufacturer’s instructions. Approximately 1 µg RNA was reversely synthesized into cDNA using the iScriptTM Synthesis Kit (Quanta BioSciences, MD). The qRT-PCR was carried out in an Eppendorf real-time PCR equipment using a 5 µl cDNA template (diluted 1/100), 5 µl primers (2.4 M), and 10 µl SYBR green mixture (Promega, Madison, WI). Histone 3 was used as the internal control, and the relative expression levels of the ORP gene were calculated by the 2−ΔΔCt method [35].
Physiological analysis
Physiological parameters, including ion leakage, chlorophyll content, excised leaf water loss, and relative leaf water content, were determined after 10 days of drought treatment. Wild type, silenced, and control plants (ten plants for each) were harvested after drought stress for oxidant and antioxidant concentration analysis. The H2O2 content, peroxidase (POD), malondialdehyde (MDA) and catalase (CAT) were determined by using the corresponding ROS content reagent kits and enzyme activity kit (Solarbio, China) according to the manufacturer’s instructions. The experiment was repeated three times.
Results
Genome-wide identification and chromosomal locations of the cotton ORP genes
To identify all ORP genes in two allotetraploid cotton, G. hirsutum (AD1), G. barbadense (AD2) and its two diploid ancestors G. arboreum (AA) and G. raimondii (DD), we used conserved domain Pfam 15,413 to retrieve ORP genes and identified 42 ORP genes. G. hirsutum, G. barbadense, G. arboreum, and G. raimondii have 14, 14, seven, and seven ORP genes, respectively. The ORP gene ID and predicted protein properties and subcellular locations are listed in Table 1. Variable distribution of ORP genes on chromosomes across all four cotton species was observed (Fig. 1). In G. hirsutum and G. barbadense, ORP genes were uniformly distributed on the At and Dt chromosome. In G. hirsutum and G. barbadense, 14 ORP genes were located on chromosomes A02, A03, A05, A06, A09, D02, D03, D05, D06 and D09. Two ORP genes were located on chromosomes A03, A05, D02 and D05. In G. arboreum, seven ORP genes were located on chromosomes A01, A03, A05, A06 and A09. In G. raimondii, seven ORP genes were located on chromosomes D03, D05, D06, D09 and D10.
Phylogenetic and collinearity analysis of cotton ORP genes
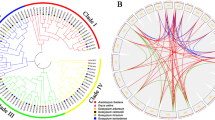
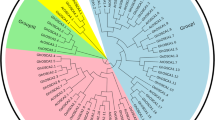
According to the phylogenetic analyses, all the ORP genes could be classified into four clades (Fig. 2A). Four, two, two, and six ORP genes from G. hirsutum were classified into Group I to Group IV. Both in G. arboreum and G. raimondii, Group I, II, III and IV have two, one, one, and three ORP genes. To analyze the evolution of the ORP genes from diploid to tetraploid species, collinearity analysis was performed among three cotton species (G. hirsutum, G. arboreum, and G. raimondii). There were seven, seven, and seven orthologous gene pairs between the A and D genomes, the At subgenome and the A genome, and the Dt subgenome and the D genome (Fig. 2B). The number and relatedness of ORP genes in the three species suggested that ORP genes were not lost during G. hirsutum speciation.
Phylogenetic and collinearity analysis of ORP genes. A Phylogenetic tree of ORP genes in G. hirsutum, G. arboreum, G. raimondii. B The synteny relationships of ORP genes among three cotton species. ORP genes in G. hirsutum, G. arboreum, and G. raimondii are indicated in pink, red and blue, respectively
Evolution of ORP genes in Gossypium species
Natural selection has no effect on gene’s Ka/Ks values during the evolutionary trend, but Ka/Ks > 1, Ks/Ka = 1, or Ka/Ks < 1, the Ka/Ks value indicates positive, neutral, or negative selection, respectively [55]. Similar results were found in the distributions of Ka, Ks, and Ka/Ks among homologous pairs of Gossypium species. The Ka/Ks ratio of most orthologous gene pairs was less than one, indicating purifying selection during evolution resulting in limiting the functional divergence after duplications and polyploidization of ORP genes (Table S2). Only two orthologous gene pairs (GH_A02G0809 and Ga3G0877, GH_D02G0824 and Gorai.005G091400) have Ka/Ks ratio exceeding one, which implies these gene pairs underwent positive selection and had relatively rapid evolution rate.
Gene structure and motif identification of ORP proteins
The gene structure of ORP genes was analyzed according to the annotation files (Fig. 3A-D). Most ORP genes have 8–10 exons and only six genes in four cotton species have two or three exons. Genes classified into the same evolutionary branch have conserved gene structure patterns in terms of exon number and exon length. The MEME search identified ten conserved motifs in ORP genes, ranging from 300 to 2100 amino acids (Fig. 3E-H). The conserved motif numbers in different genes varied from 3 to 10. Motifs 2, 5, and 7 were conserved in all ORP genes in G. hirsutum. In G. barbadense, the conserved motifs were 4, 5, and 7, while in G. arboreum and G. raimondii, the conserved motifs were 4, 6, and 7.
Identification and analysis of cis-acting elements
Cis-acting regulatory elements play a key role in molecular switches that control a dynamic gene activity network that initiates many biological processes, such as hormone responses, developmental processes, and abiotic stress responses [37]. MBS (drought inducibility), ABRE (abscisic acid-responsive), and TC-rich repeats (repeat cis-actin), which are involved in defense stress response and drought stress, could be found in all four cotton species (Figure S1). CAT-Box (meristematic cell expression), GARE-motif (Gibberellin responsive), and TGA-elements (auxin-responsive element), which are involved in the germination and regeneration stage, could also be found in all four cotton species.
Expression profiles of ORP genes in Gossypium hirsutum
The raw RNA-seq data of the 14 ORP genes in G. hirsutum were normalized to log2(FPKM), and the heatmap of the expression is presented as Fig. 4. Four genes, including GH_A02G0809, GH_D02G0824, GH_A09G2118, and GH_D05G2343, were up-regulated by the abiotic stress treatment. Especially, GH_A02G0809 (GhORP_A02) expression was induced significantly by PEG treatment. We further performed experiments to characterize the function of GhORP_A02 in drought stress.
Virus-induced gene silencing of GhORP_A02 in cotton show significant sensitivity to drought
The method for gene silencing through virus-induced was used to analyze the role of the GhORP_A02 in drought tolerance. Gossypium hirsutum acc. H177 was infected with three vectors, including pCLCrVA: PDS (positive control), pCLCrVA (negative control), and pCLCrVA: GhORP_A02. Ten days after infection, the indicator pCLCrVA: PDS showed albino color, the control plant showed a normal color without visible change, and the pCLCrVA: GhORP_A02 plants showed complete shrinkage of the leaves, which indicates that VIGS was successful (Fig. 5A). qRT-PCR was used to analyze the expression level of GhORP_A02 in silenced plants, and the result showed that the infected plant (pCLCrVA: GhORP_A02) showed a lower expression level than the control plant (Fig. 5B). The physiological analysis includes ion leakage, chlorophyll contents, excised leaf water loss, and relative leaf water content was done in silenced and controlled plants with and without drought treatment. The relative ion leakage level of the silenced plants increased by 20% compared to the control. The chlorophyll contents of the silenced plant were significantly lower in comparison to control plants. While in excised leaf water loss, the silenced plants lost more water than the control plant. The relative leaf water content of the silenced plant didn’t show a significant difference when compared to the control under drought conditions (Fig. 6A-D). Determination of antioxidant (CAT and POD) and oxidant (MDA and H2O2) enzyme concentration levels were analyzed in both control and silenced plants under drought conditions. There is a significant increase in the concentration of antioxidants and a decrease in the concentration of H2O2 in silenced plants compared with their respective control (Fig. 6E-H).
Virus-induced gene silencing of GhORP_A02 in upland cotton. A Phenotypes of wild type, negative (pCLCrVA), and silenced plants (pCLCrVA: GhORP_A02) after drought treatment. B qRT-PCR analysis of wild type, silenced and control cotton plants after 10 days of drought treatment. Different letters indicated significant difference at p < 0.05
Discussion
Drought is one of the most significant abiotic stresses, resulting in considerable yield losses in cotton [23, 40]. Plants have evolved self-defense systems to deal with abiotic stresses, which involves the transcription of stress-related genes [39]. Genetic enhancement of drought tolerance hinges on identifying genes related to drought tolerance [53]. In earlier research, drought-responsive genes were identified in many species like rice, peanut, soybean, wheat, maize and cotton [9, 21, 36, 44, 54]. Oxysterol-binding protein (ORP) and its homologs constitute a protein family in many eukaryotes, from yeast to humans, which are involved in cellular lipid metabolism, vesicle transport and signal transduction [51]. Recent studies have demonstrated that the ORP gene family was stress-responsive in various plants [31, 48, 43, 45]. Our current research has demonstrated the function of GhORP_A02 in drought stress response. This study used the protein domain PFAM 15,413 to retrieve ORP genes in the four cotton species, and G. hirsutum, G. barbadense, G. raimondii, and G. arboreum encoding 14, 14, seven, and seven ORP genes, respectively. In previous research, 12 and six ORP genes were identified in Arabidopsis and rice, respectively [48]. Gene structure and phylogenetic tree analyses indicated that all GhORP genes, classified into one group, have a similar gene structure. The evolution analysis of the ORP gene in four cotton species shows no negative selection across all the species.
Conserved domains correspond conformational changes due to binding [28, 50]. Domain rearrangement and recombination, which typically occurs due to gene duplication and fission or fusion events, are used to develop new protein functions [38]. In this study, we identified two conserved motifs, motifs 5 and 7 in G. hirsutum and G. barbadense, while motif 7 was conserved in all four Gossypium species. Subcellular localization and the transcription of a gene under stress are powerful mechanisms to explain its biological function. ORP genes have been identified in Arabidopsis, soybean, rice, and Petunia and found to be located in plasm membrane, nucleus, and endoplasmic reticulum [31, 48, 43]. According to the subcellular localization prediction result, most ORP genes are located in the nucleus. Expression analysis of GhORP genes under different stress showed four genes were strongly induced by cold and drought stress from 1 to 24 h. A similar result was reported in soybean and Arabidopsis. For example, GmOSBP was induced by salt stress, and AtORP4A and AtORP4B were induced by drought stress [31, 48]. Expression analysis showed that four GhORP genes, especially GhORP_A02, were significantly up-regulated after drought stress, and we consider this gene as the candidate gene for drought stress response.
To investigate the function of GhORP_A02, we silenced this gene by VIGS. Resultantly, silenced plants were more sensitive to PEG treatment than control. Environmental challenges such as drought, salt and temperature cause a redox imbalance in plant cells, which rises the total rate of metabolism and finally up-regulates H2O2 production [17]. There is still no relative study about the mechanism of ORP proteins to cope with abiotic stress. How ORP proteins are involved in stress response is largely unknown. When plants are subjected to abiotic stress, membrane proteins degrade, and comparative conductivity and MDA are significantly elevated [12, 25]. In this study, electrolytes in silenced plants (pCLCrVA: GhORP_A02) increase significantly under drought stress compared to control plants. Both chlorophyll content and relative water contents decrease significantly in silenced plants, and this is in agreement with many previous research findings, which indicated that plants tend to close stomata to avoid water loss and decrease photosynthesis in drought conditions [22, 29, 46]. The reactive oxygen system produces substances such as POD and CAT, which are accompanied by an increase in reactive oxygen to limit and regulate the damage of reactive oxygen to plants, but also as a signal molecule to activate the plant body to respond to the external adverse environment [2, 3]. Our present work showed that Both CAT and POD decreased in the pCLCrVA: GhORP_A02 plants when compared to control plants, and this signifies the signaling role of the ORP gene in enzymatic activity in cotton, which is consistent with previous research [33, 42].
Conclusion
In this research, we carried out genome-wide identification, and a total of 42 ORP genes were distributed in G. hirsutum, G. barbadense, G. arboreum, and G. raimondii. All genes showed one-to-one homology relationships among G. hirsutum, G. arboreum, and G. raimondii. Gene structure and phylogenetic analysis indicated that ORP genes classified into one clade have similar structures. Analysis of ORP genes in four Gossypium species revealed that most proteins are localized in the nucleus. The Ka/Ks ratio between orthologous gene pairs revealed that ORP genes had undergone purifying and positive selection during evolution. We also identified ABA, GA, auxin and drought stress response elements in promoter regions. Further expression analysis using transcriptome data indicated that four GhORP genes were highly expressed after abiotic stress treatment. Characterization of GhORP_A02 through virus-induced gene silencing found that GhORP_A02 participated in drought stress by inducing various physiological and biochemical changes. Our study provided a useful reference for further functional investigation of GhORP genes.
Availability of data and materials
Genome sequences of G. hirsutum acc. TM-1 (ZJU_V2.1), G. barbadense acc.3–79 (HAU_V2.0), G. arboreum (CRI_V3.0) and G. raimondii (JGI_V2.1) are available in the CottonFGD website (https://cottonfgd.net/about/download.html). Transcriptome data of TM-1 was downloaded from NCBI Sequence Read Archive under the accession number SRA180756.
Abbreviations
- ORP:
-
Oxysterol-binding protein-related proteins
- VIGS:
-
Virus-induced gene silencing
- ROS:
-
Reactive oxygen species
- G. arboreum :
-
Gossypium arboreum
- G. hirsutum :
-
Gossypium hirsutum
- G. barbadense :
-
Gossypium barbasense
- G. raimondii :
-
Gossypium raimondii
- MW:
-
Molecular weight
- NJ:
-
Neighbor-joining
- ML:
-
Maximum likelihood
- Ka:
-
Non-synonymous
- Ks:
-
Synonymous
- FPKM:
-
Fragments per kilobase of exon per million fragments mapped
- qRT-PCR:
-
Quantitative real-time PCR
- ABA:
-
Abscisic acid
- JA:
-
Jasmonic acid
- POD:
-
Peroxidas
- MDA:
-
Malondialdehyde
- CAT:
-
Catalase
References
Abdurakhmonov IY, Kohel RJ, Yu JZ, Pepper AE, Abdullaev AA, Kushanov FN, et al. Molecular diversity and association mapping of fiber quality traits in exotic G. hirsutum L. germplasm. Genomics. 2008;92:478–87.
Ahmed IM, Dai H, Zheng W, Cao F, Zhang G, Sun D, et al. Genotypic differences in physiological characteristics in the tolerance to drought and salinity combined stress between tibetan wild and cultivated barley. Plant Physiol Biochem. 2013;63:49–60.
Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006;141(2):391–6.
Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Res. 2015;43:W39–49.
Beasley JO. The production of polyploids in Gossypium. J Hered. 1940;31:39–48.
Bita CE, Gerats T. Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci. 2013;4:1–18.
Boyer JS. Plant productivity and environment. Science. 1982;218:443–8.
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an integrative Toolkit developed for interactive analyses of big Biological Data. Mol Plant. 2020;13:1194–202.
Chen XJ, Chen G, Li JP, Hao XY, Tuerxun Z, Chang XC, et al. A maize calcineurin B-like interacting protein kinase ZmCIPK42 confers salt stress tolerance. Plant Physiol. 2021;171:161–72.
Comas LH, Becker SR, Cruz VM, Byrne PF, Dierig DA. Root traits contributing to plant productivity under drought. Front Plant Sci. 2013;4:1–16.
Danquah A, Zelicourt A, Colcombet J, Hirt H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv. 2014;32:40–52.
Demirevska K, Simova-Stoilova L, Vassileva V, Feller U. Rubisco and some chaperone protein responses to water stress and rewatering at early seedling growth of drought sensitive and tolerant wheat varieties. Plant Growth Regul. 2008;56:97–106.
Dong T, Park Y, Hwang I. Abscisic acid: biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015;58:29–48.
Fang Y, Xiong L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell Mol Life Sci. 2015;72:673–89.
Field CB, Barros V, StockerV TF, Qin D, Dokken DJ, Ebi KL, et al IPCC. “Summary for policymakers,”. In: Managing the Risks of Extreme events and disasters to Advance Climate Change Adaptation. A Special Report of Working Groups I and II of the Intergovernmental Panel on Climate Change, eds. Cambridge: Cambridge University Press; 2012. pp. 1–19.
Fryxell PA. A revised taxonomic interpretation of Gossypium L. (Malvaceae). Rheedea. 1992;2:108–65.
Gechev TS, Hille J. Hydrogen peroxide as a signal controlling plant programmed cell death. J Cell Biol. 2005;168:17–20.
Grant CK, Kaoru U, Serge D, Mario P, Kazuo S. Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 2011;11:163.
Gu ZH, Huang CJ, Li FF, Zhou XP. A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol J. 2014;12(5):638-49.
Gu LJ, Wei HL, Wang HT, Su JJ, Yu SX. Characterization and functional analysis of GhWRKY42, a group IId WRKY gene, in upland cotton (Gossypium hirsutum L.). BMC Genet. 2018;19:1–14.
Hajheidari M, Eivazi A, Buchanan BB, Wong JH, Majidi I, Salekdeh GH. Proteomics uncovers a role for redox in drought tolerance in wheat. J Proteome Res. 2007;6:1451–60.
Hasheminasab H, Assad MT, Aliakbari A, Sahhafi SR. Evaluation of some physiological traits associated with improved drought tolerance in Iranian wheat. Annals Biol Res. 2012;3:1719–25.
Hu W, Liu Y, Loka DA, Zahoor R, Wang S, Zhou Z. Drought limits pollen tube growth rate by altering carbohydrate metabolism in cotton (Gossypium hirsutum) pistils. Plant Sci. 2019;286:108–17.
Kawakami EM, Oosterhuis DM, Snider JL. Physiological effects of 1-methylcyclopropene on well-watered and water-stressed cotton plants. J Plant Growth Regul. 2010;29:280–8.
Kocheva KV, Landjeva SP, Georgiev GI. Variation in ion leakage parameters of two wheat genotypes with different Rht-B1 alleles in response to drought. J Biosci. 2014;39:753–9.
Kohli A, Nick P. Exploring jasmonates in the hormonal network of drought and salinity responses. Front Plant Sci. 2015;6:1–16.
Kumar B, Pandey DM, Goswami CL, Jain S. Effect of growth regulators on photosynthesis, transpiration and related parameters in water stressed cotton. Biol Plant. 2001;44:475–8.
Lawlor MA, Alessi DR. PKB/Akt: a key mediator of cell proliferation, survival and insulin responses ? J Cell Sci. 2001;114(16):2903–10.
Lawlor DW, Cornic G. Photosynthetic carbon assimilation and associated metabolism in relation to water deficits in higher plants. Plant Cell Environ. 2002;25:275–94.
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van De Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–7.
Li DY, Inoue H, Takahashi M, Kojima T, Shiraiwa M, Takahara H. Molecular characterization of a novel salt inducible gene for an OSBP (oxysterol-binding protein)-homologue from soybean. Gene. 2008;407:12–20.
Li LB, Yu DW, Zhao FL, Pang CY, Song MZ, Wei HL, et al. Genome-wide analysis of the calcium-dependent protein kinase gene family in Gossypium raimondii. J Integr Agric. 2015;14:29–41.
Linyerera SM, Odongo MR, Cai XY, Nyangasi KJ, Xu YC, Gereziher MT, et al. Knockdown of 60S ribosomal protein L14–2 reveals their potential regulatory roles to enhance drought and salt tolerance in cotton. J Cott Res. 2021;4:27.
Liu R, Jiao T, Zhang Z, Yao Z, Li Z, Wang S, et al. Ectopic expression of the Allium cepa1-SST gene in cotton improves drought tolerance and yield under drought stress in the field. Front Plant Sci. 2022;12:783134.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆Ct method. Method. 2001;408:402–8.
Manna M, Thakur T, Chirom O, Mandlik R, Deshmukh R, Salvi P. Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Plant Physiol. 2021;172:847–68.
Mao H, Li S, Wang Z, Cheng X, Li F, Mei F, et al. Regulatory changes in TaSNAC8-6A are associated with drought tolerance in wheat seedlings. Plant Biotechnol J. 2020;18:1078–92.
Marchler-bauer A, Anderson JB, Cherukuri PF, Deweese-scott C, Geer LY, Gwadz M, et al. CDD: a conserved domain database for protein classification. Nucleic Acid Res. 2005;33:192–6.
Mehari TG, Xu Y, Magwanga RO, Umer MJ, Kirungu JN, Cai X, et al. Genome wide identification and characterization of light-harvesting Chloro a/b binding (LHC) genes reveals their potential role in enhancing drought tolerance in Gossypium hirsutum. J Cott Res. 2021;4,15.
Niu J, Zhang SP, Liu SD, Ma HJ, Chen J, Shen Q, et al. The compensation effects of physiology and yield in cotton after drought stress. J Plant Physiol. 2018;224:30–48.
Raychaudhuri S, Prinz WA. The diverse functions of oxysterol-binding proteins. Annu Rev Cell Dev Biol. 2010;26:157–77.
Sadau SB, Ahmad A, Tajo SM, Ibrahim S, Kazeem BB, Wei H, et al. Overexpression of GhMPK3 from cotton enhances cold, drought, and salt stress in Arabidopsis. Agronomy. 2021;11:1–18.
Saravanan RS, Slabaugh E, Singh VR, Lapidus LJ, Haas T, Brandizzi F. The targeting of the oxysterol-binding protein ORP3a to the endoplasmic reticulum relies on the plant VAP33 homolog PVA12. Plant J. 2009;58:817–30.
Shiraku ML, Magwanga RO, Zhang YY, Hou YQ, Kirungu JY, Mehari TG, et al. Late embryogenesis abundant gene LEA3 (Gh_A08G0694) enhances drought and salt stress tolerance in cotton. Int J Biol Macromol. 2022;207:700–14.
Skirpan AL, Dowd PE, Sijacic P, Jaworski CJ, Gilroy S, Kao TH. Identification and characterization of PiORP1, a Petunia oxysterol-binding-protein related protein involved in receptor-kinase mediated signaling in pollen, and analysis of the ORP gene family in Arabidopsis. Plant Mol Biol. 2006;61:553–65.
Sonone MP, Rathod TH, Dhage PS. Effect of moisture stress on cotton genotypes. Int J Chem Stud. 2020;8:232–5.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Umate P. Oxysterol binding proteins (OSBPs) and their encoding genes in Arabidopsis and rice. Steroids. 2011;76(5):524–9.
USDA-ERS. Cotton and wool outlook. 2017.
Vanhaesebroeck B, Leevers SJ, Timms J, Katso R, Driscoll PC, Woscholski R, et al. Synthesis and function of 3-phosphorylated inositol lipids. AnnuRev Biochem. 2001;70:535–602.
Yan D, Olkkonen VM. Characteristics of oxysterol binding proteins. Int Rev Cytol. 2008;265:253–85.
Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol. 2014;21:133–9.
Zhang W, Xu H, Duan X, Hu J, Li J, Zhao L, et al. Characterizing the leaf transcriptome of Chrysanthemum rhombifolium (Ling et C. Shih), a drought resistant, endemic plant from China. Front Genet. 2021;12:625985.
Zhao XB, Li CJ, Wan SB, Zhang TT, Yan CX, Shan SH. Transcriptomic analysis and discovery of genes in the response of Arachis hypogaea to drought stress. Mol Biol Rep. 2018;45:119–31.
Zhao L, Lv Y, Chen W, Yao J, Li Y, Li Q, et al. Genome-wide identification and analyses of the AHL gene family in cotton (Gossypium). BMC Genomics. 2020;21:1–14.
Zheng JY, Oluoch G, Riaz Khan MK, Wang XX, Cai XY, Zhou ZL, et al. Mapping QTLs for drought tolerance in an F2: 3 population from an inter-specific cross between Gossypium tomentosum and Gossypium hirsutum. Genet Mol Res. 2016;15:1–14.
Acknowledgements
We thank the Gene Bank of Institute of Cotton Research of Chinese Academy of Agricultural Sciences for providing the germplasm seeds. We thank Teame Gereziher (Institute of Cotton Research, Chinese Academy of Agricultural Science), Tajo Sammani (University of Maiduguri), Mustapha Dansabo Hauwa (Usman Danfodiyo University), Sani Ibrahim (Research Institute of Oil Crops, Chinese Academy of Agricultural Science), and Rabiu Sani Shawai (Institute of Crop Science, Chinese Academy of Agricultural Science) for providing valuable assistance and revising the original manuscript.
Funding
This work was supported by Research on key technologies of cotton germplasm resource collection and excellent gene mining, Major science and technology projects of Xinjiang Uygur Autonomous Region (2022A03004-2) and the Ministry of Agriculture and Rural Affairs of China’s Purchase Service (19221957).
Author information
Authors and Affiliations
Contributions
XG and XD conceived and designed the experiments. SMT, BC, and SBS performed the experiments. SMT, ZP, YJ, and SH analyzed the data. SMT and XG drafted the manuscript. YK, AAA, MFN, and UA help to revise the paper. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All experiments using plant materials in this study were carried out in accordance with relevant guidelines and regulations of the Ministry of Agriculture and Rural Affairs of China.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Table 1.
Primers used in this study.
Additional file 2: Supplementary Table 2.
Ka/Ks analysis of duplicated ORP gene pairs of G. hirsutumm, G.raimondii and G. arboretum.
Additional file 3: Figure S1.
Cis-acting elements identified in promoter regions of ORP genes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tajo, S.M., Pan, Z., Jia, Y. et al. Silencing of GhORP_A02 enhances drought tolerance in Gossypium hirsutum. BMC Genomics 24, 7 (2023). https://doi.org/10.1186/s12864-022-09099-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-022-09099-y