Abstract
Background
Dissolved oxygen (DO) in the water is a vital abiotic factor in aquatic animal farming. A hypoxic environment affects the growth, metabolism, and immune system of fish. Glycolipid metabolism is a vital energy pathway under acute hypoxic stress, and it plays a significant role in the adaptation of fish to stressful environments. In this study, we used multi-omics integrative analyses to explore the mechanisms of hypoxia adaptation in Genetically Improved Farmed Tilapia (GIFT, Oreochromis niloticus).
Results
The 96 h median lethal hypoxia (96 h-LH50) for GIFT was determined by linear interpolation. We established control (DO: 5.00 mg/L) groups (CG) and hypoxic stress (96 h-LH50: 0.55 mg/L) groups (HG) and extracted liver tissues for high-throughput transcriptome and metabolome sequencing. A total of 581 differentially expressed (DE) genes and 93 DE metabolites were detected between the CG and the HG. Combined analyses of the transcriptome and metabolome revealed that glycolysis/gluconeogenesis and the insulin signaling pathway were down-regulated, the pentose phosphate pathway was activated, and the biosynthesis of unsaturated fatty acids and fatty acid metabolism were up-regulated in GIFT under hypoxia stress.
Conclusions
The results show that lipid metabolism became the primary pathway in GIFT under acute hypoxia stress. Our findings reveal the changes in metabolites and gene expression that occur under hypoxia stress, and shed light on the regulatory pathways that function under such conditions. Ultimately, this information will be useful to devise strategies to decrease the damage caused by hypoxia stress in farmed fish.
Similar content being viewed by others
Background
Fish growth is affected by environmental elements such as dissolved oxygen (DO), temperature, salinity, the ammonia nitrogen concentration, and pH. In recent years, the effects of hypoxia stress on aquatic organisms have become increasingly severe [1]. This is because the DO in water is prone to decrease as a result of high-density farming, excessive feeding, and blue algae blooms that occur in fish farmed for breeding or for the commercial market [2]. Low DO affects the growth and feed utilization of aquatic organisms and disrupts their morphology, physiology, and behavioral adaptability [3, 4]. Compared with mammals, some aquatic animals have a strong ability to tolerate low oxygen levels [5,6,7]. However, in general, the normal life processes and body development of aquatic animals can be guaranteed when the DO concentration is higher than 4 mg/L [8].
Fish adapt to hypoxia stress through a series of complex physiological and biochemical processes, including a reduction in the metabolic rate, changes in metabolic pathways, and an improvement in the oxygen transport capacity [9, 10]. Many studies have found that when an organism is in an anoxic environment, glycolipid metabolism undergoes a series of changes [11,12,13,14]. Anaerobic metabolism can reduce oxygen consumption and quickly provide energy under hypoxia stress [15]. Previous studies have shown that turbot (Scophthalmus maximus) and largemouth bass (Micropterus salmoides) switch to anaerobic metabolism under hypoxia stress, causing a decrease in liver glycogen levels and an increase in lactic acid in the blood [16, 17]. Genetically Improved Farmed Tilapia (GIFT, Oreochromis niloticus) preferentially use triglyceride (TG) and serum glucose (GLU) to provide energy under acute hypoxia stress [18]. The compensation mechanism of fish under hypoxia stress is connected with increased lipid metabolism [19]. In obese mice, hypoxia stimulates the lipolysis of adipocytes and inhibits the absorption of free fatty acids (FFA) by adipocytes, leading to increased fatty acid concentrations in the plasma [20]. Other studies have found that chronic hypoxia inhibits glycolysis and increases lipolysis, revealing that lipid metabolism is the main energy supply pathway under chronic hypoxia stress [21, 22].
Tilapia, as the main freshwater fish in southern China, has a rapid growth rate and a high economic value. Tilapia is an ideal model to explore the effects of hypoxia stress on fish, because it is mainly cultivated in high-density ponds and shows strong adaptability to hypoxia [23]. There are different types of metabolism under acute hypoxia and long-term hypoxia stress. In Nile tilapia, different energy supply pathways function under short-term and long-term hypoxic conditions; carbohydrates supply energy during short-term hypoxia stress, and lipids become the primary energy source during long-term hypoxia stress [21]. Mahfouz et al. [2] found that glycolysis decreases and gluconeogenesis increases in the liver and muscle of tilapia under acute hypoxia stress. Long-term hypoxia stress has also been shown to affect the fatty acid composition and blood biochemical indicators of GIFT [24].
In the post-genomic era, the widespread use of transcriptome, proteome, and metabolome analyses has led to new insights into biology [25]. Transcriptomics is the study of gene expression at the RNA level, and provides information about differentially expressed (DE) genes and gene function [26]. Metabolomics explains the metabolic processes that occur organisms after stimulation or disruption by providing information about the types and metabolites and changes in their concentrations [27]. In recent years, multi-omics analysis methods have been used to study the stress responses of aquatic animals. For example, combined metabolome and transcriptome analyses of Litopenaeus vannamei revealed the relationships between certain secondary metabolites and the transcript levels of DE genes, thereby clarifying the mechanism of nitrite tolerance [28]. Combined metabolome and transcriptome analyses of tilapia under hypoxic stress have provided information about changes in liver metabolism and the cause of death. In the present study, we used a multi-omics method to analyze how glycolipid metabolism changes in GIFT liver tissues under hypoxia stress, and used qRT-PCR to verify the transcriptional patterns of eight DE genes related to glycolipid metabolism. This is the first report of combined transcriptome and metabolome analyses of the GIFT liver. Our results reveal changes in energy supply pathways under hypoxia stress, and provide information about the mechanisms of hypoxia adaptation and hypoxia tolerance in GIFT.
Results
Determination of 96 h-LH50 in GIFT
A decrease in DO (from 3.2 to 0.2 mg/L) significantly increased the mortality of GIFT (Table 1). When the DO was 3.2 or 1.6 mg/L, no fish death occurred within 96 h. However, GIFT began to die at 96 h when the DO level was 0.8 mg/L. When the DO level was 0.4 mg/L, the cumulative mortality at 12 h was 20%; and it gradually increased over time to 36.7 and 56.7% at 48 h and 96 h, respectively. When the DO level was 0.2 mg/L, the cumulative mortality at 96 h was 86.7%. The 96 h-LH50 calculated by linear interpolation was 0.536 mg /L. The linear regression equation was y = 18.696 + 58.406x (R = 0.868, P < 0.0001). On the basis of this result, we chose 0.55 mg/L as the 96 h-LH50 of GIFT for further experiments.
Hepatic biochemical indicators
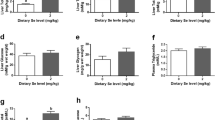
Figure 1 shows the changes in hepatic cholesterol (TC), triglyceride (TG), free fatty acids (FFA), and glycogen levels and lactate dehydrogenase (LDH) activity in GIFT after 96 h at DO 0.55 mg/L. At 96 h, the TG was significantly higher (P=0.0373< 0.05) in the hypoxia-stressed group (HG) than in the control group (CG) (Fig. 1a). Compared with the CG, the HG showed increased TC levels (Fig. 1b), but the difference was not significant (P=0.0786> 0.05). Hepatic glycogen (P=0.0187< 0.05) (Fig. 1c) and FFA levels (P=0.0145< 0.05) (Fig. 1d) were significantly higher in the HG than in the CG at 96 h. Compared with the CG, the HG showed significantly lower (P=0.0312< 0.05) hepatic LDH activity at 96 h (Fig. 1e).
Glucose and lipid metabolism indicators in liver of GIFT under 96 h acute hypoxia stress (n = 9 replicates per group). a triglyceride, TG; b total cholesterol, TC; c glycogen; d free fatty acids, FFAs; e lactate dehydrogenase, LDH. Asterisk (*) indicates significant difference (P < 0.05) between hypoxia stress group (HG) and control group (CG)
Metabolome profiles in liver of hypoxia stressed GIFT
The liver tissues were collected from the HG and CG for LC-MS analysis. The total ion chromatograms, m/z peak width, and retention time peak width of metabolites detected in liver samples from CG or HG in positive (POS) and negative (NEG) modes are shown in Fig. S1 and Fig. S2, respectively. These figures show the reliability of the metabolomics data in this trial and the stable performance of the UPLC-MS analyses.
A total of 13,765 and 10,308 features were obtained in the POS and NEG mode, respectively. After the data were processed and filtered, 11,303 and 8353 high-quality features were obtained in the POS and NEG mode, respectively (Table 2). The high-quality features were analyzed using multiple statistical methods, including principal component analysis (PCA) and partial least square discriminant analysis (PLSDA). Unsupervised PCA was performed prior to PLSDA. The PCA score plot (Fig. 2) revealed tight clustering for the quality control (QC) sample, indicative of good stability of the metabolic profiles. The first two primary components (PC1 and PC2) explained 53.54 and 54.55% of the PCA model in the POS and NEG modes, respectively, indicating that the high-quality features of the CG and HG were naturally separated and clustered (Fig. 2). The PLSDA models were further used to distinguish differences in metabolites among samples detected in the CG and HG in POS and NEG modes (Fig. 3). After 200 permutation tests, the R2 values were 0.9890 or 0.9913 and the corresponding Q2 values were 0.9654 or 0.9481 in the POS and NEG mode, respectively, indicating good credibility of the PLSDA models.
High-quality metabolites were selected for the DE metabolites analysis. The DE metabolites were identified on the basis of multiple statistical analyses. Metabolites with a variable importance in the projection (VIP) value> 1, fold change (FC) > 2 or < 0.5, and an adjusted P-value < 0.05 were considered to be significant. A total of 3028 DE features were identified in the comparative analysis between the HG and the CG, of which 1596 were up-regulated and 1432 were down-regulated in the HG compared with the CG (Table 2). We matched the obtained DE features at online databases such as Kyoto Encyclopedia of Genes and Genomes (KEGG) and the human metabolome database (HMDB), and further validated them by comparison with our in-house fragment spectrum library. In total, 93 DE metabolites were identified by MS2, with 76 and 17 in the POS and NEG modes, respectively. We detected 15 DE metabolites (Table 3) in the glucose and lipid metabolism pathways, of which five metabolites were down-regulated and eight were up-regulated.
Gene expression profiles in liver of hypoxia-stressed GIFT
We established and sequenced six mRNA libraries, three from the DO 5.00 mg/l control groups (CG-1, CG-2, and CG-3) and three from the 96 h-LH50 groups (HG-1, HG-2, and HG-3). The biological replicates had good repeatability. After removing low-quality raw sequences, there were 57,905,522, 46,781,934, 42,604,052, 38,581,554, 44,305,838, and 51,230,978 clean reads for the CG-1, CG-2, CG-3, HG-1, HG-2, and HG-3 libraries, respectively (98.05–98.89% valid data; Q20 values of 99.65–99.79%; Q30 values of 95.33–96.54%, and GC contents of 46.5–47%) (Table 4). The number of reads that mapped to the Nile tilapia genome was 44,797,588 (CG-1), 37,629,408 (CG-2), 32,951,044 (CG-3), 31,304,433 (HG-1), 35,303,322 (HG-2), and 41,293,836 (HG-3). More reads mapped to exon regions than to intron and intergenic regions in the genome (Fig. S3).
Genes with P< 0.05, fold-change≥2 or ≤0.5, and fragments per kilobase of exon model per million mapped reads (FPKM) > 10 were considered to be DE genes. We identified 2375 DE genes between the CG and HG libraries, of which 1201 were up-regulated and 1174 were down-regulated (Fig. 4a). A list of DE genes is provided in Table S1. The enrichment analysis of DE genes was conducted using tools at the Metascape database. In the Gene Ontology (GO) enrichment analysis, the GO pathways most enriched with DE genes were carboxylic acid metabolic process, oxidation-reduction process, small molecule catabolic process, dioxygenase activity, lipid metabolic process, and monosaccharide metabolic process. These results show that acute hypoxia stress strongly affects the immune regulation and metabolism of GIFT (Fig. 4b).
Differentially expressed (DE) genes and related Gene Ontology (GO) terms in liver of GIFT under hypoxia stress. a DE gene volcano plot graph. b Enriched GO terms based on DE genes. Red and blue dots in volcano plot graph represent up-regulated and down-regulated DE genes, respectively. X and y-axes represent log2(FC) value and -log10(P) value, respectively
We identified 581 DE genes using tools at the KEGG database. The KEGG pathway enrichment analysis of DE genes identified 20 pathways enriched with DE genes under hypoxia stress (P< 0.05, Table S2) (Fig. 5). The DE genes were mainly enriched in the metabolism, organism system, and immune regulation categories, and the main pathways were the glucose, lipid, amino acid, and vitamin metabolic pathways. The specific pathways enriched with DE genes were the insulin signaling pathway, glycolysis/gluconeogenesis, and fatty acid metabolism.
Integrated transcriptome and metabolome analysis
We identified the DE metabolites and genes in the same biological pathway. Five representative pathways of glucose and lipid metabolism are shown in Fig. S4, S5, S6, S7, S8. The main DE metabolites and top 40 DE genes in the glucose and lipid metabolism pathways were selected. Pearson’s correlation coefficient analyses were performed using the screened DE metabolites and genes. The selected DE metabolites and genes in the glucose and lipid metabolism pathways were subjected to correlation analyses (Fig. 6).
Correlation analysis between differentially expressed (DE) genes and DE metabolites in liver of GIFT under hypoxia stress. Blue squares represent DEGs; red circles represent DE metabolites; solid red line represents positive correlation between metabolites and genes; blue dotted line represents negative correlation between metabolites and genes
Validation of selected DE mRNAs by qRT-PCR
We conducted qRT-PCR analyses to validate the transcriptional patterns of eight DE genes (Table 5) involved in lipid metabolism under hypoxic stress. The changes in gene expression detected by qRT-PCR were consistent with those detected from the sequencing results. The transcript levels of ELOVL6 (encoding elongation of the very long chain fatty acid protein 6; ELOVL6) and ACAT2 (encoding acyl-coenzyme A: cholesterol acyltransferase 2; ACAT2) were significantly higher in the HG than in the CG. The transcript levels of genes encoding phosphoenolpyruvate carboxykinase 1 (PCK1), insulin receptor (INSR), heat shock protein family B (small) member 1 (HSPB1), myoglobin (MB), glyceraldehyde-3-phosphate dehydrogenase (GAPDHS), and lactate dehydrogenase A (LDHA) were significantly lower in HG than in CG (Fig. 7).
Discussion
A hypoxic environment can cause respiratory and metabolic disorders in fish, leading to increased mortality [29, 30]. Fish have evolved a variety of adaptation mechanisms, and adapt to the hypoxic environment by altering their energy supply and metabolic pathways [8, 31]. In this study, we researched responses in the GIFT liver to acute hypoxia stress through transcriptome and metabolome sequencing. We screened eight DE genes involved in the GIFT response to acute hypoxia stress and verified their transcriptional patterns by qRT-PCR. Relevant biochemical indexes and the activity of key enzymes were determined to clarify the effects of hypoxia on the glycolipid metabolic pathways of GIFT.
Carbohydrate metabolism is the main energy pathway for animals in an unstable environment [32, 33]. Previous studies have shown that fish have increased anaerobic metabolism and inhibited aerobic metabolism in the early stage of acute hypoxia stress [18, 21, 34]. For example, Trichogaster microlepis exposed to 12 h of hypoxia stress showed a decline in blood glucose and an increase in glucose metabolism to meet the body’s energy requirements [35]. In this study, we found that DE genes under hypoxia stress were enriched in several vital glucose metabolism pathways, including glycolysis/gluconeogenesis, the insulin signaling pathway, and the pentose phosphate pathway. Glycolysis is the main pathway of anaerobic metabolism [36]. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a multifunctional enzyme in glycolysis [37, 38]. It can catalyze the mutual conversion of 1,3-diphosphoglyceric acid and 3-phosphoglycerate and it participates in multiple biological processes, including DNA repair, membrane fusion and transport, RNA binding, autophagy, cell death cytoskeleton dynamics, and cytoskeleton dynamics [39]. Inactivation of GAPDHS can convert the carbohydrate decomposition process in cells from glycolysis to the pentose phosphate pathway, leading to the generation of NADPH and protection of cells against oxidative stress [40]. Lactate dehydrogenase (LDH), a marker enzyme of anaerobic metabolism, catalyzes the conversion of pyruvate and lactic acid in vivo. Its activity represents the level of anaerobic metabolism to a certain extent [41]. LDHA is a subunit of LDH, which is mainly expressed in anaerobic tissues of the liver [42]. A high concentration of pyruvate can increase the transcriptional levels and activity of LDHA. The oxygen partial pressure in tissue cells affects LDH activity. Differences in MB content reflect the oxygen supply to tissue cells, and thus indirectly reflect the LDH activity [43]. In this study, we detected down-regulation of GAPDHS, LDHA, and MB and decreased hepatic LDH activity in the GIFT liver at 96 h of hypoxia stress. Metabolome analyses showed that the level of alpha-D-glucose in the glycolytic pathway was significantly increased in hypoxia-stressed GIFT. Alpha-D-glucose is an important initial metabolite in the glycolysis pathway. In this study, the significant increase in alpha-D-glucose was consistent with the down-regulation of important regulatory genes in the glycolysis pathway, indicating that the glycolysis pathway was weakened under 96 h hypoxia stress. This would reduce the content of lactic acid, thereby reducing the damage caused by lactic acid accumulation in GIFT under acute hypoxia stress and reducing acidosis. Down-regulation of GAPDHS may be related to reducing oxidative stress injury. The results of this study are inconsistent with those of Su et al. [44], who detected a negative correlation between LDH activity and MB expression. It may be that the severe oxidative stress weakened the liver regulation capacity of GIFT, leading to decreases in both anaerobic metabolism and oxygen transport capacity after 96 h in a hypoxic environment (DO, 0.55 mg/L). Under acute hypoxic stress, the gluconeogenesis pathway in the fish liver may be inhibited [8, 45]. PCK1 can catalyze the production of phosphoenolpyruvate from oxaloacetic acid, releasing GDP and carbon dioxide. PCK1 is a rate-limiting enzyme that regulates gluconeogenesis, which participates in maintaining blood glucose levels and is considered indispensable for glucose homeostasis [46, 47]. The down-regulation of PCK1 detected in this study may lead to the inhibition of the gluconeogenic pathway and reduced generation of glucose and glycogen.
The insulin signaling pathway is a key pathway for the maintenance of glucose homeostasis in fish. Insulin is an essential hormone that controls critical energy functions such as glucose and lipid metabolism, and it is perceived by the insulin receptor, INSR. Only when insulin and INSR are combined can they play a role in lowering blood sugar. Any impairment of the relationship between INS and INSR will lead to insulin resistance [48]. HSPB1 encodes the heat shock protein HSP27, which is a member of the heat shock protein (HSP) family [49, 50]. This protein plays pivotal roles in apoptosis, insulin resistance, and inflammatory responses [51]. We detected significant down-regulation of INSR and HSPB1 in the liver of GIFT under hypoxia stress. We speculate that this may inhibit glucose consumption in GIFT, and may result in hypoxia-induced damage to GIFT by triggering inflammation and insulin resistance. These results confirmed that in GIFT under hypoxia stress, anaerobic glucose metabolism is weakened and glucose homeostasis is imbalanced, which may be related to the increased death rate at 96 h.
The metabolome analyses revealed increased contents of D-gluconate, N-acetyl-D-glucosamine, and N-acetyl-D-glucosamine-6-phosphate in the pentose phosphate pathway, indicating that this pathway was activated under acute hypoxia stress, similar to the results reported elsewhere [52]. Besides providing energy, the pentose phosphate pathway is also the main source of NADPH and ribose in the body [53], and it can provide raw materials for the synthesis of fatty acids, ribonucleic acids, and cholesterol. A previous study showed that NADPH functions as a reducing agent to reduce ROS and alleviate oxidative damage in the tilapia liver caused by acute hypoxia stress [54]. In this study, the pentose phosphate pathway was activated in GIFT under hypoxia stress. This pathway may consume glucose, which would help to maintain glucose homeostasis and, at the same time, produce NADPH to reduce oxidative damage.
Studies have shown that under long-term hypoxia stress in fish, lipid metabolism becomes the primary energy metabolism pathway [19, 34]. In this study, some of the DE genes in the liver of GIFT in response to acute hypoxia stress were related to the biosynthesis of unsaturated fatty acids and fatty acid degradation. Therefore, we verified the transcript levels of ACAT2 and ELOVL6 by qRT-PCR, and measured TC, TG, and FFA concentrations as indicators of liver function. The elongase of very long-chain fatty acids gene family comprises ELOVL1–7, and different members show catalytic activity towards different fatty acid substrates [55, 56]. In animals, ELOVL6 is one of the key genes in the biosynthesis of unsaturated fatty acids and its expression levels are related to regulation of this pathway. It catalyzes the extension of saturated fatty acids and monounsaturated fatty acids, and is an essential rate-limiting enzyme for long-chain fatty acids synthesis [57]. ACAT2 is a key enzyme that regulates cholesterol homeostasis in cells by catalyzing the synthesis of cholesterol esters, and it is specifically expressed in the liver [58]. It can also participate in β-oxidation and lipid metabolism [59]. In this study, key lipid metabolism genes such as ELOVL6 and ACAT2 were up-regulated, indicating that lipid metabolism-related pathways were activated in GIFT under hypoxia stress. This is consistent with the results of Sun [60] and others who found that lipid metabolism was enhanced in largemouth bass (Micropterus salmoides) under acute hypoxia stress. The transcriptional upregulation of ELOVL6 may activate the long-chain fatty acid synthesis pathway and increase the synthesis of n-3 and n-6 fatty acids. Interestingly, the content of stearic acid decreased, which may be due to the simultaneous increase in fatty acid synthesis and degradation. A greater metabolic rate than biosynthesis rate would lead to a decrease in the fatty acid content. Recent studies reported that the TG content in the liver and lipid peroxidation were increased in goby (Gillichthys mirabilis) and carp (Cyprinus carpio) under hypoxia stress [14, 61]. We obtained similar results in this study, in that the TG and FFAs contents were increased in the liver of GIFT at 96 h of acute hypoxic stress, while the TC content was not significantly changed. Similar results have been reported for largemouth bass under hypoxia stress [34]. These results indicate that hypoxia stress activated the unsaturated fatty acid biosynthetic pathway in the GIFT liver, leading to increased lipid production. Our results show that the biosynthesis of unsaturated fatty acids and the fatty acid degradation pathway were up-regulated under 96 h acute hypoxic stress, indicating that GIFT rely on lipid metabolism to provide energy under acute hypoxic stress.
Conclusions
We reported for the first time the changes in the liver metabolism of GIFT under hypoxia stress based on combined transcriptome and metabolome analyses. We detected 2375 DE genes between the HG and CG groups, of which 581 DE genes were annotated by KEGG. We verified the transcript levels of eight DE genes by quantitative real-time PCR. We screened 15 DE metabolites related to glucose and lipid metabolism and conducted correlation analysis of DE metabolites and DE genes related to glucose and lipid metabolism. These analyses revealed that glucose metabolism and lipid metabolism change markedly in GIFT under hypoxia stress, and lipid metabolism becomes the main metabolic mode (Fig. 8). These findings provide vital information about the regulatory mechanisms of GIFT as they adapt to hypoxic conditions.
Methods
Experimental fish
The experimental fish were obtained from Yixing, a base of the Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences. Before the start of experiment, the fish were cultured for 15 days in an indoor water-cycling system with DO > 5 mg/L, 28±1 °C, and pH 7.6±0.2. During this period, fish were fed twice a day with feed containing 8% (w/w) crude fat and 30% (w/w) crude protein at 7:00 and 16:00. Before the start of experiment, the fish were fasted for 24 h.
Determination of 96-h median lethal hypoxia for GIFT
We selected five low DO levels (3.2, 1.6, 0.8, 0.4, and 0.2 mg/L) to determine the 96 h median lethal hypoxia (96 h-LH50) for GIFT. The preliminary experiment was performed in 400-L plastic tanks (10 fish per tank, a total of 150 fish), and each tank was filled with 150 L aerated tap water. According to the method of Jiang et al. [62], real-time readings using a DO meter (LD0101 probe, range 0.1–20.0 mg/L, Hach, Loveland, USA) were used to detect and control the DO in water. At the beginning of the experiment, the DO in the experiment tanks reached oxygen requirement within 2 h by filling with nitrogen [34]. In the experiment, we observed the conditions of the experimental fish every hour, and caught the dead fish in time and recorded it. After the experiment, the surviving experimental fish were temporarily kept in the tank for a week, and then put back into the pond. We calculated the cumulative mortality of each treatment group within 96 h, and obtained the 96 h-LH50 of GIFT through linear interpolation.
Treatment and sampling
A total of 240 experimental fish were used for the acute hypoxic stress experiment. The GIFT were randomly sorted into six tanks with 40 fish per tank; there were three hypoxic stress groups (HG) and three 5 mg/L control groups (CG). The DO of the three treatment groups was rapidly reduced to 96 h-LH50 using a nitrogen gas cylinder to pump nitrogen into the water [34]. The DO in water was detected by a DO meter and controlled by adjusting the oxygen intake. At the end of the 96-h acute hypoxia stress experiment, all GIFT were rapidly deep anesthetized by the following methods. Three grams Tricaine methanesulfonate (MS-222, Argent Chemical Laboratories, Redmond, WA, USA) was dissolved in 30 L aquaculture water to prepare 100 mg L− 1 MS-222 anesthetics in a plastic bucket. Then we soaked 10 GIFT each time with the above anesthetic for 5 min. After 5 min, fish body remained still, the abdomen facing up and the gill movement is not continuous. Then the GIFT were considered under deep anesthesia. After this, the GIFT were euthanized by rapid cooling method. The specific operation method was to soak the experimental fish in 30 L 0 °C ice water mixture (25 L of ice and 5 L of water) for 10 min until the gill cap did not open or close within 60 s. It was identified that the fish had been euthanized and then the subsequent sampling experiment was conducted [63]. Three fish were randomly taken from each tank to sample liver tissues. The liver samples were divided into two parts: one part of the liver was used for high-throughput transcriptome analysis; and the other part was used to analyze gene transcript levels by qRT-PCR. Another 13 fish were caught from each tank and their liver tissues were sampled. Ten of those samples were used for LC-MS metabolome analysis and three were used for biochemical indicator analysis. All the samples were frozen quickly in liquid nitrogen and stored at − 80 °C until subsequent experiments.
Measurement of hepatic biochemical indicators
Liver samples (about 0.1 g) were homogenized in precooled phosphate buffer (50 mmol L− 1, pH 7.4) and then centrifuged for 20 min (4 °C, 3000 g). Test kits purchased from the Nanjing Jiancheng Bioengineering Institute (Nanjing, China) were used to measure the hepatic TC, TG, and FFA contents. Enzyme-linked immunosorbent assays were used to measure liver glycogen content and LDH activity, with kits from the Shanghai Longton Biotechnology Co., Ltd. (Shanghai, China).
Metabolite extraction and parameter settings
The liver tissue samples (about 50 mg) were homogenized in 120 μL pre-cooled 50% methanol buffer using a high-throughput tissue lyser (Ningbo, China). The solutions were placed on ice for 10 min and then centrifuged for 20 min (4 °C, 4000 g). The supernatants were collected and used for metabolomic analysis.
All chromatographic separations were performed using an ultra-performance liquid chromatography (UPLC) system (SCIEX, Warrington, UK) equipped with an ACQUITY UPLC BEH Amide column (100 mm × 2.1 mm, 1.7 μm, Waters, Manchester, UK). The sample injection volume was 4 μl, and the flow rate was 0.4 mL/min. The solvent system consisted of solvent A (25 mM ammonium acetate + 25 mM NH4H2O) and solvent B (IPA: ACN=9:1 + 0.1% formic acid) and the elution gradient program was as follows: hold at 95% B for 0.5 min, decrease linearly from 95 to 65% B (0.5–9.5 min), decrease linearly from 65 to 40% B (9.5–10.5 min), hold at 40% B (10.5–12 min), linear increase from 40 to 95% B (12–12.2 min) and hold at 95% B until 15 min.
Mass spectrometry data acquisition was performed using a high-resolution tandem mass spectrometer TripleTOF5600plus (SCIEX). The Q-TOF was manipulated in both POS and NEG ion modes using the following parameters: curtain gas and nebulizer pressure set to 30 PSI and 60 PSI, respectively; interface heater temperature, 650 °C. In POS and NEG ion modes, the detector voltage was 5 kV and − 4.5 kV, respectively. The data were collected in IDA mode, and the mass spectra scan range was set to m/z 60 to 1200. The QC samples were prepared by mixing the supernatant mixture of all eight individual liver samples. These QC samples were injected at the beginning, middle, and end of the analytical process to ensure stability of the LC–MS analyses [64].
Identification and quantification of metabolites
After LC−MS analysis, raw data were converted into mzXML format using ProteoWizard (version 3.0.18199). Then, XCMS software (http://xcmsonline.scripps.edu) was used for peak alignment, extraction, and quantification [65]. The peak data were further processed by the MetaX toolbox implemented with R software and CAMERA. Detectable features were first processed to remove low-quality features (detected in less than 50% of QC samples or 80% of biological samples). The remaining features with missing values were then filled using the K-nearest neighbor algorithm [66]. Before PCA, data normalization was conducted using the probabilistic quotient normalization method [67] and the quality control-base robust loess signal correction method [68]. After data normalization, features with a coefficient of variation > 30% in all QC samples were removed. The remaining features were considered high-quality and used for further analyses. We used PCA to visualize the datasets of the live samples in the HG and CG. PLSDA was then used for cluster analysis and to build a discriminant model. Permutation tests (n = 200) were used to evaluate the validity of PLSDA models [69].
Analysis of DE metabolites
The DE metabolites were preliminarily screened based on variable importance in the projection (VIP) values generated by the PLSDA model in this study. The VIP cut-off value was set at 1.0 and VIP values > 1.0 were chosen as significant discriminatory features between the CG and HG. Then, these features were confirmed by Wilcoxon rank sum test [70]. The P-value was adjusted by Benjamini–Hochberg’s approach. The fold change (FC) of each feature between these two groups was calculated as follows: FC = mean value of normalized ion intensity obtained from CG / mean value of normalized ion intensity obtained from HG. Features with a VIP value > 1, FC > 2 or < 0.5, and an adjusted P-value < 0.05 were selected as DE features and used for further analyses. These criteria were selected on the basis of previous studies [71, 72].
DE features were annotated in KEGG (http://www.kegg.jp/) [73] and HMDB (http://www.hmdb.ca/) according to Tao et al. [74]. The mass data of each feature were compared with that in the database (mass tolerance, 10 ppm). We also used an in-house fragment spectrum library of metabolites to validate the metabolite identification [72].
RNA sequencing
Total RNA was extracted from liver samples using Trizol reagent (Invitrogen, CA, USA) according to the manufacturer’s instructions. The total RNA quality and purity were analyzed using a Bioanalyzer 2100 instrument and RNA 6000 Nano Lab Chip Kit (Agilent, Palo Alto, CA, USA) for samples with RNA integrity number (RIN) > 7.0. Poly(A) mRNA was separated from approximately 10 μg total RNA with poly-T oligo-attached magnetic beads (Invitrogen, Carlsbad, CA, USA). Following purification, the mRNA was fragmented into small pieces using divalent cations under elevated temperature. According to the protocol of the mRNA Seq Sample Preparation kit (Illumina, San Diego, CA, USA), mRNA was reverse-transcribed to establish six cDNA libraries. Then, according to the vendor’s recommended protocol, paired-end sequencing (300 ± 50 bp) was performed on the Illumina Hiseq4000 platform (LC Sciences, Santiago, CA, USA) [75].
Analysis of DE genes
The mapped data set in each sample was assembled using StringTie to reconstruct the complete transcriptome. Then, Ballgown was used to estimate the differential expression levels of genes using the complete transcriptome assembled by StringTie. The transcript levels of mRNAs and genes were estimated based on FPKM values [76]; DE genes were identified to obtain a P-value for each gene between any pair of samples, according to Audic et al. [77]. We calculated the false discovery rate (FDR) using the corrected P-value. We selected DE mRNAs and genes based on log2 (fold change) > 1 or log2 (fold change) <− 1 and corrected P < 0.05 using the R package, Ballgown. GO (http://geneontology.org/) and KEGG enrichment analyses [73] were performed to determine the functions of the DE genes and the metabolic pathways associated with them, respectively, according to the method of Qiang et al. [78]. The GO enrichment analysis of DE genes was carried out using tools at the Metascape database (https://metascape.org/gp/index.html#/main/step1), and a map was drawn. KEGG enrichment analysis of DE genes was performed to make scatter plots in ggplot2 (https://ggplot2.tidyverse.org/).
Integrated analysis of metabolite and transcription profiles
On the basis of the transcriptome and metabolome results, DE metabolites and DE genes annotated in the same KEGG pathway were observed. Then, we selected the DE metabolites and top 40 DE genes (in order of |log2(fold change) | from large to small) in pathways related to glucose and lipid metabolism for subsequent analysis. Pearson’s correlation coefficient analysis was performed on the selected DE metabolites and genes using the cor program from R (version 3.5.1) [79, 80].. The correlation analysis screening conditions were |r|> 0.9 and p value< 0.01. The OmicStudio tools (https://www.omicstudio.cn/tool) were used for network map analysis of the screened pairs.
Validation of DE gene in GIFT liver after 96 h hypoxic stress by qRT-PCR
We verified the expression patterns of selected DE genes associated with changes in the glycolipid metabolism pathway under hypoxia stress by qRT-PCR, with the primers shown in Table S3. Total RNA was extracted from liver tissues from the CG and HG with Trizol reagent (Invitrogen). We used PrimeScript™ RT Master Mix (Takara, Dalian, China) for the RT reaction of the mRNAs. Each 10 μL RT mixture included RNA sample (≤ 500 ng), 2 μL 5 × PrimeScript RT Master Mix, and RNase Free dH2O to complete the volume to 10 μL. The reaction procedure was as follows: 37 °C for 15 min, 85 °C for 5 s, and the reaction was terminated by cooling to 4 °C. We used SYBR® Premix Ex Taq kits (Takara) for analyzing DE genes using a 7900HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). Each 25-μL qRT-PCR mixture included 12.5 μL SYBR (2◊) Advantage Premix, 0.5 μL ROX Dye II (5◊), 1 μL forward primer, 1 μL reverse primer, 2 μL cDNA template, and 8 μL RNase-free dH2O. The PCR reaction method was that described previously by Qiang et al. [81]. The Ct values measured for each sample were normalized against the value of β-Actin, and the relative fold change relative to β-Actin expression was calculated by the 2-ΔΔCt method.
Data analysis
Data were expressed as means±standard error. Independent-sample t tests was used to detect differences between the CG and HG. P values < 0.05 were considered to indicate significance.
Availability of data and materials
The raw sequencing data generated and analyzed in this study have been deposited at the Gene Expression Omnibus (GEO) repository under the accession number (GSE146142) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146142). The hepatic metabolome data in this study have been deposited at Dryad: https://doi.org/10.5061/dryad.qjq2bvqdw.
Abbreviations
- GIFT:
-
Genetically improved farmed tilapia
- DO:
-
Dissolved oxygen
- 96 h-LH50:
-
96 h median lethal hypoxia
- CG:
-
Control group
- HG:
-
Hypoxia stress group
- DE:
-
Differentially expressed
- GLU:
-
Glucose
- TC:
-
Cholesterol
- FFA:
-
Free fatty acids
- TG:
-
Triglyceride
- LDH:
-
Lactate dehydrogenase
- UPLC:
-
Ultra-performance liquid chromatography
- QC:
-
Quality control
- HMDB:
-
Human metabolome database
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- GO:
-
Gene ontology
- FPKM:
-
Fragment per kilobase of exon per million fragments mapped
- FC:
-
Fold change
- FDR:
-
False discovery rate
- ELOVL6:
-
Elongation of the very long chain fatty acid protein 6
- ACAT2:
-
Acyl-coenzyme A: cholesterol acyltransferase 2
- PCK1:
-
Phosphoenolpyruvate carboxykinase 1
- INSR:
-
Insulin receptor
- HSPB1:
-
Heat shock protein family B (small) member 1
- LDHA:
-
Lactate dehydrogenase A
- MB:
-
Myoglobin
- GAPDHS:
-
Glyceraldehyde-3-phosphate dehydrogenase
- PCA:
-
Principal component analysis
- PLSDA:
-
Partial least square discriminant analysis
References
Diaz RJ, Rosenberg R. Spreading dead zones and consequences for marine ecosystems. Science. 2008;321(5891):926–9.
Mahfouz ME, Hegazi MM, El-Magd MA, Kasem EA. Metabolic and molecular responses in Nile tilapia, Oreochromis niloticus during short and prolonged hypoxia. Mar Freshw Behav Physiol. 2015;48(5):319–40.
Abdel-Tawwab M, Hagras AE, Elbaghdady HAM, Monier MN. Effects of dissolved oxygen and fish size on Nile tilapia, Oreochromis niloticus (L.). growth performance, whole-body composition, and innate immunity. Aquac Int. 2015;23(5):1261–74.
Robertson CE, Wright PA, Koblitz L, Bernier NJ. Hypoxia-inducible factor-1 mediates adaptive developmental plasticity of hypoxia tolerance in zebrafish, Danio rerio. Proc Biol. 2014;281(1786):130–43.
Hardy KM, Follett CR, Burnett LE, Lema SC. Gene transcripts encoding hypoxia-inducible factor (HIF) exhibit tissue- and muscle fiber type-dependent responses to hypoxia and hypercapnic hypoxia in the Atlantic blue crab, Callinectes sapidus. Comp Biochem Physiol A-Mol Integr Physiol. 2012;163(1):137–46.
Kodama K, Rahman MS, Horiguchi T, Thomas P. Assessment of hypoxia-inducible factor-1 alpha mRNA expression in mantis shrimp as a biomarker of environmental hypoxia exposure. Biol Lett. 2012;8(2):278–81.
Brouwer M, Brown-Peterson NJ, Larkin P, Patel V, Denslow N, Manning S, et al. Molecular and whole animal responses of grass shrimp, Palaemonetes pugio, exposed to chronic hypoxia. J Exp Mar Biol Ecol. 2007;341(1):16–31.
Richards JG. Physiological, behavioral and biochemical adaptations of intertidal fishes to hypoxia. J Exp Biol. 2011;214(2):191–9.
Terova G, Rimoldi S, Cora S, Bernardini G, Gornati R, Saroglia M. Acute and chronic hypoxia affects HIF-1 alpha mRNA levels in sea bass (Dicentrarchus labrax). Aquaculture. 2008;279(1–4):150–9.
Chabot D, Claireaux G. Environmental hypoxia as a metabolic constraint on fish: The case of Atlantic cod, Gadus morhua. Mar Pollut Bull. 2008;57(6–12):287–94.
Moulton TL, Chapman LJ, Krahe R. Effects of hypoxia on aerobic metabolism and active electrosensory acquisition in the African weakly electric fish Marcusenius victoriae. J Fish Biol. 2020;96(2):496–505.
Ding J, Liu C, Luo SY, Zhang YB, Gao XM, Wu XF, et al. Transcriptome and physiology analysis identify key metabolic changes in the liver of the large yellow croaker (Larimichthys crocea) in response to acute hypoxia. Ecotox Environ Safe. 2020;109957:189.
Kumar A, Gopesh A. Effect of Hypoxia and Energy Conservation Strategies in the Air-Breathing Indian Catfish, Clarias batrachus. Natl Acad Sci Lett-India. 2015;38(2):135–7.
Gracey AY, Lee TH, Higashi RM, Fan T. Hypoxia-induced mobilization of stored triglycerides in the euryoxic goby Gillichthys mirabilis. J Exp Biol. 2011;214(18):3005–12.
Heinrichs-Caldas W, Campos DF, Paula-Silva MN, Almeida-Val VMF. Oxygen-dependent distinct expression of hif-1 alpha gene in aerobic and anaerobic tissues of the Amazon Oscar, Astronotus crassipinnis. Comp Biochem Physiol B-Biochem Mol Biol. 2019;227:31–8.
Pichavant K, Maxime V, Thebault MT, Ollivier H, Garnier JP, Bousquet B, et al. Effects of hypoxia and subsequent recovery on turbot Scophthalmus maximus: hormonal changes and anaerobic metabolism. Mar Ecol-Prog Ser. 2002;225:275–85.
Yang S, Wu H, He K, Yan T, Zhou J, Zhao LL, et al. Response of AMP-activated protein kinase and lactate metabolism of largemouth bass (Micropterus salmoides) under acute hypoxic stress. Sci Total Environ. 2019;666:1071–9.
Sheng Y, Hua ZY, Yang Z, Wei XL, Sheng YJ, Jia HL, et al. Effects of acute hypoxic stress on biochemical parameters, immune regulation and metabolic capacity of the blood in genetically improved farmed tilapia (GIFT, Oreochromis niloticus). J Appl Ichthyol. 2019;35(4):978–86.
Zhao LL, Sun JL, Liang J, Liu Q, Luo J, Li ZQ, et al. Enhancing lipid metabolism and inducing antioxidant and immune responses to adapt to acute hypoxic stress in Schizothorax prenanti. Aquaculture. 2020;519:734933.
Yin J, Gao ZG, He Q, Zhou DQ, Guo ZK, Ye JP. Role of hypoxia in obesity-induced disorders of glucose and lipid metabolism in adipose tissue. Am J Physiol-Endocrinol Metab. 2009;296(2):E333–42.
Li MX, Wang XD, Qi CL, Li EC, Du ZY, Qin JG, et al. Metabolic response of Nile tilapia (Oreochromis niloticus) to acute and chronic hypoxia stress. Aquaculture. 2018;495:187–95.
Meer DLMVD, Witte F, Bakker MAGD, Besser J, Richardson MK, Spaink HP. Gene expression profiling of the long-term adaptive response to hypoxia in the gills of adult zebrafish. Am J Phys. 2005;289(5):R1512–9.
Lague SL, Speers-Roesch B, Richards JG, Farrell AP. Exceptional cardiac anoxia tolerance in tilapia (Oreochromis hybrid). J Exp Biol. 2012;215(8):1354–65.
Chen DJ, Qiang J, Tao YF, Bao JW. zhu HJ, Hu LJ et al. Effects of different dissolved oxygen levels on the growth, blood biochemistry, fatty acid composition and against Streptococcus iniae infection of GIFT juvenile (Oreochromis niloticus). Freshwater Fisheries. 2019;49(04):83–9 (in chinese).
Saito K, Matsuda F. Metabolomics for Functional Genomics, Systems Biology, and Biotechnology. Annu Rev Plant Biol. 2010;61(1):463–89.
Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63.
Fiehn O. Metabolomics - the link between genotypes and phenotypes. Plant MolBiol. 2002;48(1–2):155–71.
Xiao J, Liu QY, Du JH, Zhu WL, Li QY, Chen XL, et al. Integrated analysis of physiological, transcriptomic and metabolomic responses and tolerance mechanism of nitrite exposure in Litopenaeus vannamei. Sci Total Environ. 2020;134416:711.
Anttila K, Lewis M, Prokkola JM, Kanerva M, Seppanen E, Kolari I, et al. Warm acclimation and oxygen depletion induce species-specific responses in salmonids. J Exp Biol. 2015;218(10):1471–7.
McBryan TL, Healy TM, Haakons KL, Schulte PM. Warm acclimation improves hypoxia tolerance in Fundulus heteroclitus. J Exp Biol. 2016;219(4):474–84.
Mandic M, Todgham AE, Richards JG. Mechanisms and evolution of hypoxia tolerance in fish. Proc R Soc B. 2009;276(1657):735–44.
Bacca H, Huvet A, Fabioux C, Daniel JY, Delaporte A, Pouvreau S, et al. Molecular cloning and seasonal expression of oyster glycogen phosphorylase and glycogen synthase genes. Comp Biochem Physiol B-Biochem Mol Biol. 2005;140(4):635–46.
Sun JL, Liu Q, Zhao LL, Cui C, Wu H, Liao L, et al. Potential regulation by miRNAs on glucose metabolism in liver of common carp (Cyprinus carpio) at different temperatures. Comp Biochem Physiol D-Genomics Proteomics. 2019;100628:32.
Sun JL, Zhao LL, Wu H, Liu Q, Liao L, Luo J, et al. Acute hypoxia changes the mode of glucose and lipid utilization in the liver of the largemouth bass (Micropterus salmoides). Sci Total Environ. 2020;135157:713.
Huang CY, Lin HC, Lin CH. Effects of hypoxia on ionic regulation, glycogen utilization and antioxidative ability in the gills and liver of the aquatic air-breathing fish Trichogaster microlepis. Comp Biochem Physiol A-Mol Integr Physiol. 2015;179:25–34.
Mizock BA. Alterations in carbohydrate-metabolism during stress - a review of the literature. Am J Med. 1995;98(1):75–84.
Seidler NW. Basic Biology of GAPDH. Adv Exp Med Biol. 2013;985:1–36.
Camacho-Jimenez L, Peregrino-Uriarte AB, Martinez-Quintana JA, Yepiz-Plascencia G. The glyceraldehyde-3-phosphate dehydrogenase of the shrimp Litopenaeus vannamei: Molecular cloning, characterization and expression during hypoxia. Mar Environ Res. 2018;138:65–75.
Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: Views from different subcellular compartments. Cell Signal. 2011;23(2):317–23.
Ralser M, Wamelink MM, Kowald A, Gerisch B, Heeren G, Struys EA, et al. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J Biol. 2007;6(4):10.
Everse J, Kaplan NO. Lactate dehydrogenases: structure and function. Adv Enzymol Relat Areas Mol Biol. 1973;37:61–133.
Cui XG, Han ZT, He SH, Wu XD, Chen TR, Shao CH, et al. HIF1/2 alpha mediates hypoxia-induced LDHA expression in human pancreatic cancer cells. Oncotarget. 2017;8(15):24840–52.
Wei DB, Ma JB. The investigation of comparison between contents of myoglobin and activity of lactate dehydrogenase in heart musule and skeletal Musule of plateau zokor and mouse. J Qinghai Univ. 2001;02:20–1.
Su YJ. Molecular cloning & purification of myoglobin in yak and corresponding enery metabolism pilot study. master. Southwest University for Nationalities. 2006. (in chinese).
Genz J, Jyde MB, Svendsen JC, Steffensen JF, Ramlov H. Excess post-hypoxic oxygen consumption is independent from lactate accumulation in two cyprinid fishes. Comp Biochem Physiol A-Mol Integr Physiol. 2013;165(1):54–60.
Lin JY, Yuan Y, Liu XG. The research progress of PCK1. Chem Life. 2019;39(04):637–42 (in chinese).
Beale EG, Forest C, Hammer RE. Regulation of cytosolic phosphoenolpyruvate carboxykinase gene expression in adipocytes. Biochimie. 2003;85(12):1207–11.
Aziz AUR, Farid S, Qin KR, Wang HQ, Liu B. Regulation of insulin resistance and glucose metabolism by interaction of PIM kinases and insulin receptor substrates. Arch Physiol Biochem. 2020;126(2):129–38.
Fan WB, Zhou Y, Jiang XP, Yang XR, Wang RY. Changes of heat shock protein 70 expression in gastrocnemius muscles in different recovery environments after hypoxic exercise. Chin J Appl Physiol. 2009;25(04):460–461+499 (in chinese).
Huang ZW, Zhou M, Wang Q, Zhu MJ, Chen S, Li H. Mechanical and hypoxia stress can cause chondrocytes apoptosis through over-activation of endoplasmic reticulum stress. Arch Oral Biol. 2017;84:125–32.
Furukawa S, Fujita T, Shimabukuro M, Iwaki M, Yamada Y, Nakajima Y, et al. Increased oxidative stress in obesity and its impact on metabolic syndrome. J Clin Invest. 2004;114(12):1752–61.
Qi M, Wu Q, Liu T, Hou Y, Miao Y, Hu M, et al. Hepatopancreas Transcriptome Profiling Analysis Reveals Physiological Responses to Acute Hypoxia and Reoxygenation in Juvenile Qingtian Paddy Field Carp Cyprinus carpio var qingtianensis. Front Physiol. 2020;11:1110.
Hu W, Zhi L, Zhuo MQ, Zhu QL, Zheng JL, Chen QL, et al. Purification and characterization of glucose 6-phosphate dehydrogenase (G6PD) from grass carp (Ctenopharyngodon idella) and inhibition effects of several metal ions on G6PD activity in vitro. Fish Physiol Biochem. 2013;39(3):637–47.
Jin ES, Lee MH, Murphy RE, Malloy CR. Pentose phosphate pathway activity parallels lipogenesis but not antioxidan processes in rat liver. Am J Physiol-Endocrinol Metab. 2018;314(6):E543–51.
Ohno Y, Suto S, Yamanaka M, Mizutani Y, Mitsutake S, Igarashi Y, et al. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc Natl Acad Sci U S A. 2010;107(43):18439–44.
Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog Lipid Res. 2006;45(3):237–49.
Junjvlieke Z, Khan R, Mei C, Cheng G, Wang S, Raza SHA, et al. Effect of ELOVL6 on the lipid metabolism of bovine adipocytes. Genomics. 2019;112(3):2282–90.
Chang TY, Li BL, Chang CCY, Urano Y. Acyl-coenzyme A: cholesterol acyltransferases. Am J Physiol-Endocrinol Metab. 2009;297(1):E1–9.
Sodhi SS, Ghosh M, Song KD, Sharma N, Kim JH, Kim NE, et al. An Approach to Identify SNPs in the Gene Encoding Acetyl-CoA Acetyltransferase-2 (ACAT-2) and Their Proposed Role in Metabolic Processes in Pig. PLoS One. 2014;9(7):e102432.
Sun JL, Zhao LL, He K, Liu Q, Luo J, Zhang DM, et al. MiRNA-mRNA integration analysis reveals the regulatory roles of miRNAs in the metabolism of largemouth bass (Micropterus salmoides ) livers during acute hypoxic stress. Aquaculture. 2020;526:735362.
Mustafa SA, Karieb SS, Davies SJ, Jha AN. Assessment of oxidative damage to DNA, transcriptional expression of key genes, lipid peroxidation and histopathological changes in carp Cyprinus carpio L. following exposure to chronic hypoxic and subsequent recovery in normoxic conditions. Mutagenesis. 2015;30(1):107–16.
Jiang LX, Pan LQ, Fang B. Effect of dissolved oxygen on immune parameters of the white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 2005;18(2):185–8.
Koerber AS, Kalishman J. Preparing for a Semiannual IACUC Inspection of a Satellite Zebrafish (Danio rerio) Facility. J Amer Assoc Lab Anim Sci. 2009;48(1):65–75.
Zelena E, Dunn WB, Broadhurst D, Francis-McIntyre S, Carroll KM, Begley P, et al. Development of a Robust and Repeatable UPLC-MS Method for the Long-Term Metabolomic Study of Human Serum. Anal Chem. 2009;81(4):1357–64.
Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using Nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78(3):779–87.
Hrydziuszko O, Viant MR. Missing values in mass spectrometry based metabolomics: an undervalued step in the data processing pipeline. Metabolomics. 2012;8(1):S161–74.
Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in H-1 NMR metabonomics. Anal Chem. 2006;78(13):4281–90.
Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6(7):1060–83.
Qiu LG, Song QQ, Jiang X, Zhao HW, Chen H, Zhou HL, et al. Comparative gonad protein and metabolite responses to a binary mixture of 2,4 '-DDT and benzo(a) pyrene in the female green mussel Perna viridis. Metabolomics. 2016;12(8):11.
Degu A, Hochberg U, Sikron N, Venturini L, Buson G, Ghan R, et al. Metabolite and transcript profiling of berry skin during fruit development elucidates differential regulation between Cabernet Sauvignon and Shiraz cultivars at branching points in the polyphenol pathway. BMC Plant Biol. 2014;14:188.
Cao MS, Li CJ, Liu YL, Cai KL, Chen L, Yuan CF, et al. Assessing Urinary Metabolomics in Giant Pandas Using Chromatography/Mass Spectrometry: Pregnancy-Related Changes in the Metabolome. Front Endocrinol. 2020;11:215.
Wang H, Ding J, Ding SY, Chang YQ. Metabolomic changes and polyunsaturated fatty acid biosynthesis during gonadal growth and development in the sea urchin Strongylocentrotus intermedius. Comp Biochem Physiol D-Genomics Proteomics. 2019;32:100611.
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–4.
Tao Y-F, Qiang J, He J, Zhu H-J, Bao J-W, Xu P. Untargeted LC-MS metabolomics approach reveals metabolic changes in genetically improved farmed tilapia (Oreochromis niloticus) with fatty liver induced by a high-fat diet. Aquac Res. 2020;52(2):724–35.
Qiang J, Bao WJ, Tao FY, He J, Li XH, Xu P, et al. The expression profiles of miRNA-mRNA of early response in genetically improved farmed tilapia (Oreochromis niloticus) liver by acute heat stress. Sci Rep. 2017;7:8705.
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–8.
Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7(10):986–95.
Qiang J, Khamis OAM, Jiang HJ, Cao ZM, He J, Tao YF, et al. Effects of dietary supplementation with apple peel powder on the growth, blood and liver parameters, and transcriptome of genetically improved farmed tilapia (GIFT, Oreochromis niloticus). PLoS One. 2019;14(11):e0224995.
Ma DM, Wang ZL, Wang LJ, Alejos-Gonzales F, Sun MA, Xie DY. A Genome-Wide Scenario of Terpene Pathways in Self-pollinated Artemisia annua. Mol Plant. 2015;8(11):1580–98.
Hsu HH, Araki M, Mochizuki M, Hori Y, Murata M, Kahar P, et al. A Systematic Approach to Timeseries Metabolite Profiling and RNA-seq Analysis of Chinese Hamster Ovary Cell Culture. Sci Rep. 2017;7:43518.
Qiang J, Tao YF, He J, Xu P, Bao JW, Sun YL. miR-122 promotes hepatic antioxidant defense of genetically improved farmed tilapia (GIFT, Oreochromis niloticus) exposed to cadmium by directly targeting a metallothionein gene. Aquat Toxicol. 2017;182:39–48.
Acknowledgements
We thank Jennifer Smith, PhD, from Liwen Bianji, Edanz Group China (http://www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
The study was supported financially by Central Public-interest Scientific Institution Basal Research Fund, CAFS (NO. 2019JBFC01; 2020TD37; 2020JBFR04). The funding agency did not participate in study design, data collection and analysis, decision to publish, and preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
PX conceived and designed the experiment. JLM, YFT, and LGL conducted sampling and experimental analyses. JLM and JQ preformed transcriptome and metabolome sequencing and analyzed the data. YFT and JWB extracted RNA and verified mRNA levels by qRT-PCR. HJZ and LGL measured hepatic biochemical indicators. JLM and JQ wrote the manuscript with input from all authors. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The experimental protocols were approved by the Institutional Animal Care and Use Committee of Nanjing Agriculture University (Nanjing, China). The GIFT were maintained in well-aerated water and deep anesthetized by treating with 100 mg L− 1 MS-222 (Argent Chemical Laboratories, Redmond, WA, USA). The GIFT were euthanized using rapid cooling method by soaking the experimental fish in 30 L 0 °C ice water mixture (25 L of ice and 5 L of water). The livers were extracted from GIFT based on the Guide for the Care and Use of Laboratory Animals in China.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Total ion chromatograms of LC-MS data from profiles of GIFT hepatic metabolites detected in POS ion mode (A) and NEG ion mode (B). CL1-3_1-3 N. mzXML, HL1-3_1-3 N. mzXML, and QC1-8 N. mzXML represent samples in CG group, HG group and quality control group, respectively. X-axis represents retention time and y-axis represents total ion chromatograms in MS.
Additional file 2: Figure S2.
m/z peak width and retention time peak width of metabolites detected in liver samples from CG or HG groups in POS and NEG modes. (A) Width of m/z peak in POS mode. (B) Width of m/z peak in NEG mode. (C) Width of retention time peak in POS mode. (D) Width of retention time peak in NEG mode.
Additional file 3: Figure S3.
Regional distribution of reference genome alignment of valid data.
Additional file 4: Figure S4.
Glycolysis/gluconeogenesis pathway annotated by KEGG. Red: significantly up-regulated transcript annotated to a ko node; blue: significantly down-regulated transcript annotated to a ko node; orange: transcript annotated to a ko node that is both up-regulated and down-regulated. Boxes represent genes or enzymes; open circles represent small molecule compounds; solid arrows represent the direction of biochemical reactions; dotted arrows connect other related metabolic pathways. (Same below for Fig. S5, S6, S7, S8) Reproduced with the permission of ref. 73, copyright@Kyoto Encyclopedia of Genes and Genomes (KEGG).
Additional file 5: Figure S5.
Insulin signaling pathway annotated by KEGG. Reproduced with the permission of ref. 73, copyright@Kyoto Encyclopedia of Genes and Genomes (KEGG).
Additional file 6: Figure S6.
Pentose phosphate pathway annotated by KEGG. Reproduced with the permission of ref. 73, copyright@Kyoto Encyclopedia of Genes and Genomes (KEGG).
Additional file 7: FigureS7.
Biosynthesis of unsaturated fatty acids pathway annotated by KEGG. Reproduced with the permission of ref. 73, copyright@Kyoto Encyclopedia of Genes and Genomes (KEGG).
Additional file 8: Figure S8.
Fatty acid degradation pathway annotated by KEGG. Reproduced with the permission of ref. 73, copyright@Kyoto Encyclopedia of Genes and Genomes (KEGG).
Additional file 9: Table S1.
Differentially expressed genes in GIFT liver under hypoxia stress.
Additional file 10: Table S2.
P values of top 20 enriched pathways under hypoxia stress.
Additional file 11: Table S3.
Sequences of primers used to amplify differentially expressed mRNAs.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ma, JL., Qiang, J., Tao, YF. et al. Multi-omics analysis reveals the glycolipid metabolism response mechanism in the liver of genetically improved farmed Tilapia (GIFT, Oreochromis niloticus) under hypoxia stress. BMC Genomics 22, 105 (2021). https://doi.org/10.1186/s12864-021-07410-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-021-07410-x