Abstract
Backgrounds
Cyclic nucleotide gated channels (CNGCs) play multifaceted roles in plant physiological processes, especially with respect to signalling processes, plant development, and responses to environmental stresses. However, little information is known about the CNGC family in the large cosmopolitan family Rhamnaceae, which has strong tolerance to biotic and abiotic stresses.
Results
In the current study, a total of 15 ZjCNGCs which located on 7 chromosomes were firstly identified in Chinese jujube (Ziziphus jujuba Mill.), the most important species of Rhamnaceae in terms of economic and ecological values. Phylogenetic analysis showed that these ZjCNGCs could be classified into four groups, ZjCNGC12 belonged to group IVA, and ZjCNGC13, 14, 15 belonged to group IVB. In addition, the paralogous and orthologous homology duplication of ZjCNGC15 occurred during the evolutionary process. The characteristics of ZjCNGCs regarding to exon-intron numbers and post-translational modifications showed diversified structures and functions. Motif composition and protein sequence analysis revealed that the phosphate-binding cassette and hinge regions were conserved among ZjCNGCs. Prediction of the cis-acting regulatory elements and expression profiles by real-time quantitative PCR analysis showed that some of the ZjCNGCs responded to environmental changes, especially ZjCNGC2, which was significantly downregulated in response to cold stress, and ZjCNGC4 was highly induced in response to cold, salt and alkaline stresses. ZjCNGC13 and 14 were highly induced in the phytoplasma-resistant cultivar and downregulated in the susceptible cultivar. Furthermore, ZjCNGC2 could be regulated by cAMP treatment, microtubule changes and interact with ZjMAPKK4, which suggested that cAMP and microtubule might play important roles in ZjCNGC2 mediated ZjMAPKK4 signalling transduction involved in cold stress.
Conclusions
The identification and classification analysis of ZjCNGCs were firstly reported, and some key individual ZjCNGCs might play essential roles in the response to biotic and abiotic stresses, especially ZjCNGC2 mediated ZjMAPKK4 signalling transduction involved in cold stress. This systematic analysis could provide important information for further functional characterization of ZjCNGCs with the aim of breeding stress-resistant cultivars.
Similar content being viewed by others
Background
Calcium (Ca2+), as an important second messenger, functions as a signalling molecule in many biological processes, including plant development, pollen fertility and tip growth, various biotic and abiotic stresses [1, 2]. The Ca2+ signal is modulated by the intimate interplay of channels and transporters as a so-called ‘on’ and ‘off’ mechanisms [3]. In Arabidopsis, five families of Ca2+-permeable channels have been identified, including 20 members of cyclic nucleotide gated channels (CNGCs) [2, 4]. In the plant kingdom, CNGCs are cation transport channels that enable sodium, calcium and potassium to cross cellular membranes. CNGCs localize on the cell membrane or different organelle membranes and can be regulated by inside second messengers, such as cyclic nucleotide monophosphates (3′,5′-cAMP and 3′,5′-cGMP) and Ca2+/calmodulin (CaM) [5,6,7,8,9]. Plant CNGCs can be divided into groups I, II, III, IVA, and IVB according to their phylogenetic relationship and are mainly characterized by six hexa-transmembrane (TM) domains, a pore-forming region between the fifth and sixth TM domains, a calmodulin-binding domain (CaMBD) and a cytosolic C-terminus containing a cyclic nucleotide-binding domain (CNBD). Among these structures, the CNBD domain, which is the most conserved domain, contains a phosphate-binding cassette (PBC) that can bind to cNMP and a hinge region that promotes ligand binding efficacy and selectivity [10,11,12]. These special properties are necessary for CNGC functions and are widely used to identify CNGCs in plant systems.
With informatics studies, the CNGC gene family has been identified in various plants; 20 members have been identified in Arabidopsis [4], 16 in rice [13], 18 in tomato [14], 21 in pear [15], 47 in wheat [16], 35 in tobacco [17], 12 in maize [18] and 26 in Brassica oleracea [19]. However, little is known about the biological information of the CNGC family in Chinese jujube.
In numerous physiological and molecular processes, such as plant development, symbiosis, circadian rhythm and in response to environmental stresses, including cold, heat and salt [20, 21], plant CNGCs play important roles in signal transduction. For example, MdCNGC1 was identified in the apple genome which was highly induced by Botryosphaeria dothidea infection. With the transformation to tobacco, MdCNGC1 negatively regulates resistance to bacterial and fungal pathogens [22]. In Arabidopsis, AtCNGC7 and AtCNGC8 are essential for male reproductive fertility, and AtCNGC16 and AtCNGC18 participate in pollen development [23,24,25]. AtCNGC2 is involved in jasmonic acid (JA)-induced apoplastic Ca2+ influx, which is further activated by cAMP [26], and AtCNGC2, similar to CNGC20 and CNGC12, can bind CaM via its IQ domain [27]. In addition, the AtCNGC2 orthologues (Arabidopsis Defense No Death 1) in potato and tomato negatively regulate resistance to late blight and powdery mildew, resulting in dwarfing and necrosis in tomato but not in potato [28]. In wheat, TaCNGC14 and TaCNGC16 play a negative role in resistance against pathogens [16]. In tomato, SlCNGC7 and SlCNGC14 play a negative role in drought tolerance [29]. However, the biological function of ZjCNGCs is still elusive.
Chinese jujube (Ziziphus jujuba Mill.) is the most important economic species and multipurpose fruit tree in the family of Rhamnaceae. It can adapt to multiple abiotic stresses, especially alkaline and salinity. Moreover, the whole genome of this plant species has been sequenced [30, 31], providing a foundation for us to identify the CNGC family in Chinese jujube. In the current study, the phylogenetic analysis, gene structure and conserved motifs of CNGCs in Chinese jujube (ZjCNGCs) were predicted. Furthermore, the expression profiles of these CNGCs in response to abiotic and abiotic stresses were investigated by real-time quantitative PCR (qPCR), and the ZjCNGC2 mediated signalling cascades involved in cold stress were studied.
Results
Identification of CNGC genes in the jujube genome
With 20 Arabidopsis and 21 pear CNGC protein sequences as queries to search against the jujube genome, 15 ZjCNGCs were retrieved as potential candidate genes. The domain composition analysis demonstrated that all of the candidate genes were conformed as CNGC proteins (see additional files S1 and S2). Among them, 14 ZjCNGC protein sequences contained both an ion transporter (ITP) domain and a cyclic nucleotide-binding domain (CNBD) [32], which are bona fide CNGC genes. However, ZjCNGC3 only had an ITP domain based on SMART analysis, and the amino sequence of ZjCNGC3 was significantly shorter than that of other members (see additional files S3 and S4), but the annotation of this gene was cyclic nucleotide-gated ion channel 1-like in the NCBI database. Thus, with comprehensive analysis, we suspected that a mistake might have occurred during sequencing, and ZjCNGC3 should actually be a full-length CNGC protein. Finally, a total of 15 ZjCNGCs were identified, and this number was smaller than that the number of CNGCs identified in Arabidopsis and pear. In addition, the identified ZjCNGC genes were designated as ZjCNGC1 to ZjCNGC15 based on their subfamily classification (Table 1).
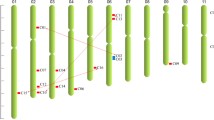
The detailed physiological and biochemical properties of the ZjCNGC genes are shown in Table 1. The ZjCNGCs were located on 7 chromosomes (Chr), including Chr 1, 2, 3, 4, 5, 8 and 11 although ZjCNGC11 could not match to a corresponding chromosome. In addition, five of the ZjCNGC genes were located on chromosome 1. The CDS length ranged from 1755 bp (ZjCNGC3) to 2361 bp (ZjCNGC12), with an average of 2127 bp. The length of amino acids ranged from 584 to 786 aa with an average of 708 aa. The predicted molecular weight (Mw) of these proteins ranged from 66.49 to 89.77 kDa, and the theoretical isoelectric point (pI) ranged from 7.83 (ZjCNGC3) to 9.57 (ZjCNGC15). Moreover, according to the instability index (II), only two proteins were stable in test tubes, namely, ZjCNGC2 and ZjCNGC3. The subcellular localization analysis of all the ZjCNGCs indicated that all of them were located in the cell membrane.
Phylogenetic analysis of ZjCNGCs
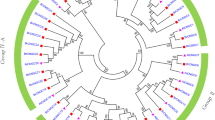
To analyse the relationship among ZjCNGC proteins, a phylogenetic tree was generated by using the available full-length amino acid sequences of jujube, Arabidopsis and pear CNGCs. As shown in Fig. 1, all the proteins could be clustered into four groups as described by Mäser et al. (2001) [4]. Group IV could be divided into another two subgroups (group IVA and group IVB). For each group of AtCNGCs and PbrCNGCs, the corresponding homologous genes were found in jujube, and the number of genes identified was different among groups. Among them, six members of ZjCNGCs (6–11) were clustered into group III, which formed the largest group, and the number of members in group III was basically similar to those in pear and Arabidopsis. In addition, group I contained three members (ZjCNGC1–3), group II contained ZjCNGC4 and 5, and the remaining three ZjCNGC (13–15) members belonged to group IVB, while ZjCNGC12 belonged to group IVA. Moreover, some ZjCNGCs located on the same chromosome, such as ZjCNGC4 and ZjCNGC5, ZjCNGC6 and ZjCNGC7 showed little divergence and thus clustered into the same group, which might indicate that some segmental duplication of ZjCNGCs might have occurred during the genome evolutionary process of Chinese jujube.
Phylogenetic analysis of the protein sequences of ZjCNGCs, AtCNGCs and PbrCNGCs. MEGA 6.0 was used to construct the phylogenetic tree with the neighbour-joining (NJ) method, and 1000 bootstrap replications were performed to show the reliability. The CNGCs from jujube, pear and Arabidopsis were distinguished with different shapes and colours
The phylogenetic tree and line charts for a lineage of gene groups for ZjCNGC15
ZjCNGC15, belonging to group IVB, is homologous to AtCNGC2. AtCNGC2 has been demonstrated to participate in multiple biological processes; thus, ZjCNGC15 could be used to perform evolutionary analysis. As shown in Fig. 2a, 20 other genes with high homology indices (HIs) in different species were clustered together with ZjCNGC15 (red colour). ZjCNGC15 was more homologous to Prunus members and three paralogous events that presumably occurred in a group of three genes (Pyrus bretschneideri XP_009365836, XP_009365836, and Malus domestica XP_008365415), a group of two genes (Citrus clementina XP_006431246 and Citrus sinensis XP_006482709) and a group of two genes in Ziziphus jujuba (XP_015879866 and XP_015879865). In addition, the.
The phylogenetic tree and line charts for a lineage of gene groups for ZjCNGC15. a The tree contains a gene of interest and 20 genes with the highest HI values. The horizontal axis represents HI. b Red, blue, and green lines represent the numbers of genes (sequences), species, and families contained in individual gene groups, respectively. The horizontal axis represents HI
HIs in the tree were all above 0.8, showing that they have similar amino acid sequences and might have conserved functions. Based on the evolutionary time analysis with the lineage of gene groups for ZjCNGC15, the gene numbers (red line) were 260 at 0.528 of HI and 0.567 at 138 for a large decrease; however, the species number (blue line) showed no decrease at this time, indicating a paralogous event. At an HI of 0.792, the decrease occurred in both the number of sequences and species, indicating that this event could be presumably orthologous (Fig. 2b). Therefore, the paralogous and orthologous homology of ZjCNGC15 occurred during the evolutionary process, and the other ZjCNGCs might have experienced a similar evolutionary process.
Gene structure and motif composition analysis
The divergence of gene structure, including exon-intron diversity, provides potential insights into the gene function during evolution. As shown in Fig. 3, the number of introns in all ZjCNGCs varied from 5 to 11, with an average number of 6. Interestingly, 9 of 15 ZjCNGCs indeed had 6 introns, and they were distributed in groups I, II, III and IVB (see additional file S5). However, ZjCNGC12 had 11 introns that were totally different from the others. Moreover, the intron phase (0, 1 and 2) could indicate that the codons in the exon parts were interrupted by introns at different positions. Similar to AtCNGCs, most ZjCNGCs were in intron phase 0 and phase 2, while in ZjCNGC2, 8, 9 and 12, intron phase 1 could be observed. These results demonstrated the changes in terms of loss or gain of exons, intron phases and their shuffling, even in the same subgroup, which promoted functional divergences in the process of evolution.
The motif divergence of proteins could also provide insights into the evolutionary history of proteins [33]. Therefore, five motifs in ZjCNGCs were predicted with the MEME database. Except for ZjCNGC3 and 12, the other ZjCNGCs all contained the five motifs (Fig. 4). In addition, motif 2 represented the IQ domain (QWRTWAA [CV] FIQ [AL] AW [RH]RY), and motif 3 was the cyclic nucleotide-binding (CNB) domain, which was located in the middle. Motifs 1, 4 and 5 were the transmembrane domains located at the N- and C-terminal. These results indicated that the transmembrane domain and cNMP-binding domain were specific to plant CNGCs. Among them, the CNBD domain, which could bind cAMP/cGMP, was the main structure of plant CNGCs [34]. The CNBD domain contains two regions, including the PBC and hinge regions. As shown in Fig. 5, a > 90% conserved motif was found in 15 ZjCNGCs, [LIV]-X(2)-[GD]-[DHNG]-[FIV]-X-G-[EGD]-E-LL-X-W-X-[LE]-X-X-S-E-[AGV]-F-X-[LIV], which consisted of the PBC and hinge regions. In addition, the conserved glycine (G) and aliphatic leucine (L) residues in the PBC motif were 100% conserved inside the PBCs. This conserved motif was consistent with the analysis in wheat, maize and rice.
The putative PBC and hinge motifs within the CNBDs of all the ZjCNGCs. The diagram at the top represents three regions of plant CNGCs: the six transmembrane domains (TM), a CNBD containing a PBC and the hinge, and a CaMBD. The ZjCNGC-specific amino acid motif is shown below the cartoon. The square brackets in different colours indicate the highly conserved amino acids
Prediction of Cis-acting regulatory elements
To better elucidate the function of ZjCNGCs, which are regulated by their corresponding transcriptional factors in response to biotic and abiotic stresses, 1.0 kb noncoding sequences upstream of the ZjCNGC genomic sequences that belong to the promoter region were used to perform cis-regulatory element analysis with the PLACE database. The results showed that most of the promoter sequences of ZjCNGCs contained several cis-regulatory element regulatory sites for biotic and abiotic factors, such as BOXLCOREDCPAL, GT1GMSCAM4, SEBFCONSSTPR10A, MYCCONSENSUSAT, and MYCATERD1, suggesting that ZjCNGCs might be involved in regulating diverse stress responses (Table 2). However, the ZjCNGC14 promoter did not have responsive cis-regulatory elements involved in biotic stresses and only contained 6 MYCCONSENSUSAT sequences in response to abiotic stress. Additionally, different WRKY transcription factor binding sites were found in all the promoter sequences of ZjCNGCs, including WBBOXPCWRKY1, BOXATNPR1, WBOXHVISO1, WBOXNTERF3, WRKY71OS and WBOXNTCHN48, which indicates that all ZjCNGCs were regulated by WRKY transcriptional factors. More importantly, Ca2+/calmodulin-binding cis-regulatory elements existed in the promoter regions of ZjCNGC9 and ZjCNGC11, indicating that these two genes might be regulated by the Ca2+/calmodulin signal transduction pathway.
Posttranslational modification and phosphorylation of ZjCNGC proteins
After analyzing the cis-regulatory elements of ZjCNGCs in the promoter region, the posttranslational modification and phosphorylation of ZjCNGC proteins were analyzed by using ScanProsite. As shown in Table 3, protein kinase C, casein kinase II, cAMP/cGMP kinases, N-myristoylation sites and N-glycosylation sites were widely found in all ZjCNGCs. Among them, the casein kinase II sites were evenly distributed in all the ZjCNGC members, which have important functions in Arabidopsis in response to stresses [35]. Protein kinase C was most abundant in the group II and III subfamilies, which belong to a family of ten isoenzymes that play vital roles in cellular signal transduction [36]. Additionally, 1 to 4 cAMP/cGMP-binding motifs were found in all ZjCNGCs, indicating that all ZjCNGCs were targets of cAMP/cGMP involved in signal transduction. However, the leucine zipper patterns and amidation sites were not conserved and randomly distributed in some ZjCNGCs.
Expression patterns of ZjCNGCs in response to biotic and abiotic stresses
To determine whether ZjCNGCs were associated with abiotic stresses, the expression profiles of ZjCNGCs in response to cold, salt and alkaline stresses were investigated. Unfortunately, the expression level of ZjCNGC5 was not detected in any of the treatments. Therefore, the remaining fourteen ZjCNGCs were used for qPCR analysis. As shown in Fig. 6, some ZjCNGCs, such as ZjCNGC2, 8, 10 and 15, were downregulated in response to cold stress within 24 h of treatment. The expression levels of ZjCNGC4 and 12 increased approximately 4- and 2-fold after 1 h of cold treatment, respectively. The expression level of the others showed no significant changes or increased/decreased at one time point.
Relative expression profiles of ZjCNGCs in response to cold stress. The relative expression level of the representative members of ZjCNGCs in three independent replications, and the error bar represents the standard deviation (SD). Asterisks indicate that the corresponding gene was significantly up- or downregulated at different time points compared with the gene expression at the 0 h time point (*P < 0.05, **P < 0.01)
In response to salt stress, most of the ZjCNGCs were downregulated, especially the genes from group III and group IVB (Fig. 7). The same changes in ZjCNGCs in response to alkaline stress were observed (Fig. 8). However, ZjCNGC2, 4 and 6 were highly induced at the early time point (1 h) under these two treatments. In summary, some ZjCNGC genes were significantly induced or repressed by the above treatments.
The response of ZjCNGCs to phytoplasma infection was also investigated. As shown in Fig. 9, most of the ZjCNGCs showed no significant difference between the phytoplasma-resistant cultivar “T11” and the susceptible cultivar “Dongzao”. In addition, the expression level of ZjCNGC10 decreased in the resistant cultivar but not in the susceptible cultivar, while ZjCNGC13 and 14 were highly induced in the resistant cultivar and downregulated in the susceptible cultivars, indicating that these four genes could have important functions in the process of phytoplasma infection.
ZjCNGC2 mediated ZjMAPK cascades involved in cold stress
Some ZjCNGCs were downregulated or upregulated in response to cold stress, indicating that they might play important roles in cold stress. Thus, to test whether the ZjCNGCs could regulate the ZjMAPK cascades during cold-induced signal transduction, the amino acids of 15 ZjCNGCs, 10 ZjMAPKs and 5 ZjMAPKKs were submitted to the STRING database for protein-protein interaction prediction. As shown in Fig. 10a, AtCNGC13, which is orthologous to ZjCNGC2, might interact with AtMAPK11, which is orthologous to ZjMAPK3/4, indicating that a protein-protein interaction existed between ZjCNGCs and ZjMAPK cascades. Then, the prediction was tested by yeast two-hybrid analysis, and ZjCNGC2 interacted with ZjMAPKK4 but not with ZjMAPKs (Fig. 10b). Moreover, to test whether cAMP or changes in microtubules (MTs) could regulate the activity of ZjCNGC2, the expression level of ZjCNGC2 was analysed in the presence of cAMP or the MTs-disrupting drug oryzalin, respectively. The results showed that ZjCNGC2 rapidly (in 1 h) responded to these two drug treatments (Fig. 10c and d). All these results might indicate that cAMP and MTs play important roles in ZjCNGC2-mediated ZjMAPKK4 signal transduction under cold stress.
ZjCNGC2-mediated signalling cascades in response to cold stress. a Protein-protein interaction analysis among 15 ZjCNGCs, 10 ZjMAPKs and 5 ZjMAPKKs by the STRING database. ZjCNGC2 is orthologous to AtCNGC13, and ZjMAPK3/4 are orthologous to AtMAPK11. b Yeast two-hybrid screening of ZjCNGC2 and ZjMAPKK4. Relative expression profiles of ZjCNGC2 in response to 50 μM cAMP (c) and 10 μM oryzalin (d) treatments at normal temperature. e Schematic and supposed model of the cAMP- and microtubules (MTs) regulated ZjCNGC2-ZjMAPKK4 signalling pathway involved in cold stress
Discussion
Plant cyclic nucleotide-gated channels (CNGCs) have important functions in Ca2+ signal transduction, including pollen tube growth, thermosensing, pathogen resistance [1, 9], and have been widely identified in plant systems. However, the genome-wide identification and functional analysis of CNGCs in Chinese jujube have not been reported. In the current study, 15 ZjCNGCs were identified. Among them, ZjCNGC3 might be controversial because it only contains the ITP domain based on SMART analysis. However, based on InterProScan analysis, ZjCNGC3 contains a cyclic nucleotide-binding-like signature. In addition, based on domain comparative analysis, the ZjCNGC3 protein contains a PBC and hinge region in the CNBD domain with only several amino acid discrepancies. These results could support that ZjCNGC3 should be bona fide member of ZjCNGCs. The same observation could be found in the OsCNGC family in which the amino acid sequence of LOC_Os06g33610 was shorter than those of the other OsCNGCs; however, this protein contained the CNGC characteristic phosphate-binding cassettes and hinge regions and was finally designated as bona fide OsCNGC3 [13]. For accuracy purposes, the ZjCNGC3 gene should be cloned and sequenced for further analysis. Moreover, the phylogenetic and gene structure analyses showed that ZjCNGCs have similar structures and gathered with Arabidopsis and pear counterparts, and they all could be divided into five groups with significant bootstrap values. Together with the evolutionary analysis of ZjCNGC15, paralogous and orthologous homology duplication processes might have taken place during the evolutionary process of ZjCNGCs, implying that duplication events play a principal role in gene evolution.
Protein posttranslational modification could modulate protein activity and affect protein functions through different mechanisms in response to stresses, including protein-protein interactions, protein localization, degradation, cleavage or allosterically regulating enzyme activity [37]. Among them, cAMP/cGMP could target ZjCNGCs; of all reported plant species, cAMP had the highest concentration in Chinese jujube, and CNGCs have been confirmed to be the target of cAMP [6]. Therefore, the cAMP-regulated ZjCNGC signalling pathway might have important functions in response to various stresses. This conclusion has been demonstrated by our results that ZjCNGC2 was rapidly induced with cAMP treatment, indicating that CNGC2 might be the target of cAMP in the jujube system. In addition, cyclic nucleotides have been suggested to be involved in plant responses to biotic and abiotic stresses, while based on cis-regulatory element analysis, nearly all ZjCNGCs contain several cis-regulatory element sites for biotic and abiotic factors. More interestingly, ZjCNGC9 and ZjCNGC11 have Ca2+/calmodulin-binding cis-regulatory elements, which could be interesting for further research on Ca2+/calmodulin- and ZjCNGC-mediated signalling pathways in Chinese jujube.
Based on the localization prediction analysis, all ZjCNGCs are located on the cell membrane, including the plasma membrane, chloroplast thylakoid membrane or mitochondrial inner membrane, as reported by Nawaz et al. (2014) [13] in the analysis of rice. In addition, regarding the CNGC proteins in Arabidopsis, 11 out of the 20 CNGC proteins were localized in the plasma membrane, while the other 9 were located in the chloroplast thylakoid membrane [13]. These results suggested that CNGC proteins were mainly localized in the cell membrane, and CNGC proteins might have important functions in the regulation of Ca2+ influx in response to biotic and abiotic stresses. Furthermore, calcium influx into the cytoplasm in response to cold stress has been demonstrated [38,39,40], and CNGCs could be one of the potential Ca2+-permeable channels [1, 2]. In the current study, ZjCNGC4 was upregulated at an early time point, and ZjCNGC2 was downregulated after 24 h of cold treatment, demonstrating that these two genes play different roles in the response to cold stress. Moreover, ZjCNGC4 was also highly induced with salt and alkaline treatments, indicating that it might have an important function in calcium influx into the cytoplasm in response to abiotic stresses. ZjCNGC4 was more homologous to AtCNGC6, while AtCNGC6 could induce Ca2+ influx in response to heat shock and facilitate the expression of heat shock protein (HSP) genes to achieve thermotolerance [41]. In addition, MTs have been demonstrated to play an important role in the response to cold stress, and modulation of MTs with their corresponding drugs could improve plant cold resistance [40]. Our results showed that ZjCNGC2 could be regulated by cAMP and oryzalin treatments, indicating that cAMP and MTs could regulate ZjCNGC2 activity for early signal transduction. Furthermore, ZjCNGC2 could interact with ZjMAPKK4, and ZjMAPKKs have been demonstrated to interact with ZjWRKYs [42]. Finally, we arrived at a supposed model of cAMP- and MTs - regulated ZjCNGC2 signalling pathways involved in cold stress (Fig. 10e). Under cold stress, Ca2+ influx into the cytosol was triggered by cold stress through the CNGC channel. However, the expression of ZjCNGC2 was repressed by cold stress, indicating that it might play a negative role in calcium influx. The activity of ZjCNGC2 was regulated by cAMP and MTs depolymerization. Then, ZjCNGC2 could interact with ZjMAPKK4, which might further interact with ZjWRKYs to induce the cold response.
In comparison with their function in abiotic stress, some more valuable clues about the functional role of ZjCNGCs in plant immunity were obtained. Many studies have demonstrated that CNGCs play an important role in biotic stress. For example, AtCNGC2 could control the Ca2+ concentration to regulate the hypersensitive response (HR) to avirulent pathogens [43]. Moreover, the AtCNGC2 orthologue (Arabidopsis Defense No Death 1) in potato and tomato negatively regulated the resistance to late blight and powdery mildew and resulted in dwarfing and necrosis in tomato but not in potato [28]. In addition to the crucial role of CNGC2 in plant immunity, dnd1 (null mutant of CNGC2) could lead to heat stress with the accumulation of heat shock proteins [44, 45]. According to our results, ZjCNGC15 was downregulated in the phytoplasma-susceptible cultivar “Dongzao”, but there was no significant difference compared to ZjCNGC15 in the resistant cultivar “T11”. However, in group IVB, ZjCNGC13 and 14 were highly induced in the resistant cultivar but downregulated in the susceptible cultivar, demonstrating that these two genes might be the main channels to induce calcium influx into the cytoplasm in the resistant cultivar to obtain resistance to phytoplasma infection. Again, this hypothesis should be confirmed at the protein expression level for ZjCNGC13 and 14. Overall, these results imply that these genes play crucial roles in the response to biotic and abiotic stresses in Chinese jujube.
Conclusions
Altogether, systematic informatics analyses of 15 CNGCs in Chinese jujube were firstly carried out, including phylogenetic, conserved motif and intron/exon analyses. Moreover, the expression profiles of ZjCNGCs were detected by qPCR in response to cold, salt and alkaline stresses and phytoplasma infection. ZjCNGC2 was significantly downregulated in response to cold stress, and ZjCNGC4 was highly induced in response to cold, salt and alkaline stresses. ZjCNGC13 and 14 were highly induced in phytoplasma-resistant cultivars and downregulated in susceptible cultivars. Furthermore, ZjCNGC2 could be regulated by cAMP and MTs disruptions and could interact with ZjMAPKK4, which suggested that cAMP and MTs might play important roles in ZjCNGC2-mediated ZjMAPKK4 signalling transduction involved in cold stress.
Methods
Identification of CNGC genes in Chinese jujube
The reference genome and genome annotation of Chinese jujube (Ziziphus jujuba Mill.) was used for the identification of the candidate CNGC genes (https://www.ncbi.nlm.nih.gov/genome/15586). First, a total of 20 Arabidopsis and 21 pear protein sequences were obtained from the TAIR database (https://www.arabidopsis.org/) and pear genome database (http://www.peargenome.njau.edu.cn), respectively. Then, to find the potential ZjCNGC genes, BLASTP searches were performed in NCBI (http://www.ncbi.nlm.nih.gov/) against the whole jujube genome database with the 20 Arabidopsis and 21 pear CNGC proteins as queries. Moreover, all the retrieved nonredundant sequences were collected and subjected to domain analysis by using the InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan5/) and SMART (http://smart.embl-heidelberg.de/) programs. In addition, all potential ZjCNGC genes were confirmed by HMMER analysis, which contains both the CNBD [cNMP_binding family (PF00027)] and the ion transport protein domain [ITP, Ion_trans family (PF00520)] [13, 14]. The protein sequences with six membrane-spanning regions, a pore region, a CNBD, a CaMBD or an IQD were recognized as ZjCNGC proteins [15].
Protein characterization and amino acid properties
Information on protein and CDS lengths, chromosomal locations and exon numbers was obtained from the NCBI database. The molecular weight (kDa), instability indices and isoelectric points (pI) of individual amino acids were obtained by ProtParam analysis (http://web.expasy.org/protparam/) [46]. The post-translational modification (PTM) sites were predicted with the ScanProsite web server (http://prosite.expasy.org/scan prosite/) [47]. In addition, the subcellular localization of the ZjCNGC proteins was identified by the Plant-mPLoc database (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) [48].
Multiple sequence alignment and phylogenetic analysis
The protein sequences of 15 ZjCNGCs, 20 AtCNGCs and 21 PbrCNGCs were aligned by ClusterX software [49] with default parameters. The conserved CNGC-specific domains were manually checked. Then, the alignment of the above protein sequences was obtained, and phylogenetic analysis was performed by using MEGA 6.0 with the neighbour-joining (NJ) method. The number of bootstrap replications was 1000 with a Poisson distribution [50]. In addition, the phylogenetic tree and line chart for a lineage of gene groups for ZjCNGC15 were analyzed in the Gcorn plant database (http://www.plant.osakafu-u.ac.jp/~kagiana/gcorn/p/) [51].
Gene structure, conserved motif domain and cis-acting regulatory element analysis
The intron/exon structure information of ZjCNGCs was obtained from the NCBI database, and the detailed exon/intron structures of the ZjCNGCs were constructed with Gene Structure Display Server (GSDS 2.0; http://gsds.cbi.pku.edu.cn/) [52]. To confirm the structural divergence of the ZjCNGC protein sequences, the conserved motifs in the ZjCNGC protein sequences were identified with the MEME (Multiple Expectation Maximization for Motif Elicitation) web server (http://meme-suite.org/), with the following parameters: the optimal motif width was set between 6 and 200, and the maximum number of different motifs was set at 5 [53]. The 1000 bp DNA sequences upstream of ZjCNGCs were used to perform cis-acting regulatory element analysis by the “Signal Scan Search” program in the PLACE database (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en&pj=640&action=page&page=newplace) [54].
Plant materials and treatments
To investigate the function of ZjCNGCs in response to abiotic stresses, cold, salt and alkaline treatments were conducted on the callus tissues of Ziziphus jujuba Mill. ‘Guanyangchangzao’. For cold treatment, the calluses were transferred to 4 °C and then collected within 0, 1, 6 and 24 h. Calluses incubated at 25 °C were collected as a negative control. For the salinity and alkaline treatments, the callus tissues were subjected to 150 mM NaCl and NaHCO3-NaOH solution (pH 9.5) [55] for 0, 1, 6 and 24 h, respectively. The samples with sterile water treatment rather than NaCl and NaHCO3-NaOH solution were used as a negative control. In addition, to gain insight into whether ZjCNGC2 could be regulated by cyclic adenosine monophosphate or microtubules, the same callus tissues were treated with 50 μM cAMP and 10 μM oryzalin at normal temperature (25 °C) for 0, 1, 3, 6 and 24 h. The samples with sterile water and the same amount of DMSO treatments were collected as a negative control.
For biotic stress, the leaves from the phytoplasma-susceptible cultivar Ziziphus jujuba Mill. ‘Dongzao’ and the resistant cultivar Ziziphus jujuba Mill. ‘T11’ that were grafted on rootstocks with JWB and healthy rootstocks [56] were collected in June 2018. Three independent replicates were performed for each treatment, and all the treated tissue samples were immediately frozen in liquid nitrogen and stored at − 80 °C for RNA extraction and qPCR analysis.
RNA extraction and qPCR analysis
Total RNA extraction from the calluses or leaves and subsequent cDNA synthesis were performed as described by Liu et al. (2017) [57]. qPCR was carried out on a Bio-Rad iQ™ 5 instrument using TransStart Top Green qPCR SuperMix AQ131 (TransGen Biotech, China). The 20 μL reaction system contained 10 μL of 2 × SYBR Premix ExTaq™, 0.4 μL of each of 10 μM primers, 1 μL of diluted cDNA and 8.2 μL of ddH2O. The reactions were performed as follows: preincubation for 3 min at 94 °C, followed by 40 cycles of 5 s at 94 °C, 15 s at 55~63 °C and 15 s at 72 °C. Relative expression levels of ZjCNGCs were calculated with the 2-ΔΔCt method [58] using ZjActin as an endogenous control for normalization [59]. The primer sequences of ZjCNGCs used for qPCR are shown in additional file S6.
Heatmap construction
The expression profiles of all ZjCNGCs in response to salt, alkaline and phytoplasma stresses are illustrated by a colour gradient heatmap. The heatmap was constructed by heatmap software Heml 1.0 with Log2-based expression fold-changes.
Protein-protein interaction prediction and yeast two-hybrid analysis
The amino acid sequences of 15 ZjCNGCs, 10 ZjMAPKs and 5 ZjMAPKKs were submitted to the STRING database (https://string-db.org/) for protein-protein interaction analysis. The orthologues of Arabidopsis thaliana were selected as references.
For the yeast two-hybrid analysis, ZjCNGC2 was fused to the Gal4 activation domain (AD), and ZjMAPKs/ZjMAPKKs were fused to the Gal4 DNA-binding domain (BD). Then, the constructs were cloned into the pGADT7 and pGBKT7 vectors, respectively, and the constructive vectors were co-transformed into the AH109 strain. Finally, the yeast clones were selected on synthetically defined medium lacking tryptophan and leucine (−LW) and medium lacking leucine, tryptophan, histidine (−LWH), −LWH medium supplemented with 7 mM 3-amino-1,2,4-triazole (3-AT; Sigma Aldrich) (−LWH + 3AT) and medium lacking leucine, tryptophan, histidine, adenine (−LWAH) as described by Xue et al. (2019) [42].
Availability of data and materials
All data and materials are presented in the main paper and additional file.
Abbreviations
- AMD:
-
Aidation site
- ASN:
-
N-glycosylation site
- Ca2+ :
-
Calcium
- CaM:
-
Calmodulin
- CaMBD:
-
Calmodulin binding domain
- cAMP/cGMP:
-
Cyclic adenosine monophosphate/Cyclic guanosine monophosphate
- CK2:
-
Csein kinase II phosphorylation site
- CNBD:
-
Cyclic nucleotide-binding domain
- CNGC:
-
Cyclic nucleotide gated channel
- HIs:
-
Hmology indices
- HSP:
-
Hat shock protein
- LEU:
-
Lucine zipper pattern
- MTs:
-
Microtubules
- MYR:
-
N-myristoylation site
- PBC:
-
Posphate-binding cassette
- PKC:
-
Potein kinase C phosphorylation site
- TM:
-
Transmembrane
- TYR:
-
Trosine kinase phosphorylation site
References
Zelman AK, Dawe A, Gehring C, Berkowitz GA. Evolutionary and structural perspectives of plant cyclic nucleotide-gated cation channels. Front Plant Sci. 2012;3:95.
Kudla J, Becker D, Grill E, Hedrich R, Hippler M, Kummer U, Parniske M, Romeis T, Schumacher K. Advances and current challenges in calcium signaling. New Phytol. 2018;218(2):414–31.
Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Bio. 2003;4(7):517–29.
Maser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJM, Sanders D, et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126(4):1646–67.
Borsics T, Webb D, Andeme-Ondzighi C, Staehelin LA, Christopher DA. The cyclic nucleotide-gated calmodulin-binding channel AtCNGC10 localizes to the plasma membrane and influences numerous growth responses and starch accumulation in Arabidopsis thaliana. Planta. 2007;225(3):563–73.
Kaplan B, Sherman T, Fromm H. Cyclic nucleotide-gated channels in plants. FEBS Lett. 2007;581(12):2237–46.
Yuen CCY, Christopher DA. The group IV-A cyclic nucleotide-gated channels, CNGC19 and CNGC20, localize to the vacuole membrane in Arabidopsis thaliana. Aob Plants. 2013;5:plt012.
Charpentier M, Sun JH, Martins TV, Radhakrishnan GV, Findlay K, Soumpourou E, Thouin J, Very AA, Sanders D, Morris RJ, et al. Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science. 2016;352(6289):1102–5.
DeFalco TA, Marshall CB, Munro K, Kang HG, Moeder W, Ikura M, Snedden WA, Yoshioka K. Multiple calmodulin-binding sites positively and negatively regulate arabidopsis CYCLIC NUCLEOTIDE-GATED CHANNEL12. Plant Cell. 2016;28(7):1738–51.
Young EC, Krougliak N. Distinct structural determinants of efficacy and sensitivity in the ligand-binding domain of cyclic nucleotide-gated channels. J Biol Chem. 2004;279(5):3553–62.
Cukkemane A, Seifert R, Kaupp UB. Cooperative and uncooperative cyclic-nucleotide-gated ion channels. Trends Biochem Sci. 2011;36(1):55–64.
Almoneafy AA, Kakar KU, Nawaz Z, Li B, Saand MA, Chun-lan Y, Xie GL. Tomato plant growth promotion and antibacterial related-mechanisms of four rhizobacterial Bacillus strains against Ralstonia solanacearum. Symbiosis. 2014;63(2):59–70.
Nawaz Z, Kakar KU, Saand MA, Shu QY. Cyclic nucleotide-gated ion channel gene family in rice, identification, characterization and experimental analysis of expression response to plant hormones, biotic and abiotic stresses. BMC Genomics. 2014;15(1):853.
Saand MA, Xu YP, Li W, Wang JP, Cai XZ. Cyclic nucleotide gated channel gene family in tomato: genome-wide identification and functional analyses in disease resistance. Front Plant Sci. 2015;6:303.
Chen JQ, Yin H, Gu JP, Li LT, Liu Z, Jiang XT, Zhou HS, Wei SW, Zhang SL, Wu JY. Genomic characterization, phylogenetic comparison and differential expression of the cyclic nucleotide-gated channels gene family in pear (Pyrus bretchneideri Rehd.). Genomics. 2015;105(1):39–52.
Guo J, Islam MA, Lin HC, Ji CG, Duan YH, Liu P, Zeng QD, Day B, Kang ZS, Guo J. Genome-wide identification of cyclic nucleotide-gated ion channel gene family in wheat and functional analyses of TaCNGC14 and TaCNGC16. Front Plant Sci. 2018;9:18.
Nawaz Z, Kakar KU, Ullah R, Yu SZ, Zhang J, Shu QY, Ren XL. Genome-wide identification, evolution and expression analysis of cyclic nucleotide-gated channels in tobacco (Nicotiana tabacum L.). Genomics. 2019;111(2):142–58.
Hao LD, Qiao XL. Genome-wide identification and analysis of the CNGC gene family in maize. Peer J. 2018;6:e5816.
Kakar KU, Nawaz Z, Kakar K, Ali E, Almoneafy AA, Ullah R, Ren XL, Shu QY. Comprehensive genomic analysis of the CNGC gene family in Brassica oleracea: novel insights into synteny, structures, and transcript profiles. BMC Genomics. 2017;18(1):869.
Jammes F, Hu HC, Villiers F, Bouten R, Kwak JM. Calcium-permeable channels in plant cells. FEBS J. 2011;278(22):4262–76.
Zhu YX, Jia JH, Yang L, Xia YC, Zhang HL, Jia JB, Zhou R, Nie PY, Yin JL, Ma DF, Liu LC. Identification of cucumber circular RNAs responsive to salt stress. BMC Plant Biol. 2019;19(1):164.
Zhang WW, Dong CH, Zhang YG, Zhu J, Dai HY, Bai SH. An apple cyclic nucleotide-gated ion channel gene highly responsive to Botryosphaeria dothidea infection enhances the susceptibility of Nicotiana benthamiana to bacterial and fungal pathogens. Plant Sci. 2018;269:94–105.
Tunc-Ozdemir M, Rato C, Brown E, Rogers S, Mooneyham A, Frietsch S, Myers CT, Poulsen LR, Malho R, Harper JF. Cyclic nucleotide gated channels 7 and 8 are essential for male reproductive fertility. Plos One. 2013;8(2):e55277.
Tunc-Ozdemir M, Tang C, Ishka MR, Brown E, Groves NR, Myers CT, Rato C, Poulsen LR, McDowell S, Miller G, et al. A cyclic nucleotide-gated channel (CNGC16) in pollen is critical for stress tolerance in pollen reproductive development. Plant Physiol. 2013;161(2):1010–20.
Gao QF, Gu LL, Wang HQ, Fei CF, Fang X, Hussain J, Sun SJ, Dong JY, Liu HT, Wang YF. Cyclic nucleotide-gated channel 18 is an essential Ca2+ channel in pollen tube tips for pollen tube guidance to ovules in Arabidopsis. P Natl Acad Sci USA. 2016;113(11):3096–101.
Lu M, Zhang YY, Tang SK, Pan JB, Yu YK, Han J, Li YY, Du XH, Nan ZJ, Sun QP. AtCNGC2 is involved in jasmonic acid-induced calcium mobilization. J Exp Bot. 2016;67(3):809–19.
Fischer C, DeFalco TA, Karia P, Snedden WA, Moeder W, Yoshioka K, Dietrich P. Calmodulin as a Ca2+ −sensing subunit of Arabidopsis cyclic nucleotide-gated channel complexes. Plant Cell Physiol. 2017;58(7):1208–21.
Sun K, Wolters AMA, Loonen AE, Huibers RP, van der Vlugt R, Goverse A, et al. Down-regulation of Arabidopsis DND1 orthologs in potato and tomato leads to broad-spectrum resistance to late blight and powdery mildew. Transgenic Rec. 2016;25(2):123–38.
Saand MA, Xu YP, Munyampundu JP, Li W, Zhang XR, Cai XZ. Phylogeny and evolution of plant cyclic nucleotide-gated ion channel (CNGC) gene family and functional analyses of tomato CNGCs. DNA Res. 2015;22(6):471–83.
Liu MJ, Zhao J, Cai QL, Liu GC, Wang JR, Zhao ZH, Liu P, Dai L, Yan GJ, Wang WJ, et al. The complex jujube genome provides insights into fruit tree biology. Nat Commun. 2014;5(1):1–12.
Huang J, Zhang CM, Zhao X, Fei ZJ, Wan KK, Zhang Z, Pang XM, Yin X, Bai Y, Sun XQ, et al. The jujube genome provides insights into genome evolution and the domestication of sweetness/acidity taste in fruit trees. Plos Genet. 2016;12(12):e1006433.
Gehring C. Cyclic nucleotide signaling in plants : methods and protocols. New York: Humana Press; Springer; 2013.
Seoighe C, Gehring C. Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 2004;20(10):461–4.
DiFrancesco D, Tortora P. Direct activation of cardiac pacemaker channels by intracellular cyclic AMP. Nature. 1991;351(6322):145–7.
Mulekar JJ, Bu QY, Chen FL, Huq E. Casein kinase II alpha subunits affect multiple developmental and stress-responsive pathways in Arabidopsis. Plant J. 2012;69(2):343–54.
Leppanen T, Tuominen RK, Moilanen E. Protein kinase C and its inhibitors in the regulation of inflammation: inducible nitric oxide synthase as an example. Basic Clin Pharmacol. 2014;114(1):37–43.
Webster DE, Thomas MC. Post-translational modification of plant-made foreign proteins; glycosylation and beyond. Biotechnol Adv. 2012;30(2):410–8.
Knight MR, Campbell AK, Smith SM, Trewavas AJ. Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature. 1991;352(6335):524–6.
Wang LX, Nick P. Cold sensing in grapevine-which signals are upstream of the microtubular "thermometer". Plant Cell Environ. 2017;40(11):2844–57.
Wang LX, Sadeghnezhad E, Nick P. Upstream of gene expression: what is the role of microtubules in cold signalling? J Exp Bot. 2020;71(1):36–48.
Gao F, Han XW, Wu JH, Zheng SZ, Shang ZL, Sun DY, Zhou RG, Li B. A heat-activated calcium-permeable channel - Arabidopsis cyclic nucleotide-gated ion channel 6 - is involved in heat shock responses. Plant J. 2012;70(6):1056–69.
Xue C, Li H, Liu Z, Wang L, Zhao Y, Wei X, Fang H, Liu M, Zhao J. Genome-wide analysis of the WRKY gene family and their positive responses to phytoplasma invasion in Chinese jujube. BMC Genomics. 2019;20(1):464.
Wang Y, Kang Y, Ma CL, Miao RY, Wu CL, Long Y, Ge T, Wu ZN, Hou XY, Zhang JX, et al. CNGC2 is a Ca2+ influx channel that prevents accumulation of apoplastic Ca2+ in the leaf. Plant Physiol. 2017;173(2):1342–54.
Katano K, Kataoka R, Fujii M, Suzuki N. Differences between seedlings and flowers in anti-ROS based heat responses of Arabidopsis plants deficient in cyclic nucleotide gated channel 2. Plant Physiol Bioch. 2018;123:288–96.
Finka A, Cuendet AFH, Maathuis FJM, Saidi Y, Goloubinoff P. Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell. 2012;24(8):3333–48.
Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–52.
de Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–5.
Chou KC, Shen HB. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. Plos One. 2010;5(6):e11335.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9.
Ogata Y, Kimura N, Sano R. Gcorn plant: a database for retrieving functional and evolutionary traits of plant genes. Plant Physiol. 2019;180(2):732–42.
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–7.
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren JY, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8.
Higo K, Ugawa Y, Iwamoto M, Higo H. PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 1998;26(1):358–9.
Guo M, Li S, Tian S, Wang B, Zhao X. Transcriptome analysis of genes involved in defense against alkaline stress in roots of wild jujube (Ziziphus acidojujuba). PLoS One. 2017;12(10):e0185732.
Liu ZG, Zhao ZH, Xue CL, Wang LX, Wang LL, Feng CF, Zhang LM, Yu Z, Zhao J, Liu MJ. Three Main genes in the MAPK Cascade involved in the Chinese jujube-Phytoplasma interaction. Forests. 2019;10(5):392.
Liu ZG, Zhang LM, Xue CL, Fang H, Zhao J, Liu MJ. Genome-wide identification and analysis of MAPK and MAPKK gene family in Chinese jujube (Ziziphus jujuba mill.). BMC Genomics. 2017;18(1):855.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–8.
Bu JD, Zhao J, Liu MJ. Expression Stabilities of Candidate Reference Genes for RT-qPCR in Chinese Jujube (Ziziphus jujuba Mill.) under a Variety of Conditions. Plos One. 2016;11(4): e0154212.
Acknowledgements
Not applicable.
Ethical standards
This research does not contain any studies with human participants or animals.
Funding
This research was funded by the Funds for Young Talent Project of Hebei Agricultural University Foundation (grant number YJ201853); Green Channel Fund of Hebei Province Natural Science Foundation (C2019204308); Youth Fund of Hebei Province Natural Science Foundation (grant number C2016204157); Hebei Distinguished Young Scholar (grant number C2016204145); Significant Fund of Hebei Province Natural Science Foundation (grant number C2017204114); National Science and Technology Support Plan of China (grant number 2013BAD14B03); Hebei Agricultural University of Hebei Foundation for Leaders of Disciplines in Science Technology (CEFF-PXM2019_014207_000099). These funding bodies took part in the design of the study and collection, analysis, and interpretation of data, and the writing of the manuscript, as well as in the open access payment.
Author information
Authors and Affiliations
Contributions
LW (Lixin Wang), JZ and ML (Mengjun Liu) designed the experiments, LW (Lixin Wang) and ML (Min Li) performed the experiments. ZL, LD, MZ and LW (Lili Wang) participated in the data analysis, LW (Lixin Wang) and ML (Mengjun Liu) wrote the manuscript and JZ substantively revised it. All authors read and approved the final the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The healthy and diseased jujube trees used in this study were from the Experimental Station of Chinese Jujube, Hebei Agricultural University, in Baoding, Hebei. Chinese jujube is one of traditional and widespread fruit trees in China, and it is not an endangered species. No specific permits are required for sample collection on Chinese jujube.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file S1.
The protein sequences of the CNGC genes from Ziziphus jujuba Mill.
Additional file S2.
The CDS sequences of ZjCNGCs.
Additional file S3.
Domain analysis of ZjCNGCs by using the InterProScan program.
Additional file S4.
Domain analysis of ZjCNGCs by using the SMART program.
Additional file S5.
The number of introns and exons of ZjCNGCs.
Additional file S6.
The primer sequences for amplification of the ZjCNGC genes for qPCR.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, L., Li, M., Liu, Z. et al. Genome-wide identification of CNGC genes in Chinese jujube (Ziziphus jujuba Mill.) and ZjCNGC2 mediated signalling cascades in response to cold stress. BMC Genomics 21, 191 (2020). https://doi.org/10.1186/s12864-020-6601-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-020-6601-5