Abstract
Background
Grape buds exhibit non-uniform, or delayed, break in early spring in subtropical regions because the accumulation of chilling is insufficient. Hydrogen cyanamide (H2CN2, HC) can partially replace chilling to effectively promote bud sprouting and is used widely in warm winter areas. However, the exact underlying mechanism of grape bud release from endodormancy induced by HC remains elusive.
Results
In this study, the transcriptome of grape winter buds under in vitro conditions following HC and water treatment (control) was analyzed using RNA-seq technology. A total of 6772 differentially expressed genes (DEGs) were identified. Furthermore, the gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that starch and sucrose metabolism and plant hormone signaling transduction were most enriched out of the 50 total pathways. HC treatment induced the upregulated expression of sucrose synthase (SUS), sucrose phosphate synthase (SPS), α-amylase (AM), and β-amylase (BM) and downregulated expression of sucrose invertase (INV), hexokinase (HK), fructokinase (FK), soluble starch synthase (SS), and granule-bound starch synthase (GBSS). Hence, the starch concentration in the HC-treated group was significantly lower than that in control, whereas soluble sugar content in the HC-treated group increased quickly and was higher than that in control between 0 and 8 d. The concentration of indoleacetic acid (IAA) and zeatin (ZT) increased, whereas that of abscisic acid (ABA) and gibberellin (GA) decreased in HC treated group, which coincided with the expression level of genes involved in above hormone signals. The content of hydrogen peroxide (H2O2) and enzyme activity of superoxide dismutase (SOD) and peroxidase (POD) were increased in grape buds with HC treatment, whereas catalase (CAT) activity was decreased. HC treatment increased the expression of POD, SOD, primary amine oxidase (PAO), polyamine oxidase (PAOX), and glutathione peroxidase (GSH-Px).
Conclusion
Based on these results, it is possible to propose a mechanistic model that underlies the regulation of endodormancy release in grapevine buds by exogenous HC application.
Similar content being viewed by others
Background
Grape (Vitis vinifera L.) is an important deciduous fruit tree worldwide and table grapes are the main horticultural crop in China. According to data from the United Nations Food and Agriculture Organization (FAO), in the past decade, China’s grape cultivation area increased by more than 336,919 ha and reached 778,585 ha in 2017 and the yield reached 13,160,811 tons. China is the biggest contributor to the world’s table grape industry [1]. Currently, the grape planting area has expanded from the eastern and northern regions to the western and southern regions in China. Provinces in these regions have become important areas for China’s grape industry, such as Sichuan, Yunnan, Zhejiang, and Guangxi [2]. However, these regions climate is a mid-subtropical, damp-heat eco-type zone with a warm winter [2]; thus, effective chilling accumulation is insufficient in these areas, which leads to non-uniform, or delayed, bud break in early spring. This is a major obstacle for the commercial production of table grapes in the above regions.
A certain amount of chilling accumulation is required in perennial species for the proclamation of the buds from endodormancy. Insufficient cold accumulation during chilling accumulation generally results in non-uniform flowering and a reduced fruit set [3]. In agronomic practices, some chemical reagents are often used instead of low temperature to complete natural dormancy. Hydrogen cyanamide (H2CN2, HC) is an effective chemical agent for releasing dormancy and has been successfully applied to commercial crops such as grape (Vitis vinifera L.) [4], apple (Malus domestica Borkh.) [5], sweet cherry (Prunus avium L.) [6], and kiwifruit (Actinidia deliciosa) [7]. There is evidence that HC has various influences on dormancy release in grape. HC treatment causes an abrupt increase in starch hydrolysis and the transient accumulation of soluble sugars in the bud coincides with a transient induction of α-amylase activity [8]. Exogenous HC application increases the expression level of sucrose synthase, pyruvate decarboxylase, a sucrose non-fermenting (SNF)-like protein kinase, grape dormancy breaking-related protein kinase, and alcohol dehydrogenase [9, 10]. Abscisic acid (ABA) also inhibits dormancy release in grape buds and attenuates the advancing effect of HC [11]. HC can promote dormancy release by increasing the content of ethylene and cytokinin [10, 12]. Differentially expressed genes and proteins involved in metabolic, ribosomal, and hormonal signaling pathways are significantly enriched between HC treatment and a control treatment [13]. In addition, HC application stimulates the temporary elevation in H2O2 levels and rapidly upregulates certain genes associated with oxidative stress [4, 9, 10], which causes a sharp decrease in catalase (CAT) activity and transient stimulation of peroxidase (POD) and ascorbate peroxidase (APX) activities [14]. Moreover, some reports have illustrated that HC-induced bud break is enhanced by calcium (Ca2+) signaling and stimulates changes in phosphorylation and transcription regulators [15].
Despite the extensive global use of HC by growers for breaking dormancy in grape, the underlying mechanism that explains the role of HC in dormancy breaking is still not clear. The aim of this study was to investigate the underlying molecular processes behind grape bud sprouting using the application of HC. In this study, RNA-seq was used to categorize and characterize the differentially expressed genes (DEGs) and pivotal pathways in different grape bud dormancy release stages. In addition, the content of products and expression of genes involved in candidate pathways was determined. This study will help us understand the mechanisms in HC-induced grape bud sprouting in regions with warm winters.
Results
Bud break rate
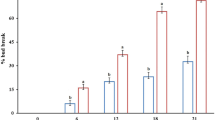
To achieve controlled endodormancy release, dormant grape buds were treated with HC and the percentage of bud break was recorded. This is shown in Fig. 1a. There were no significant differences between the HC-treated and control (CK) group for the first 4 d (Fig. 1b). Sprouting was observed 8 d and 12 d after incubation by HC and water, respectively. On 16 d, the bud breaking rate was 97.85 and 57.62% in the HC treatment group and CK group, respectively. HC effectively promoted dormant release and brought sprouting time forward.
Analysis of transcriptional sequence data
RNA sequencing analysis was used to explore how HC could break grape bud dormancy. For RNA-seq, three biological replicates from each of the following time points were selected: buds of 0 d, 4–12 d mixed sample with water treatment (CK4–12 d), and HC treatment (HC4–12 d). After the elimination of low-quality reads and adaptor sequences, a total of 240.57 million clean reads were recorded and mapped to the grape genome. Finally, of the high-quality reads generated from the three samples, uniquely mapped reads were 65.91–71.52%, whereas total mapped reads were 67.04–72.56% (Table 1).
Identification and functional classification of differentially expressed genes
The expression level of a gene is usually measured by FPKM (expected number of Fragments Per Kilobase of transcript sequence per Millions of base pairs sequenced). The screening criteria for a DEG was Fold Change > 4 and False Discovery Rate < 0.001. Nine samples were detected with 29,551 genes expressed. For the pairwise comparisons among the three stages, 951 (0 d vs CK4–12 d), 4047 (0 d vs HC4–12 d), and 1774 (CK4–12 d vs HC4–12 d) DEGs were detected (Table 2). It was observed that the greater abundance of DEGs in the comparison between the 0 d and HC4–12d stages were more distinct than other comparisons, which indicated that the transcriptome profiles of these two stages were much more distinct than those of the CK4–12d stages. In summary, the number of upregulated genes was greater than that of downregulated genes by pairwise comparison.
For gene ontology (GO), annotated genes were mainly divided into three ontologies: molecular functions (MF), biological processes (BP), and cellular components (CC). Because some genes have multiple functions, the total number of GO clustered genes will be greater than the total number of RNA-seq. The BP category contained the majority of GO annotations (222,191; 62.78%) followed by the CC category (67,034, 18.94%) and MF category (64,682, 18.28%). The major subcategories, along with the analysis of transcripts among the buds in three different stages, are shown in Fig. 2. In the transcripts annotated in GO major categories, the 0d vs CK4–12d, 0d vs HC4–12d, and CK4–12d vs HC4–12d comparisons represent 10,116, 40,080, and 16,997 transcripts, respectively.
The DEGs between each pair of stages were enriched in genes related to distinct Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The top 15 KEGG pathways corresponding to the most abundant DEGs are presented in Additional file 1: Figure S1. The shared KEGG pathways for the 0 d vs CK4–12 d, 0d vs HC4–12 d, and CK4–12 d vs HC4–12 d comparisons included plant hormone signal transduction, starch and sucrose metabolism, protein processing in the endoplasmic reticulum, phenylpropanoid biosynthesis, pentose and glucuronate interconversions, photosynthesis, photosynthesis-antenna proteins, and biosynthesis of amino acids. Based on KEGG analysis, photosynthesis (56 transcripts), starch and sucrose metabolism (113 transcripts), and plant hormone signal transduction (91 transcripts) were the enriched pathways. Therefore, starch and sucrose metabolism and plant hormone signal transduction were mainly studied regarding HC breaking dormancy.
Starch and sucrose metabolism
Soluble sugar plays an important role in plant growth and development. It provides energy for plant growth and acts as a signal molecule to regulate plant growth. In this study, soluble sugar and starch contents were determined and the expression of important DEGs involved in starch and sucrose metabolic pathways were detected by qRT-PCR. After HC treatment, the soluble sugar content peaked at 8 d and then decreased sharply. The starch content continued to decrease from 0 to 16 d (Fig. 3a). During CK sprouting, the content of soluble sugar increased from 0 to 12 d and then declined. The starch concentration in the CK was always higher than that in HC treatment. In the starch and sucrose metabolic pathways, soluble starch synthase (SS), granule-bound starch synthase (GBSS), α-amylase (AM), and β-amylase (BM) were important enzymes for starch synthesis and degradation. In HC treatment, the expression level of VvSS and VvGBSS was downregulated and that of VvAM and VvBM were upregulated at 8–16 d (Fig. 3b). Sucrose invertase (INV), hexokinase (HK), sucrose synthase (SUS), and sucrose phosphate synthase (SPS) play important roles in sucrose catabolism. The expression of VvHK, VvSPS, and VvSUS were significantly upregulated after HC treatment, especially at 8–16 d and was higher than that in the CK group. Nevertheless, the expression of VvINV and VvFK3 was downregulated by HC treatment and was lower than that in the CK (Fig. 3b). Figure 3c summarizes the different factors affected by HC treatment in the starch and sucrose metabolism pathways.
Effect of HC on starch and sucrose metabolism. Bars indicate ± SEM of three biological replicates and different letters indicate significant difference. Different letters in the figure indicate significant difference (P < 0.05) Red color: up-regulation; Green color: down-regulation. a Concentration of soluble sugar and starch in two different treatments. b Transcripts related to sucrose and starch metabolism analyzed by qRT-PCR. c Different factors in starch and sucrose metabolism pathway affected by HC treatment. AM, α-amylase; AGPase, ADP- glucose pyrophosphorylase; BM, β-amylase; FK, fructokinase; GBSS, granule-bound starch synthase; HK, hexokinase; INV, sucrose invertase; PGI, phosphoglucose isomerase; PGM, phosphoglucomutase; SBE, starch-branching enzyme; SPS, sucrose phosphate synthase; SUS, sucrose synthase; UGPase, UDP-glucose pyrophosphorylase
Indoleacetic acid (IAA) signal transduction
In the IAA signal transduction pathway, auxin/indoleacetic acids proteins-auxin response factors (AUX/IAA-ARF) dimers disintegrate when IAA is present and AUX/IAA is degraded by ubiquitination to release ARFs. ARFs form homologous and heterologous dimers after transcriptional activation to regulate the expression of downstream genes and cause the auxin-related response. In the bud breaking dormancy process, IAA was accumulated by HC treatment (Fig. 4a). Although the IAA concentration was also increased in the CK group, it was significantly lower in the CK group than that with HC treatment. In the IAA signal transduction pathway, HC treatment induced VvAUX3 and VvAUX5 downregulation at 12–16 d. VvARF5, VvIAA6, VvIAA29, VvGH3, and VvARG7 are response factors that directly act on plant growth in the IAA signal pathway. We observed that HC treatment induced the upregulation of these genes (Fig. 4b). Figure 4c summarizes the different factors affected by HC treatment in the IAA transduction pathway.
Effect of HC on IAA signals transduction. Bars indicate ± SEM of three biological replicates and different letters indicate significant difference. Different letters in the figure indicate significant difference (P < 0.05). Red color: up-regulation; Green color: down-regulation. a Concentration of IAA in two different treatments. b Transcripts related to IAA signal transduction analyzed by qRT-PCR. c Different factors in IAA signal transduction pathway affected by HC treatment. ARF, auxin response factors; AUX1, auxin / indoleacetic acids proteins; GH3, gretchen hagen 3; SAUR, small auxin-up RNA
Cytoplasmic protein-tyrosine kinase (CTK) signal transduction
CTKs promote dormancy release. We observed that the zeatin (ZT) concentration increased significantly with HC treatment. At 12 d, the ZT concentration reached its peak (90.4 ng/g) in the HC-treated group and this was 1.5 times higher than that in the CK group. The ZT concentration decreased at 16 d (Fig. 5a). In the CTK signal transduction pathway, a cytokinin receptor (CRE1) binds to CTK and is self-phosphorylated. This signal is transferred to a phosphate transporter (AHP). The phosphorylated AHP enters the nucleus and transfers the phosphate group to a series of response regulators (ARR), thereby regulating the downstream cytokinin reaction and plant growth and development. Compared with the expression in the CK group, HC treatment induced VvHPK4, VvHPT4, and VvARR6 upregulation at the sprouting stage (8–12 d) (Fig. 5b). Figure 5c summarizes the different factors affected by HC treatment in the CTK signal transduction pathway.
Effect of HC on CTK signals transduction. Bars indicate ± SEM of three biological replicates and different letters indicate significant difference. Different letters in the figure indicate significant difference (P < 0.05). Red color: up-regulation; Green color: down-regulation. a. Concentration of CTK in two different treatments. b. Transcripts related to CTK signal transduction analyzed by qRT-PCR. c. Different factors in CTK signal transduction pathway affected by HC treatment. ARR6, two-component response regulator 6; HK4, histidine kinase 4; HPT4, histidine-containing phosphotransfer protein 4
Gibberellin (GA) signal transduction
GA is a hormone that is indispensable for breaking seed dormancy. In the HC-treated group, GA1 content decreased sharply and was lowest at 8 d, then increased gradually until buds spouted, whereas GA1 in the CK group was stable up to 8 d and decreased markedly to 16 d (Fig. 6a). GA1 content in the HC treatment group was significantly lower than that in the CK group, in almost the whole process of HC-induced dormancy release. In the GA signal transduction pathway, DELLA protein inhibits the growth and development of plants in the absence of GA. When a GA signal is received by the GA signal sensing region on the DELLA protein, receptor GA-insensitive dwarf 1 (GID1) binds to DELLA to promote the release of the repression function of the DELLA protein and the plant shows normal GA reaction and growth. In this study, HC treatment downregulated the expression of VvGID1B and VvDELLA, but upregulated the expression of VvPIF3, which induced the expression of downstream genes related to GA anabolism to promote bud breaking (Fig. 6b). Figure 6c summarizes the different factors affected by HC treatment in the GA signal transduction pathway.
Effect of HC on GA signals transduction. Bars indicate ± SEM of three biological replicates and different letters indicate significant difference. Different letters in the figure indicate significant difference (P < 0.05). Red color: up-regulation; Green color: down-regulation. a. Concentration of GA3 in two different treatments. b. Transcripts related to GA signal transduction analyzed by qRT-PCR. c. Different factors in GA signal transduction pathway affected by HC treatment. DELLA, DELLA protein; GID1, GA-insensitive dwarf1; PIF3: phytochrome interacting factors 3; TF, Transcription factor
ABA signal transduction
ABA is closely related to plant dormancy. In the ABA signal transduction pathway, ABA combined with ABA receptor (PYR/PYL) can inhibit the enzyme activity of protein phosphatase 2C (PP2C). Serine/thread-protein kinase 2 (SNPK2) activates its own activity via autophosphorylation and acidifies different downstream targets, which triggers ABA-induced physiological reactions. In this study, the concentration of ABA increased at 4 d and then sharp decreased (Fig. 7a). ABA content began to decline after 8 d in the CK group. However, the content of ABA in the HC group decreased from 38.56 ng/g (4 d) to 1.80 ng/g (16 d) and was consistently lower than that in the CK group. In the ABA signal transduction pathway, ABA receptor VvPYL4 was upregulated and VvPP2C8, VvPP2C37, and VvPP2C51 were downregulated by HC treatment. Subsequently, VvSAPK2 was activated by autophosphorylation and downregulated by HC, which induced the decreased expression of VvABAIP5 and VvbZIP (Fig. 7b). Figure 7c summarizes the different factors affected by HC treatment in the ABA signal transduction pathway.
Effect of HC on ABA signals transduction. Bars indicate ± SEM of three biological replicates and different letters indicate significant difference. Different letters in the figure indicate significant difference (P < 0.05). Red color: up-regulation; Green color: down-regulation. a. Concentration of ABA in two different treatments. b. Transcripts related to ABA signal transduction analyzed by qRT-PCR. c. Different factors in GA signal transduction pathway affected by HC treatment. ABAIP, abscisic acid-insensitive 5-like protein; ABF, ABRE binding factors; bZIP, basic leucine zipper transcription factors; PP2C, protein phosphatase 2C; PYR/PYL, abscisic acid receptor; SAPK2, serine/threonine-protein kinase 2; SnRK2, SNF1-related kinase 2
H2O2 accumulation and antioxidative enzyme assay
The accumulation of H2O2 in grapevine buds was determined in this study. For all treatments, H2O2 increased rapidly, reaching its maximum level after 12 d and decreased slightly thereafter (Fig. 8a). From 0 d to 8 d, there was no significant difference in H2O2 content between HC and CK. However, the averaged amount of H2O2 in the HC treated group was higher than that in control from 8 d to 16 d. CAT activity decreased sharply in the two groups with the prolongation of treatment time, but the value in the HC-treated group was lower than that in the CK group from 4 d to 16 d (Fig. 8a). When the buds sprouted, POD and SOD activity increased gradually in all treatments, reaching the peak at 16 d (Fig. 8a). There were no significant changes in POD activity between the two treatments at 0–8 d, but the POD activity in the HC-treated group was higher than that in the CK group at 8–16 d (Fig. 8a).
Effect of HC on oxidation system. Bars indicate ± SEM of three biological replicates and different letters indicate significant difference. Different letters in the figure indicate significant difference (P < 0.05). Red color: up-regulation; Green color: down-regulation. a. The changes of H2O2 Concentration and CAT, POD and SOD activity in two different treatments. b. Transcripts related to oxidation system analyzed by qRT-PCR. c. Different factors in oxidation system pathway affected by HC treatment. CAT, catalase; GSH-Px, glutathione peroxidase; FSD, superoxide dismutase [Fe]; PAO, primary amine oxidase; PAOX, polyamine oxidase; POD, peroxidase; SOD, superoxide dismutase
In the process of reactive oxygen species (ROS) metabolism, the HC-treated group induced the upregulation of ROS-generating genes (VvFSD2, VvFSD3, VvPAO, and VvPAOX) and led to the H2O2 accumulation. To maintain normal growth, the antioxidant defense system of the plant plays a role: ROS-scavenging gene (VvPOD3, VvPOD29, and VvGSH-Px) expression was upregulated by HC treatment (Fig. 8b). Figure 8c summarizes the different factors affected by HC treatment in the ROS metabolism pathway.
Discussion
Carbohydrate metabolism plays an important role in regulating dormancy release processes in plants. A ratio of hexoses to starch content (10:1) appears to be associated with the end of endodormancy in sweet cherry (Prunus avium L.) [16]. During endodormancy, a decrease in the shoot starch concentration and an increase of soluble sugars in both the flower buds and shoots are observed in the Japanese pear (Pyrus pyrifolia Nakai) [17]. HC treatment also causes an abrupt increase in starch hydrolysis and a transient accumulation of soluble sugars in the bud and the internode tissues of superior seedless grapevines during the first 5 days following HC treatment [8]. In this study, the starch content declined remarkably at 0–12 d and this was followed by a slight increase with HC treatment, whereas soluble sugar content increased significantly at 0–8 d followed by a remarkable decrease from 8 to 16 d (Fig. 3a). These results indicated that the increase in soluble sugar (for example, sucrose in grape) and decrease in starch content played key roles in the dormancy release process and can be impacted by HC application. The results of this study indicated that HC treatment upregulated the expression of VvSUS and VvSPS and downregulated the expression of VvSS and VvGBSS (Fig. 3b). HC treatment also upregulated the expression of VvAM and VvBM related to starch degradation (Fig. 3b). In addition, the expression of VvINV was downregulated at 4–16 d after HC treatment and this downregulation was lower than that after CK treatment (Fig. 3b). Hence, the starch content declined at 0–12 d and this was followed by a slight increase with HC treatment. The trend in soluble sugar content was the opposite of this trend in starch content (Fig. 3a). We speculate that the decrease of starch along with the accumulation of soluble sugars during the bud dormancy release stage, which is attributed to the hydrolyzation of starch to soluble sugar, provides energy for the plant to resist low temperatures in winter and maintain normal growth [18]. When dormancy was relieved completely, the starch content did not change significantly and the soluble sugar content decreased significantly. This was because the plant needed a lot of energy to resume growth and soluble sugar was utilized [19].
Endogenous hormones play key roles in the promotion and inhibition of growth in a dynamic balance. A low concentration of IAA promotes bud breaking and a high concentration of IAA has no significant effect on sprouting [20]. In this experiment, although the content of endogenous IAA increased after HC treatment (Fig. 4a), it was still in the low concentration range that can promote grape bud breaking. After HC treatment, the level of trans-zeatin riboside increased sharply during grape sprouting [21]. HC breaks the dormancy of sweet cherry flower buds by increasing trans-zeatin riboside and dihydrozeatin content [6]. Hence, in this experiment, the content of ZT in the HC-treated group was significantly higher than that in the CK group (Fig. 5a).
In many cases, the determination of dormancy or sprouting mainly depends on the balance between ABA and GA levels. HC treatment leads to a decrease in the endogenous ABA level by promoting ABA degradation and inhibiting ABA synthesis, as shown in grape [11] and sweet cherry [6]. In this study, ABA concentration decreased gradually with bud breaking in the HC treatment group (Fig. 7a). This can be explained by HC treatment downregulating the expression of the key genes (VvNCED and VvXERICO) involved in ABA biosynthesis and upregulating the expression of the gene (VvA8H3) that encodes the ABA catabolic enzyme [22, 23]. In addition, HC treatment produced respiratory stress, which led to anaerobic respiration in cells and increased H2O2 content (Fig. 8). Studies have shown that ABA 8′-hydroxylase (ABA8’OH) mediates H2O2 and NO to break dormancy and catalyzes the degradation of ABA [24, 25]. GA has this same antagonistic relationship with ABA [26]. HC treatment causes GA biosynthesis and the degradation genes of grape to increase and decrease, respectively [27]. GA displays a rapid increase when sweet cherry buds enter the burst stage and is associated with the promotion of budburst and blooming by HC treatment [28]. Recent studies have shown that the expression of the GA biosynthesis genes VvGA3ox2, VvGA20ox3, and VvGA20ox6 are significantly downregulated after HC treatment for 24 h and the expression of VvGA3ox2 and VvGA20ox6 are upregulated after HC treatment for 96 h [26]. In this study, GA content decreased at 0–8 d and then increased, but this change was not significant (Fig. 6a). It has been proposed that during the initial activation of the dormant bud meristem, the level of GA must be restricted, but after meristem activation, an increase in its level enhances primordial regrowth [26].
Plant growth is regulated not only by endogenous hormone content, but also by signaling transduction pathways. During chilling accumulation and subsequent germination sprouting of white spruce (Picea glauca) seeds, seed dormancy and germination sprouting may be partly mediated by the changing hormone concentrations and modulation of the interactions among central auxin-signaling pathway components (TIR1/AFB, Aux/IAA, and ARF4) [29]. The DELLA protein is an important negative regulator of the GA reaction, but it has a positive correlation with ABA [30]. ABA level is positively regulated by the DELLA protein by upregulating the expression of the XERICO gene [31]. In this study, we observed that HC treatment resulted in the upregulation (AUX/IAA, ARF, and CYCD) and downregulation (DELLA and ABF) of key genes. These genes are associated with hormone signaling pathways during grape bud breaking. These results indicated that the phytohormone signaling transduction pathway plays key roles in dormancy release in response to HC; however, the accurate regulatory mechanism is not clear even now.
In previous studies of accumulation, some researchers have linked a decrease in starch content during dormancy release to hormones. ABA is a hormone that promotes plant dormancy. Studies have shown that ABA inhibits AA activity during dormancy [32]. Rubio et al. [33] suggested that ABA content was positively correlated with starch content during dormancy in grape buds. In our study, ABA-induced the expression of VvSS1 and VvSS3 and inhibited the expression of VvSUSs, VvSPS, and VvINVs. The results of this experiment also suggested that HC treatment caused the concentration of ABA to decrease (Fig. 7a), which led to an increase in the activity of AA and promoted starch catabolism and soluble sugar accumulation (Fig. 3a). However, more studies are needed to explore how phytohormones induces gene expression and enzyme activity.
Many experiments have shown that H2O2 content increases in buds with HC treatment [4, 6, 11], which is the major factor responsible for bud break. H2O2 treatment inhibits CAT activity, which then generates respiratory stress, leading to cell hypoxia and anaerobic respiration [14]. On one hand, H2O2 is used as a signal molecule in cell wall loosening and leads to buds sprouting [4, 6]. On the other hand, anaerobic respiration produces ethylene, which activates related transcription factors to promote cell elongation and formation, then bud dormancy release [9, 10]. In our study, HC induced an increase in H2O2 content and a decrease in CAT activity than that in the CK group from 8 to 16 d; therefore, to scavenge redundant ROS, the activity of SOD and POD were upregulated, along with key genes involved in protective enzyme metabolism, such as FSD, POD, PAO, GST-Px, and PAOX (Fig. 8). The same conclusion was reached with Seedless grapevine buds [14].
Conclusion
In summary, this study provided an analysis of the metabolic changes during controlled dormancy release in grape. The results suggest a mechanism of action for HC-induced buds break that involves the activation of two pathways: sucrose and starch metabolism and plant hormone signal transduction. As shown in Fig. 9, we obtained a working model of HC breaking winter buds dormancy in grapes, which reveals the relationship between the two pathways.
Material and methods
Plant material
Four-year-old grapevine trees (Vitis vinifera L. cv. Summer black) were grown in an experimental orchard at the Sichuan Agricultural University, Chongzhou (30°24′N, 103°59′E), Sichuan province, China. The 1.5 × 3.0 m planted vines were exposed to standard horticultural practices under a rain shelter protected with polyvinyl film.
HC treatment and sampling
At the end of December 2016 (according to production experience, grape buds were in deep dormancy during this period), the detached canes, each carrying 8 buds at positions 5–12 in the node order, were transferred to the lab. Canes were cut into single-node cuttings, randomly mixed, and separated into two groups (300 single-bud cuttings per group). Two groups were treated with 2% Dormex (520 g/L HC, AlzChem, Germany) and water (as CK). The conditions in the artificial culture room were as follows: culture temperature 25 °C, light intensity 40 μmol/m2∙s, day/night: 12 h/12 h, and relative humidity 80%. Sprouting was defined as the opening of bud scales to reveal new green leaves. Percent bud break was determined at 0, 4, 8, 12, and 16 d after treatment in each treatment group. HC-treated and the CK group were sampled at 0, 4, 8, 12, and 16 days, always between 3 and 5 PM. At every sampling point, buds from 60 cuttings were homogenized and divided into 6 biological replicates for different detections. Samples were frozen immediately in liquid nitrogen and kept at − 80 °C until analysis.
Preparation of RNA-seq libraries
For RNA-seq, three biological replicates from each of the following time points were selected: buds of 0 d, 4–12 d mixed sample with water treatment and 4–12 d mixed sample with HC treatment (those equal in quality at 4 d, 8 d, and 12 d were mixed). In total, 1 μg of RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using the NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations. The library fragments were purified with an AMPure XP system (Beckman Coulter, Beverly, USA). Then, 3 μL of USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min followed by 5 min at 95 °C before PCR. Then, PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. Finally, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system.
Clustering and sequencing
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v4-cBot-HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq Xten platform and paired-end reads were generated.
Differential expression analysis
Differential expression analysis of the two groups was performed using the DESeq R package (1.10.1). DESeq provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value <0.05 by DESeq were assigned as differentially expressed.
GO and KEGG pathway enrichment analysis
GO enrichment analysis of the DEGs was implemented by the GOseq R packages based on Wallenius’ non-central hypergeometric distribution [34], which can adjust for gene length bias in DEGs. KEGG is a database resource for understanding high-level functions and utilities of a biological system, such as the cell, organism, and ecosystem. KEGG can process molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (http://www.genome.jp/kegg/) [35]. KOBAS software was used to test the statistical enrichment of DEGs in KEGG pathways.
Determination of soluble sugar and starch concentration
Anthrone colorimetry was used for the determination of soluble sugar and starch [36]. The content of soluble sugar was based on a standard curve generated with known soluble sugar concentrations. The soluble sugar standard curve was used to calculate sample sugar content. Starch content was converted into a soluble sugar content of 0.9 times. The absorbance was determined at 630 nm.
Quantitation of plant endogenous hormone
The concentrations of four hormones, ZT, IAA, ABA, and GA, were determined by high-performance liquid chromatography (HPLC) using Agilent 1260. Three biological replicates for every sample were performed for endogenous hormone extraction in buds. Buds (0.5 g) were weighed, ground in liquid nitrogen, and subjected to ultrasonic extraction with an extraction solvent (0.5% formic acid and 80% methanol) for 30 min. After centrifugation for 15 min, the supernatant was collected. The extraction was repeated twice and the supernatant was merged. After rotary vacuum evaporation at 38 °C, the water phase was retained and centrifuged for 10 min. The supernatant was concentrated with N-EVAP and dissolved with acetonitrile. All centrifugation was performed at 12000 g at 4 °C. Finally, the extract was filtered with a 0.22 μm filter for HPLC detection. The chromatographic conditions were as follows: C18 (Agilent Zorbax SB) reversed phase column (250 mm × 4.6 mm), flow velocity at 0.8 mL/min, injection volume of 15 μL, and detection wavelength of 270, 218, 200, and 270 nm for ZT, IAA, ABA, and GA, respectively. The endogenous hormone contents were calculated from the peak area of each compound to internal standards (SIGMA-ALDRICH, Oakville, ON, Canada).
Measurement of H2O2 concentration and activity of antioxidant enzymes
The determination of H2O2 content was based on the method of Lin et al. [37]. Briefly, 0.3 g of sample was ground into homogenate with 5 mL of cold acetone and centrifuged at 10000 g for 10 min at 4 °C. Then, 1 mL of supernatant was mixed with 0.1 mL 5% (w/v) of titanium sulfate and 0.2 mL of concentrated ammonia, centrifuged, and the supernatant was discarded. Acetone was used to wash the precipitation 3–5 times and the precipitation was dissolved with 5 mL of 2 M concentrated sulfuric acid. Finally, the volume was adjusted to 10 mL with distilled water. The absorbance was determined at 415 nm. The content of H2O2 was based on a standard curve generated with known H2O2 concentrations.
The activity of POD, SOD, and CAT were determined using the method of Liang et al. [38]. Sample (0.5 g) was ground into homogenate in 8 mL of cold 50 mM phosphate-buffered saline (PBS) (pH 7.8), containing 1% (w/v) polyvinylpyrrolidone (PVP), 0.1 mM ethylenediaminetetraacetic acid (EDTA), and 2 mM dithiothreitol (DTT). The homogenate was then centrifuged at 10000 g for 10 min at 4 °C. POD activity was determined by the guaiacol colorimetric method. The reaction mixtures containing 50 mM PBS (pH 5.7), guaiacol, and 30% (v/v) H2O2 were used to measure POD activity. The absorbance change in 470 nm was monitored and the result was expressed as U/g. The activity of SOD was determined by the nitro-blue tetrazolium method (NBT) and the absorbance was determined at 560 nm. The result was expressed as U/g. The reaction mixture contained 200 mM PBS (pH 7.8), 100 mM H2O2, and the enzyme extract. The activity of CAT was determined at 240 nm and the result was expressed as U/g.
Quantitative real-time PCR
Total RNA was extracted from frozen grape buds using a modified cetyltrimethylammonium bromide method and treated with RNase-free DNase I (Takara, Dalian, China) to remove genomic DNA contamination. The forward and reverse primers for candidate and reference genes (Additional file 1: Table S1) were designed using the Primer Express® Software v2.0 (ABI, USA). Quantitative real-time PCR was performed on the CFX96 Real-Time System C1000 Thermal Cycler (Bio-RAD, Hercules, CA, USA), following the manufacturer’s protocol in an SYBR Premix Ex Taq kit (TaKaRa, Dalian, China) and analyzed with 2−∆∆CT. Three replicates were performed for three separate RNA extracts from three samples.
Availability of data and materials
The datasets generated and analyzed during the current study are available in the NCBI repository [PRJNA558570]. Other supporting data can be found within the manuscript and its additional files.
Abbreviations
- ABA:
-
Abscisic acid
- ABAIP:
-
Abscisic acid-insensitive 5-like protein
- ABF:
-
ABRE binding factors
- AGPase:
-
ADP- glucose pyrophosphorylase
- AM:
-
α-amylase
- ARF:
-
Auxin response factors
- ARR6:
-
Two-component response regulator 6
- AUX1:
-
Auxin/indoleacetic acids proteins
- BM:
-
β-amylase
- bZIP:
-
Basic leucine zipper transcription factors
- CAT:
-
Catalase
- DEGs:
-
Differentially expressed genes
- DELLA:
-
DELLA protein
- FK:
-
Fructokinase
- FSD:
-
Superoxide dismutase [Fe]
- GA:
-
Gibberellin
- GBSS:
-
Granule-bound starch synthase
- GH3:
-
Gretchen hagen 3
- GID1:
-
GA-insensitive dwarf1
- GO:
-
Gene ontology
- GSH-Px:
-
Glutathione peroxidase
- H2O2 :
-
Hydrogen peroxide
- HC:
-
Hydrogen cyanamide
- HK:
-
Histidine kinase
- HPT4:
-
Histidine-containing phosphotransfer protein 4
- IAA:
-
Indoleacetic acid
- INV:
-
Sucrose invertase
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- PAO:
-
Primary amine oxidase
- PAOX:
-
Polyamine oxidase
- PGI:
-
Phosphoglucose isomerase
- PGM:
-
Phosphoglucomutase
- PIF3:
-
Phytochrome interacting factors 3
- POD:
-
Peroxidase
- PP2C:
-
Protein phosphatase 2C
- PYR/PYL:
-
Abscisic acid receptor
- SAPK2:
-
Serine/threonine-protein kinase 2
- SAUR:
-
Small auxin-up RNA
- SBE:
-
Starch-branching enzyme
- SnRK2:
-
SNF1-related kinase 2
- SOD:
-
Superoxide dismutase
- SPS:
-
Sucrose phosphate synthase
- SS:
-
Starch synthase
- SUS:
-
Sucrose synthase
- TF:
-
Transcription factor
- UGPase:
-
UDP-glucose pyrophosphorylase
- ZT:
-
Zeatin
References
OIV. Statistical report on world vitiviniculture. Paris: International Organisation of Vine and Wine; 2018.
Deng QX, Xia H, Lin LJ, Wang J, Yuan L, Li KN, et al. SUNRED, a natural extract-based biostimulant, application stimulates anthocyanin production in the skins of grapes. Sci Rep. 2019;9:2590.
Anderson JV, Horvath DP, Chao WS, Foley ME. Bud dormancy in perennial plants: a mechanism for survival. Top Curr Genet. 2010;21:69–90.
Sudawan B, Chang CS, Chao HF, Maurice SBK, Yung-fu Y. Hydrogen cyanamide breaks grapevine bud dormancy in the summer through transient activation of gene expression and accumulation of reactive oxygen and nitrogen species. BMC Plant Biol. 2016;16(1):202.
Krasuska U, Ciacka K, Debska K, Bogatek R, Gniazdowska A. Dormancy alleviation by NO or HCN leading to decline of protein carbonylation levels in apple (Malus domestica Borkh.) embryos. J Plant Physiol. 2014;171(13):1132–41.
Ionescu I, López-Ortega G, Meike B, Bayo-Canha A, Junge A, Gericke O, et al. Transcriptome and metabolite changes during hydrogen cyanamide-induced floral bud break in sweet cherry. Front Plant Sci. 2017;8:1233.
Walton EF, Fowke PJ. Effect of hydrogen cyanamide on kiwifruit shoot flower number and position. J Hortic Sci. 1993;68(4):529–34.
Ben Mohamed H, Vadel AM, Geuns JMC, Khemira H. Carbohydrate changes during dormancy release in superior seedless grapevine cuttings following hydrogen cyanamide treatment. Sci Hortic. 2012;140:19–25.
Pérez FJ, Vergara R, Rubio S. H2O2 is involved in the dormancybreaking effect of hydrogen cyanamide in grapevine buds. Plant Growth Regul. 2008;55:149–55.
Ophir R, Pang X, Halaly T, Venkateswari J, Lavee S, Galbraith D, et al. Gene-expression profiling of grape bud response to two alternative dormancy-release stimuli expose possible links between impaired mitochondrial activity, hypoxia, ethylene-ABA interplay and cell enlargement. Plant Mol Biol. 2009;71:403–23.
Khalil-Ur-Rehman M, Wang W, Xu YS, Haider MS, Li CX, Tao JM. Comparative study on reagents involved in grape bud break and their effects on different metabolites and related gene expression during winter. Front Plant Sci. 2017;8:1340.
Vergara R, Parada F, Rubio S, Pérez FJ. Hypoxia induces H2O2 production and activates antioxidant defense system in grapevine buds through mediation of H2O2 and ethylene. J Exp Bot. 2012;63:4123–31.
Khalil-Ur-Rehman M, Wang W, Dong Y, Faheem M, Xu YS, Gao ZH, et al. Comparative transcriptomic and proteomic analysis to deeply investigate the role of hydrogen cyanamide in grape bud dormancy. Int J Mol Sci. 2019;20:3528.
Ben Mohamed H, Vadel AM, Geuns JMC, Khemira H. Effects of hydrogen cyanamide on antioxidant enzymes’ activity, proline and polyamine contents during bud dormancy release in superior seedless grapevine buds. Acta Physiol Plant. 2012;34:429–37.
Pang X, Halaly T, Crane O, Keilin T, Keren-Keiserman A, Ogrodovitch A, et al. Involvement of calcium signaling in dormancy release of grape buds. J Exp Bot. 2007;58:3249–62.
Kaufmann H, Blanke M. Changes in carbohydrate levels and relative water content (RWC) to distinguish dormancy phases in sweet cherry. J Plant Physiol. 2017;218:1–5.
Ito A, Sakamoto D, Moriguchi T. Carbohydrate metabolism and its possible roles in endodormancy transition in Japanese pear. Sci Hortic. 2012;144:187–94.
Horikoshi H, Sekozawa Y, Sugaya S. Inhibition of carbohydrate metabolism by thermal fluctuations during endodormancy lead to negative impacts on bud burst and incidence of floral necrosis in “Housui” Japanese pear flower buds. Sci Hortic. 2017;224:324–31.
Bonhomme M, Rageau R, Lacointe A, Gendraud M. Influences of cold deprivation during dormancy on carbohydrate contents of vegetative and floral primordia and nearby structures of peach buds (Prunus persica L. batch). Sci Hortic. 2005;105(2):223–40.
Brady S, Mccourt P. Hormone cross-talk in seed dormancy. J Plant Growth Regul. 2003;22(1):25–31.
Lombard P, Cook N, Bellstedt D. Endogenous cytokinin levels of table grape vines during spring budburst as influenced by hydrogen cyanamide application and pruning. Sci Hortic. 2006;109(1):92–6.
Zheng CL, Halaly T, Acheampong AK, Takebayashi Y, Jikumaru Y, Kamiya Y, et al. Abscisic acid (ABA) regulates grape bud dormancy, and dormancy release stimuli may act through modification of ABA metabolism. J Exp Bot. 2015;66(5):1527–42.
Yue C, Cao H, Hao X, Zeng J, Qian W, Guo Y, et al. Differential expression of gibberellin- and abscisic acid-related genes implies their roles in the bud activity-dormancy transition of tea plants. Plant Cell Rep. 2017;37(3):425–41.
Liu Y, Zhang J. Rapid accumulation of NO regulates ABA catabolism and seed dormancy during imbibition in Arabidopsis. Plant Signal Behav. 2009;4(9):905–7.
Liu Y, Ye N, Liu R, Zhang J. H2O2 mediates the regulation of ABA catabolism and GA biosynthesis in Arabidopsis seed dormancy and germination. J Exp Bot. 2010;61(11):2979–90.
Zheng CL, Acheampong AK, Shi ZW, Halaly T, Kamiya Y, Ophir R, et al. Distinct gibberellin functions during and after grapevine bud dormancy release. J Exp Bot. 2018;69(7):1635–48.
Giacomelli L, Rota-Stabelli O, Masuero D, Acheampong AK, Moretto M, Caputi L, et al. Gibberellin metabolism in Vitis vinifera L. during bloom and fruit-set: functional characterization and evolution of grapevine gibberellin oxidases. J Exp Bot. 2013;64:4403–19.
Wang L, Zhang CX, Huang JC, Zhu LN, Yu XM, Li JF, et al. Hydrogen cyanamide improves endodormancy release and blooming associated with endogenous hormones in ‘summit’ sweet cherry trees. New Zeal J Crop Hort. 2017;45(1):14–28.
Liu Y, Müller K, El-Kassaby Y, Kermode A. Changes in hormone flux and signaling in white spruce (Picea glauca) seeds during the transition from dormancy to germination in response to temperature cues. BMC Plant Biol. 2015;15(1):292.
Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L. Gibberellic acid signaling repressor RGL2 inhibits arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell. 2009;20(1):2729–45.
Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19(10):3037–57.
Huang YQ, Cai SG, Ye LZ, Hu HL, Li CD, Zhang GP. The effects of GA and ABA treatments on metabolite profile of germinating barley. Food Chem. 2016;192(1):928–33.
Rubio S, Noriega X, Pérez FJ. ABA promotes starch synthesis and storage metabolism in dormant grapevine buds. J Plant Physiol. 2019;234-235:1–8.
Young M, Wakefield M, Smyth G, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14.
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007;36:480–4.
Hansen J, Moller IB. Percolation of starch and soluble carbohydrates from plant tissue for quantitative determination with anthrone. Biochem Anal Biochem. 1975;68:87–94.
Lin Z, Li S, Lin G, Guo J. Relationship between H2O2 accumulation and membrane lipid peroxidation in senescent leaves and chloroplasts. Plant Physiol Mol Biol. 1988;14:16–22.
Liang D, Shen YQ, Ni ZY, Wang Q, Lei Z, Xu NQ, et al. Exogenous melatonin application delays senescence of kiwifruit leaves by regulating the antioxidant capacity and biosynthesis of flavonoids. Front Plant Sci. 2018;9:426.
Acknowledgements
We would like to thank Editage (www.editage.cn) for English language editing.
Funding
The present work was supported by Sichuan Science and Technology Program (2017JY0169). The funding body had no role in the design of the study, data collection and analysis, interpretation of data, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
DL, HX and HFZ conceived and designed the experiment. DL, XJH and TS participated in sample collection, physiological index determination, performed bioinformatics and statistical analyses. YQS participated in RNA extraction, carried out the qRT-PCR, DL and XJH drafted the manuscript. LJL, JW, QXD and XLL revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
Number of DEGs up and down-regulated in most enriched pathways among three stages of dormancy. Table S1. Primers used in this study.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liang, D., Huang, X., Shen, Y. et al. Hydrogen cyanamide induces grape bud endodormancy release through carbohydrate metabolism and plant hormone signaling. BMC Genomics 20, 1034 (2019). https://doi.org/10.1186/s12864-019-6368-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-6368-8