Abstract
Background
The Type II clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) is a powerful genome editing technology, which is more and more popular in gene function analysis. In CRISPR/Cas, RNA guides Cas nuclease to the target site to perform DNA modification.
Results
The performance of CRISPR/Cas depends on well-designed single guide RNA (sgRNA). However, the off-target effect of sgRNA leads to undesired mutations in genome and limits the use of CRISPR/Cas. Here, we present OffScan, a universal and fast CRISPR off-target detection tool.
Conclusions
OffScan is not limited by the number of mismatches and allows custom protospacer-adjacent motif (PAM), which is the target site by Cas protein. Besides, OffScan adopts the FM-index, which efficiently improves query speed and reduce memory consumption.
Similar content being viewed by others
Background
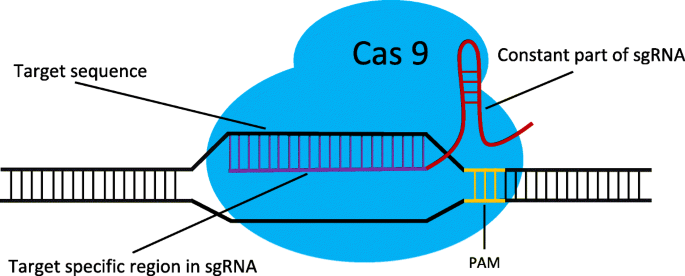
CRISPR/Cas is a powerful genome editing tool. When delivered into cells, as illustrated in Fig. 1, sgRNA will guide Cas nuclease to the desired DNA site and create a DNA double-strand break, the repair of which leads to a variety of DNA sequence modifications [1,2,3]. The performance of CRISPR/Cas is highly dependent on well-designed sgRNA. However, the off-target effect of sgRNA may lead to undesired mutations in the genome and limit the use of this technology. The off-target effect is caused by both sgRNA and Cas9. A few mismatches between the 5′ 20-nt sequence in sgRNA (the purple part in Fig. 1) and the target DNA sequence can be tolerated [4, 5]. Some studies have shown that CRISPR/Cas9 non-specifically cleave DNA sites with several mismatches, generating off-target mutations with considerable frequency [4,5,6,7,8,9]. The optimal PAM (the yellow part in Fig. 1) recognized by SpCas9 is 5′-NGG-3′. However, SpCas9 also binds 5′-NAG-3′ or 5′-NGA-3′ with low frequency [6, 10]. Accordingly, it is essential to identify potential off-target sites and improve sgRNA specificity.
Finding target sites can generally be accomplished quite easily by scanning the whole genome for the PAM sequence, such as 5′-NGG-3′ for the CRISPR/Cas9 system. Then we can obtain a set K of candidates to be sgRNA. However, we must remove the sgRNA with high off-target potential from K.
Third-party alignment tools, such as BWA [11] and Bowtie [12], are often used to search off-target sites [13,14,15]. However, alignment tools are not originally designed for off-target detection. As mentioned in CRISPR-DO [15], for each candidate sgRNA k-mer in K, alignment tools have to scan the entire genome once to identify its off-target sites, rather than searching in K, which results in a large amount of redundant computations. On the other hand, although alignment tools support pattern matching within several mismatches, the mismatch position in the query cannot be set. As a result, these alignment tools cannot consider the PAM sequence when searching mismatched sites. The off-target sites returned by alignment tools contain a number of sequences that are not adjacent to PAM. Accordingly, we have to filter the returned off-target sites and remove those false off-targets [15].
CasOT [16] is a Perl script that searches for potential off-target sites in any given genome, with user-specified PAM. It allows mismatches in the seed and non-seed regions and provides both a single-gRNA searching mode and a paired-gRNA searching mode. CasOT can identify potential off-target sites in an acceptable period of time.
Cas-OFFinder [17] can search for potential off-target sites in a given genome or set of user-defined sequences. It is not limited by the number of mismatches and allows various PAM sequences. The tool is partly written in OpenCL, enabling operations using an accelerator such as GPU, which can significantly speed up the searching process.
Perez et al. constructed a trie to store all candidate sgRNA (k-mer) in K, and thus to search off-target sites by traversing the trie rather than the entire genome [18]. It is necessary to traverse the trie once for each k-mer to detect its off-target sites within several mismatches; consequently, this involves a large amount of redundant computations. In addition, a trie is a space-consuming data structure, and the number of k-mers adjacent to PAM in the genome is usually huge [15]. Detecting off-target sites for large genomes will thus consume hundreds of GB of memory.
Accordingly, in this paper, we present OffScan, a universal and fast CRISPR/Cas off-target site detection tool. OffScan is not limited by a number of mismatches and/or PAM. It adopts FM-index [19] to assist in off-target searching, which efficiently reduces memory footprint and query time and enables the design of highly specific sgRNA.
Results and discussion
CRISPR target sites scan
The PAM scan model is used to generate candidate sgRNAs. We tested the PAM scan model of OffScan on four genomes: hg38 (human), mm10 (mouse), danRer7 (zebrafish), and ce10 (C. elegans). Since GuideScan [18] does not include a PAM scan model, we used the results of a popular sgRNA design tool, CRISPR-DO [15], for comparison. The total number of k-mers in the millions of target sequences identified by 5′-NGG-3′ is shown in Table 1. OffScan can find a comparable number of candidate target sites compared with CRISPR-DO.
Off-target detection module
We test the performance of OffScan in hg38 with 1, 3, and 10 mismatches and mm10 with 3 mismatches. The sgRNA length is 20 bp. We arbitrarily chose 1000 SpCas9 targets and run OffScan to detect off-target sites. The CPU version is E7–8890 v3. As shown in Table 2, the number of mismatches and genome size will affect the time taken to detect off-target sites.
We compared the performance of OffScan with GuideScan under different number of mismatches on four genomes used in the previous test. As shown in Fig. 2, when the number of mismatches is small, the performance is similar. As the number of mismatches increases, the performance gap between the two becomes larger and larger. In the fuzzy matching part, a bounded traversal strategy is adopted. The lower bound of the mismatch number of the query string Q and the original string X are estimated before the traversal, so that the program can return the traversal of the branch in advance, effectively reducing the search space in suffix tree. The depth of the downward extension increases the efficiency of the comparison. In addition, since OffScan is designed for general sgRNA design, the program behaves similarly on different species.
Conclusions
OffScan enables off-target site detection in any genome, without limitation of the PAM sequence or number of mismatches. Moreover, OffScan utilizes the FM-index to support fast, memory-efficient, and highly specific sgRNA design. The sgRNA design and off-target site detection can be made more convenient using OffScan. In future, we will incorporate more features into OffScan and provide useful tools for the research community.
Methods
CRISPR target sites scan
To facilitate sgRNA design, we implemented a PAM scan module in OffScan. The Cas proteins require a PAM sequence to bind to, so the PAM scan is implemented to scan the entire genome in order to find the sequences with this PAM (e.g. 5′-NGG-3′ for the Cas9 enzyme). The module supports the use of a custom PAM sequence, which can be used for general-purpose sgRNA design. In the majority of situations, we simply need to scan the entire genome once to find the candidate sgRNA with PAM; accordingly, we did not use the FM-index in this module.
Off-target sites detection
The off-target site detection problem can be divided into two problems: namely, off-target sites with and without mismatches.
We adopt the backward search algorithm [20] to find sites without mismatch, which is a pattern matching problem. Let ∑ be an alphabet. A sentinel symbol $ is not present in it and is lexicographically smaller than all the symbols in ∑. A string X = a0a1 ...an − 1 is terminated with symbol $ (i.e. an − 1 = $) and this symbol only appears at the end. The length of string X is |X| = n. X[i] = ai is the i-th symbol of X and X[i,j] is the substring ai…aj.
The suffix array data structure is a succinct representation of the lexographic ordering of all the suffixes of a string [21]. The suffix array of string X, denoted as SA(X), is actually the permutation of the indices {1, 2, …, n-1} of X that
iif X[j,n-1] is the i-th lexographically smallest suffix of X. For example, if X = banana$, the SA(X) = [6,5,3,1,0,4,2], as shown in Fig. 3a. Since the suffixes in SA are sorted in lexographic order, the start positions of all the instances of a pattern Q in X should be an interval of SA, that is called a suffix array interval, denoted as a pair of integers [sp,ep]. Thus, the pattern matching of Q in X is equivalent to find the suffix array interval of Q in X.
Ferragina and Manzini developed a data structure called FM-index [19], which can determine the suffix array interval [sp,ep] of pattern Q in O(|Q|) time and requires much less memory than a suffix array. FM-index is a careful combination of a compression algorithm Burrows-Wheeler transform (BWT) [22] and suffix array. The BWT of X, denoted as B(X), is a permutation of the string that
That is to say, B[i] is the symbol preceding the first symbol of the suffix starting at SA[i]. For example, if X = banana$, the B(X) = annb$aa, as presented in Fig. 3a.
To enable pattern matching with FM-index, Ferragina and Manzini added two more data structures: C array and Occ matrix. As illustrated in Fig. 3b, for a symbol a, C[a] is the number of occurrences of symbols that are lexographically smaller than a. Occ[a,i] records the number of occurrences of the character a in B[0,i]. For a query Q whose suffix array interval in X is [sp,ep], the interval of string aQ can be calculated with C and Occ arrays as follows:
From the above eq. (3), we notice that the pattern matching should start from the last character of Q, that is backward search algorithm. Algorithm 1 presents the details of searching procedure. Firstly, the suffix array interval of the last symbol of Q is calculated from C array. Then the interval is calculated iteratively based on Eq. (3), as shown in Fig. 3c. For the returned values of backward search algorithm, if sp < ep, Q has more than one instances in X; if sp = ep, Q has just one instance in X; if sp > ep, Q is not included in X.

The backward search algorithm updates the suffix array interval at most |Q| times, so the time complexity is O(|Q|). The time complexity of backward search is linear of query string length |Q|, equal to the exact matching in a trie; however, the FM-index is more space-efficient than a trie. Besides, the backward search algorithm actually realizes a top-down traversal of a trie.

Since sgRNA allows several mismatches when binding to DNA, an off-target detection algorithm supporting only exact match is not sufficient. OffScan also implements search with mismatches. A naïve method is to traverse the trie to search sgRNA with mismatches, but this process is rather time-consuming. To avoid unnecessary comparisons, we introduce a bounded-search for mismatches in OffScan. For a query of Q in X, we define LB[i] as the lower bound of the number of differences between Q[i,|Q|-1] and X. If the current allowing number of mismatches is smaller than LB[i], the search process will stop and backtrack to other branches. Algorithm 2 presents the procedure of calculating LB[i].
FM-index construction
The off-target site detection module used in OffScan supports any number of mismatches, including no mismatches for exact matching. In terms of the general procedure, we have to detect off-target sites for each k-mer. To avoid the redundant computations involved in scanning of the entire genome, we detect off-targets only within the candidate sgRNA set K.
To improve the query efficiency and reduce space requirements, we adopt the FM-index to aid in off-target site detection. While this data structure is originally designed for single-string queries, we want to extend it to multiple sequence queries. To solve this problem, we add a “$” character to the end of each k-mer and concatenate all k-mers as a long string. We then construct an FM-index for the concatenation.
Availability of data and materials
Please contact author for data requests.
Abbreviations
- BWT:
-
Burrows-Wheeler transform
- Cas:
-
CRISPR-associated
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- DSB:
-
Double-strand break
- PAM:
-
Protospacer-adjacent motif
- sgRNA:
-
Single guide RNA
References
Koonin EV, Makarova KS. CRISPR-Cas: an adaptive immunity system in prokaryotes. F1000 Biol Rep. 2009;1:95.
Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–70.
Memi F, Ntokou A, Papangeli I. CRISPR/Cas9 gene-editing: research technologies, clinical applications and ethical considerations. Semin Perinatol. 2018;S0146-0005(18):30081–8.
Lin Y, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42(11):7473–85.
Chen CH, Xiao T, Xu H, Jiang P, Meyer CA, Li W, Brown M, Liu XS. Improved design and analysis of CRISPR knockout screens. Bioinformatics. 2018;34(23):4095–101.
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting speci city of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–32.
Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput pro ling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease speci city. Nat Biotechnol. 2013;31:839–43.
Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting -globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–92.
Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–41.
Jiang W, Bikard D, Cox D, Zhang F, Marraf ni LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–9.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60.
Langmead B, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25.
Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–84.
Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2014;33(2):187–97.
Ma J, Köster J, Qin Q, et al. CRISPR-DO for genome-wide CRISPR design and optimization. Bioinformatics. 2016;32(21):3336–8.
Xiao A, Cheng Z, Kong L, Zhu Z, Lin S, Gao G, Zhang B. CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics. 2014;30(8):1180–2.
Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30(10):1473–5.
Perez AR, Pritykin Y, Vidigal JA, et al. GuideScan software for improved single and paired CRISPR guide RNA design. Nat Biotechnol. 2017;35(4):347–9.
Ferragina P, Manzini G. Opportunistic data structures with applications. In: FOCS 2000: proceedings of the 41st Symposium on Foundations of Computer Science. Washington: IEEE Computer Society; 2000. p. 390–8.
Lam TW, et al. Compressed indexing and local alignment of DNA. Bioinformatics. 2008;24:791–7.
Manber U, Myers G. Suffix arrays: a new method for on-line string searches. In: SODA’90: proceedings of the first annual ACM-SIAM symposium on discrete algorithms. Philadelphia: Society for Industrial and Applied Mathematics; 1990. p. 319–27.
Burrows M, Wheeler DJ. A block-sorting lossless data compression algorithm. In Technical report 124, Digital Equipment Corporation, Palo Alto: Citeseer; 1994.
Acknowledgements
We would like to thank Jiaming Xu and Minxia Cheng for the collection of related papers and the three anonymous reviewers for their thoughtful comments and valuable feedback.
About this supplement
This article has been published as part of BMC Genomics, Volume 21 Supplement 1, 2020: Selected articles from the 14th International Symposium on Bioinformatics Research and Applications (ISBRA-18): genomics. The full contents of the supplement are available at https://bmcgenomics.biomedcentral.com/articles/supplements/volume-21-supplement-1.
Funding
This work was supported by National Key R&D Program of China 2017YFB0202003, 2017YFB0202602, 2017YFC1311003, 2016YFC1302500, 2016YFB0200400, 2017YFB0202104; NSFC Grants 61772543, U1435222, 61625202, 61272056; the Fundamental Research Funds for the Central Universities; and Guangdong Provincial Department of Science and Technology under grant No. 2016B090918122. The funding bodies did not influence the design of the study, data collection, analysis, or interpretation, or writing of the manuscript.
Author information
Authors and Affiliations
Contributions
YC designed the study, analyzed, interpreted the data and wrote the manuscript. SP and TT prepared data and performed tests. XL, CH and CY revised and edited the manuscript. All authors have given final approval of the version to be published.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Cui, Y., Liao, X., Peng, S. et al. OffScan: a universal and fast CRISPR off-target sites detection tool. BMC Genomics 21 (Suppl 1), 872 (2020). https://doi.org/10.1186/s12864-019-6241-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-6241-9