Abstract
Background
Coffee production relies on plantations with varieties from Coffea arabica and Coffea canephora species. The first, the most representative in terms of coffee consumption, is mostly propagated by seeds, which leads to management problems regarding the plantations maintenance, harvest and processing of grains. Therefore, an efficient clonal propagation process is required for this species cultivation, which is possible by reaching a scalable and cost-effective somatic embryogenesis protocol. A key process on somatic embryogenesis induction is the auxin homeostasis performed by Gretchen Hagen 3 (GH3) proteins through amino acid conjugation. In this study, the GH3 family members were identified on C. canephora genome, and by performing analysis related to gene and protein structure and transcriptomic profile on embryogenic tissues, we point a GH3 gene as a potential regulator of auxin homeostasis during early somatic embryogenesis in C. arabica plants.
Results
We have searched within the published C. canephora genome and found 17 GH3 family members. We checked the conserved domains for GH3 proteins and clustered the members in three main groups according to phylogenetic relationships. We identified amino acids sets in four GH3 proteins that are related to acidic amino acid conjugation to auxin, and using a transcription factor (TF) network approach followed by RT-qPCR we analyzed their possible transcriptional regulators and expression profiles in cells with contrasting embryogenic potential in C. arabica. The CaGH3.15 expression pattern is the most correlated with embryogenic potential and with CaBBM, a C. arabica ortholog of a major somatic embryogenesis regulator.
Conclusion
Therefore, one out of the GH3 members may be influencing on coffee somatic embryogenesis by auxin conjugation with acidic amino acids, which leads to the phytohormone degradation. It is an indicative that this gene can serve as a molecular marker for coffee cells with embryogenic potential and needs to be further studied on how much determinant it is for this process. This work, together with future studies, can support the improvement of coffee clonal propagation through in vitro derived somatic embryos.
Similar content being viewed by others
Background
Coffee is a worldwide consumed commodity, mostly produced through Coffea arabica (63.21% of total production) and Coffea canephora plantations, with Brazil as the biggest producer country that corresponds to 35.7% of the global production [1]. The beverage generated from the roasted grains is mainly characterized by caffeine content and its effects as stimulant [2], but has other metabolite compounds, like flavonols, with antioxidant properties that are beneficial for human health [3].
The crop is predominantly propagated by seeds, which impair the plantation homogeneity. Rooting recalcitrance of plantlets is one of the main reasons by which common vegetative propagation has not been applied [4] yet, leading coffee researchers to the challenge of establishing an efficient alternative method for propagation. In vitro somatic embryogenesis (SE) followed by development and acclimatization of the plantlets is an interesting option for achieving efficient clonal propagation, as in 2016 around 7 million coffee plants were produced through this process in Central America [5].
Somatic embryogenesis is also an important process for genetic transformation, due to the possibility of regenerating plantlets from single cells or small cellular clusters, an alternative way to improve perennial crops breeding. One of the challenges is to understand what differentiates cells with embryogenic competence from the others, and possibly confirm or establish new molecular markers, as morphological characteristics alone are not enough to predict embryogenic capacity [6].
In the case of C. arabica, SE is achieved by the indirect pathway, that is, with an intermediate step of calli formation before embryo regeneration. The coffee leaf explants incubated on auxin-rich medium generate embryogenic-competent calli just after nearly three months, together with non-embryogenic callus production. This pattern of embryogenic calli formation seems to occur via root meristem-associated pathway, with the cellular identity being similar to root meristem cells, which is induced by incubation on auxin-rich medium and wounds, triggered mostly by auxin signaling and regulators such as ARFs and WOX11 [7].
Comprehension of metabolic pathways related to auxin homeostasis can be very informative because such hormone is among the major regulators of SE induction and embryo development [8, 9]. The balance between auxin and its conjugates with amino acids is determinant for cell responses to environment stimuli [10] and represents a deeper layer of complexity related to auxin balance influence on SE, as exemplified by the report that different conjugates are associated to specific direct somatic embryogenesis phases in C. canephora [9]. This conjugation between amino acids and auxin is catalyzed by Gretchen Hagen 3 (GH3) family proteins [10,11,12], which is a widespread family in plants [13] and have been recently characterized in some species like Solanum lycopersicum [14], Malus domestica [15] and Medicago truncatula [16].
Proteins of GH3 family catalyze amino acid conjugation to acyl substrates, mostly to auxin, jasmonic acid and benzoates, thus being associated to many plant metabolic pathways. It is reported that members of this family can be clustered into three main groups and, possibly, proteins from the same group share similarities regarding the specificity to acyl substrates [13]. Generally, specific sets of amino acid residues are related to protein interaction with specific substrates and they are different among GH3 proteins with different substrate affinity [11, 17]. Therefore, the knowledge about the relations of these specific amino acid sequences with substrate specificity is helpful in the search for a proper GH3 gene related to a specific study subject.
According to this context, our aim was to identify the members of GH3 family in C. canephora, the Coffea species with an available public genome, and analyze their phylogenetic and structural features, as well as transcriptional profile and point transcription factors related to potential SE regulators GH3 members. We have found four potential members (CcGH3.9, CcGH3.13, CcGH3.15 and CcGH3.16) that may be associated with auxin conjugation to acidic amino acids which can lead auxin to degradation [17], and some of their potential transcriptional regulators. We analyzed the transcriptional profile of these four homologous CcGH3s genes in C. arabica calli with embryogenic competence or not. CaGH3.15 expression pattern was the only correlated with embryogenic potential and also with the CaBBM expression profile, a regulator of somatic embryogenesis in coffee [18, 19] and other species [20]. These findings will help to increase the knowledge about coffee somatic embryogenesis and point to the influence of auxin homeostasis, highlighting molecular aspects that may be useful for the comprehension of this process that could be an alternative for coffee clonal propagation.
Results
Identification and distribution of GH3 members in C. canephora
The blastp analysis against C. canephora proteome resulted in 20 amino acid sequences, but three of them (Additional file 1: Data S1) lacked the domains commonly shared by GH3 proteins (PLN02247 and pfam03321). The other 17 putative GH3 members are further summarized (Table 1) with putative protein length, predicted gene position in chromosome (Additional file 3: Figure S1) and locus identification based on Coffee Genome Hub database [21]. Most of these proteins have between 530 and 630 amino acid residues and their genes are distributed along chromosomes 1, 2, 5, 7 and 10. However, almost half of the genes identified in our work are still unmapped (chromosome 0). CcGH3.10 and CcGH3.11 are localized in tandem on chromosome 2 and some other genes share high degree of similarity, like CcGH3.2 and CcGH3.5 with 98% identity on nucleotide level. In addition, some genes that are not yet anchored to any chromosome seem to be closely mapped in chromosome 0 like CcGH3.4, CcGH3.5 and CcGH3.6.
Phylogenetic and structural analysis of putative GH3 genes and proteins
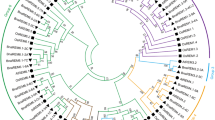
All the nucleotide sequences of putative GH3 genes found in C. canephora genome were used as input data to construct a phylogenetic tree (Fig. 1). Some genes have similar genomic structures, although no general structural pattern for GH3 genes on C. canephora was identified.
The tetrad CcGH3.2, CcGH3.4, CcGH3.5 and CcGH3.16 has four exons and three introns with similar lengths. They are similar to the pair CcGH3.9-CcGH3.14, differing only in intron length. There are other two pairs with similar structure, CcGH3.6-CcGH3.8 and CcGH3.13-CcGH3.15. The arrangement of exons and introns did not correlate with similarity at sequence level in all cases, for example, CcGH3.14 is more similar with CcGH3.12 at nucleotide sequence level than with CcGH3.9.
To discriminate CcGH3s in functional groups according to literature, a second phylogenetic tree was constructed with GH3 amino acid sequences of Arabidopsis thaliana, Zea mays and Oriza sativa (Fig. 2). This approach clustered proteins CcGH3.12 and CcGH3.14 in group I, CcGH3.2, CcGH3.3, CcGH3.4, CcGH3.5, CcGH3.6, CcGH3.8 and CcGH3.17 in group II and CcGH3.1, CcGH3.7, CcGH3.9, CcGH3.11, CcGH3.13, CcGH3.15 and CcGH3.16 in group III. The OsGH3.7 protein did not cluster with any other sequence, which made it difficult to classify in one of the previous groups. Three sister groups are formed by only CcGH3s and four C. canephora GH3 proteins formed sister groups with proteins from other species, which are CcGH3.2-CcGH3.5, CcGH3.1-CcGH3.7, CcGH3.11-CcGH3.16, CcGH3.12-AtGH3.11, CcGH3.14-AtGH3.10 and CcGH3.9-AtGH3.9.
After grouping sequences through phylogenetic relationships, the multiple alignments between all CcGH3 putative proteins were used to search for conserved patterns. Firstly, we searched for sets of amino acid sequences that could be related to acyl substrate specificity as described in literature [4] and afterwards for the sets “F(V/I/T)K” and “DKT”, commonly present in GH3 proteins that conjugate acidic amino acids to auxins [11]. These sequences were found only in CcGH3.9, CcGH3.13, CcGH3.15 and CcGH3.16 (Fig. 3) and such sequences were selected to perform a structural analysis on SWISS-MODEL software [22]. For CcGH3.9, CcGH3.13 and CcGH3.15, the models were constructed based on the crystal structure of GH3.5 of A. thaliana [17] and the identities were 55.21, 75.21 and 81.83%, respectively. For CcGH3.16, the best fitted model was based on the crystal structure of a GH3 protein from Vitis vinifera [12] with 80.76% identity. In addition to some differences among the four models (Additional file 4: Figure S2), only CcGH3.13 and CcGH3.15 presented ligands like adenosine monophosphate (AMP) and 1H-indol-3-Yacetic acid (IAC) in its tridimensional structure, as further represented for CcGH3.13 (Fig. 4).
Alignment view of the CcGH3 proteins sections containing the conserved amino acid residues involved in auxin conjugation by acidic amino acids. a Alignment of five amino acid residue sets (numbered from 1 to 5) related to acyl acid-binding specificity. The yellow shade represents key residue positions for specificity and residues written in red match with patterns for auxin binding; b Set of two amino acid sequences related to amino acid-binding specificity in which red shade represents the pattern for acidic amino acid binding and blue shade represents nonpolar amino acid binding
Transcription factors network approach
An illustrative network between target GH3 genes and their possible transcriptional regulators was constructed based on data from PlantTFDB [23] about motifs in the GH3 promoter regions, and named as transcription factors network (Sequences used for constructing the network can be found in the Additional file 2: Data S2). Some motifs are overrepresented in a given promoter and even among GH3 genes (Fig. 5, Additional file 6: Table S1 and Additional file 7: Table S1 Appendix). The transcriptional regulator with more binding possibilities is the Cc10_g07850, a gene from TALE transcription factor family. This gene has 29 binding sites in the CcGH3.15 promoter and 2 binding sites in both CcGH3.9 and CcGH3.16. Two genes in C. canephora genome, Cc02_g03700 and Cc07_g05550, have binding sites in the promoter region of all the tested GH3 genes and they are members of the MIKC-MADS and Dof transcription factor families, respectively.
Motif-binding network for selected CcGH3s genes (red ellipses) and their related transcription factors (rectangles). The color scale from white to black refers to the number of CcGH3 genes (one to four) in which a specific transcription factor can bind. Arrow width refers to the number of binding sites for one transcription factor at the promoter region of some GH3 gene (Additional file 6: Table S1)
The number of motifs found in CcGH3.9, CcGH3.13, CcGH3.15 and CcGH3.16 promoters were 21, 41, 124 and 64, and through these motifs 17, 38, 22 and 48 different transcription factors can bind, respectively. These motifs are specific for genes from 24 different transcription factor families and the ERF family is the most overrepresented. Some families have motifs specific for one out of the GH3 genes like ARF and SBP for CcGH3.15, HSF and G2-like for CcGH3.16 and HD-ZIP for CcGH3.13.
Histology analysis of C. arabica somatic cells with different embryogenic potential and GH3 gene expression patterns
Embryogenic, non-embryogenic calli and cell suspension with embryogenic potential were sampled for histological and gene expression analysis. The diameter of the different cell types varied between non-embryogenic and embryogenic calli, while the cell suspension with embryogenic potential presented no pattern for cellular length. The cytoplasmic density checked by toluidine blue staining varied regarding the cell types (Fig. 6), in which isodiametric cells were stained more.
For RT-qPCR analysis, the integrity and quality of extracted RNAs were analyzed by electrophoresis and spectrophotometry before its conversion to cDNA. Only samples with suitable characteristics were selected for expression experiments (Additional file 5: Figure S3, Additional file 8: Table S2). All the primers used in RT-qPCR experiments were previously checked for their amplification efficiency with these same samples [18] (Additional file 9: Table S3). A set of candidate reference genes were tested by their stability among the treatment conditions (Additional file 10: Table S4 and Additoal file 11: S4 appendix) and Ca24S and CaRPL39 were established as the most suitable reference genes.
For gene expression analysis, the correspondent genes to CcGH3.9, CcGH3.13, CcGH3.15 and CcGH3.16 in C. arabica (CaGH3.9, CaGH3.13, CaGH3.15 and CaGH3.16, respectively) were selected (Fig. 7), based on previous results demonstrating their possible involvement in acidic amino acid conjugation to auxin. CaGH3.9, CaGH3.13 and CaGH3.15 did not present any expression in non-embryogenic cells (NEC). CaGH3.9, CaGH3.13 and CaGH3.16 presented higher expression in embryogenic cell suspension (ECS), while CaGH3.15 had more transcript quantity in embryogenic cells (EC). The CaGH3.16 gene was the only that exhibited expression in all the cell types.
Discussion
The C. canephora GH3 putative genes identified in our work have all the conserved domains commonly found in members of this family. Such domains are required for its proper functionality and the number of putative GH3 members found herein is close to other dicotyledonous species like Malus domestica [15], Medicago truncatula [16] and Solanum lycopersicum [14]. However, C. canephora did not go through any polyploydization event after core eudicots diversification, unlike S. lycopersicum, which belongs to the same C. canephora class (asterid) [21]. Therefore, it seems some CcGH3s could have been originated from local duplications.
This hypothesis is interesting upon analysis of C. canephora GH3 genes containing similar structures, like the tetrad CcGH3.2, CcGH3.4, CcGH3.5 and CcGH3.16 and the pair CcGH3.8 and CcGH3.6 (Fig. 1). Except for CcGH3.16, the other genes are clustered in a group exclusively constituted by C. canephora GH3 proteins in the phylogenetic tree, constructed with GH3 protein sequences of A. thaliana, O. sativa and Z. mays (Fig. 2). These proteins from C. canephora are the closest in sequence similarity to those from A. thaliana apparently local duplicated in this species [13]. Further detailed syntenic studies may confirm such hypothesis and help to understand if there is a specific function evolved in C. canephora for these members of GH3 family.
Studies supported on the gene family wide analysis approach have been broadly performed recently [24,25,26,27]. These studies have been also applied to unravel GH3 gene family members characteristics in a wide perspective, usually with genic and protein structure description, gene expression patterns along plant tissues [28] or analyzed in a specific process [29]. Here, we speculate if some genes of GH3 family may influence the somatic embryogenesis in coffee tree, specifically the possible correlation with embryogenic potential of different types of calli, which is the key to understand indirect somatic embryogenesis process.
Group III from the phylogenetic tree has the most widely studied members and all the A. thaliana proteins clustered are associated to amino acid conjugation to auxin, accordingly to transcriptional activation, enzyme activity or mutant phenotype assays [13]. The involvement of some CcGH3 homologs in the conjugation of auxins to amino acids can be analyzed through reports in the literature such as for the members AtGH3.2, AtGH3.3, AtGH3.4, AtGH3.5 and AtGH3.6 [30] and AtGH3.9 [31]. These works have suggested that in the presence of GH3 family members auxins can be conjugated to different amino acids using specific approaches to analyze the conjugation product. Furthermore, studies on the 3D molecular structure of AtGH3.5 followed by in vitro and in planta biochemical analyses suggest the ability of this protein in conjugating auxins to amino acids and mediate their homeostasis [17]. This reinforces the importance of investigating some CcGH3 members clustered together with these well-studied AtGH3 proteins and to link the role of conjugating auxin with the coffee somatic embryogenesis.
Amino acid sequence alignment with functional characterized proteins revealed a correlation between substrate specificity and conserved sequence patterns [11, 17]. It allowed us to choose four CcGH3s candidates likely involved in acidic amino acid conjugation to auxin, such as the members CcGH3.9, CcGH3.13, CcGH3.15 and CcGH3.16. Although just CcGH3.13 has all conserved residues for both auxin and acidic amino acid binding sites, the CcGH3.9 and CcCGH3.15 proteins have sufficient potential to be further analyzed as well, as the present mismatches do not change the amino acid classes and, also, we decided analyze CcGH3.16, besides the absence of amino acids residues in positions of β8-β9 (Fig. 3).
The tridimensional models for the four CcGH3s proteins revealed that only CcGH3.13 and CcGH3.15 have AMP and IAC ligands (Fig. 4, Additional file 4: Figure S2). The binding of IAC ligand represents the affinity for auxin as a substrate for this enzyme and the interaction is mediated by the highlighted amino acid residues sites of α5, α6 and β8-β9 (Fig. 3a) [17]. In fact, the IAC ligand interaction in tridimensional model is supported by the presence of the correct set of amino acid residues in the interaction sites, by comparison with GH3 proteins that are already confirmed as having auxin affinity [17]. This suggests the potential of these proteins to influence auxin homeostasis dependent process in the plant.
To further speculate on whether the selected GH3 members have a possible link to cell embryogenic competence, we identified the binding sites of transcription factors in promoter regions of these four CcGH3s genes and with this, a transcription factor network was constructed aiming to analyze the transcription factor binding possibilities in each gene. The network was constructed based on information taken from Coffee Genome Hub and PlantTFDB databases [21, 23] to support the transcription factor possible connections for each selected CcGH3 gene. Genes from 24 different transcription factor families were identified as able to bind in the GH3 promoters. Out of 24, at least 15 families have members with already known influence on somatic embryogenesis process [9, 32, 33].
The Ethylene Responsive Factor (ERF) family, the most overrepresented transcription factor family in the network with 9 genes, has members already reported to influence on somatic embryogenesis [9], like SERF in Medicago truncatula and ERF022 in A. thaliana. Furthermore, a decrease in ethylene concentration was observed in calli from a maize lineage with high embryogenic potential, likely regulated by an ERF gene, which did not occur with the low embryogenic potential lineage [34].
Another interesting feature in the network is that some genes contain multiple binding sites at certain promoters, what could influence still more upon its probability of regulating specific gene expression, like TALE-1 (Cc10_g07850), that have 29 predicted binding sites in CcGH3.15 and just two in CcGH3.9 and CcGH3.16 promoters. This transcription factor belongs to TALE family, subfamily KNOX, which is reported to have members involved in the shoot apical meristem maintenance [35, 36]. Although we cannot speculate at this moment if this transcription factor is an activator or repressor of CcGH3.15, a regulatory influence is reinforced by the number of binding sites in the CcGH3.15 promoter, which may be associated to meristem-like features of embryogenic cells. Furthermore, in japonica rice subspecies, the downregulation of a KNOX-type transcription factor improved the regeneration and development of somatic embryos, highlighting the additional role of such subfamily on the somatic embryogenesis process [37].
Taking into account the differences in regulation profile of each CcGH3 proposed by the network, we performed an RT-qPCR experiment to analyze the expression pattern of these genes in C. arabica cell types contrasting in its embryogenic potential. The cell types used are clearly different; those with embryogenic potential are isodiametric, have larger nucleus and high cytoplasmic density, which are the expected characteristics for embryogenic coffee cells [19, 38, 39]; whereas the non-embryogenic ones are larger, with undefined shape and low cytoplasmic density [40]. The embryogenic cell suspension is composed by these two cell types, an interesting material for correlating embryogenic potential to gene expression pattern.
It is reported that AtGH3.17 protein inhibits hypocotyl cell elongation by means of auxin conjugation with glutamate [10], indirectly influencing over hypocotyl cell expansion through H+- ATPase proton efflux [41]. Accordingly, we speculate that the embryogenic and non-embryogenic cell shapes presented in histological analysis could be a consequence of GH3-mediated auxin conjugation, what would reinforce the influence of this protein over coffee early somatic embryogenesis process.
Although we selected just GH3 genes similar in amino acid sequence level, they exhibited a different expression pattern among the samples, possibly due to different roles performed by these enzymes in metabolic pathways and their requirements within the cellular environment. Even being descendent from a common ancestral, paralog genes tend to perform distinctive functions in a given organism. It is consistent with the patterns of gene family evolution, which is generally owing to duplications (either local or derived from whole genome duplication), neo/subfunctionalization and retention according or not to environment selective pressure [42].
CaGH3.9 and CaGH3.13 had similar expression patterns among cell types (ρ = 0,9393, correlation by Spearman method, Additional file 12: Table S5), but CaGH3.13 does not share many transcription factor binding sites with CaGH3.9 like it shares with CaGH3.16 (Fig. 5). The predicted transcription factor binding sites are condition-independent, thus not all of them necessarily regulate the GH3 genes at certain tissue or timing. We speculate that CaGH3.9 and CaGH3.13 expression pattern similarities in our embryogenic cell types may be owing to shared binding sites, even if a greater number of unshared motifs can influence transcriptional regulation in other conditions. The common binding sites of these two genes are for the transcription factors Cc11_g17110 and Cc10_g13750, both belonging to MIKC-MADS family, which has already characterized members related to many regulatory functions in plant kingdom [43], including some that influence over somatic embryogenesis [44].
CaGH3.15 gene has the most correlated expression pattern with embryogenic potential of cell types tested: high transcript levels in embryogenic cell, average values in embryogenic cell suspension (which is composed by both embryogenic and non-embryogenic cells) and lower expression in non-embryogenic cells. This gene also exhibited the most evident relationships with some regulators in the network, due to high number of binding sites for a specific transcription factor. While other GH3 genes have in maximum four binding sites for a given transcription factor, CcGH3.15 have 29, 20, 15, 14, 14 and 13 binding sites for the transcription factors TALE-1, BBR-BPC-2, MIKC-MADS-2, BBR-BPC-1, AP2–1 and C2H2–8, respectively.
The protein families TALE, MIKC-MADS, AP2 and C2H2 have well characterized TFs involved in somatic embryogenesis while BBR-BPC, despite being a poorly explored TF family in plants, has a member reported to control floral meristem size and seed development by regulating other transcription factors like LEC2 and WUSCHEL [20, 45]. These last two genes have been considered key regulators of somatic embryogenesis [9], thus BBR-BPC genes might also be involved in SE process.
The Cc09_g04020 gene belongs to AP2 family and is the most suitable transcription factor that can explain the CaGH3.15 expression pattern in cells with embryogenic potential. This C. canephora gene is the most similar to the previously characterized BBM gene from C. arabica, which was already studied during embryogenic process [18, 19]. Recently, this gene was reported to be the major regulator of somatic embryogenesis, being associated to cellular totipotency induction through the control of LAFL (LEC1, ABI3, FUS3, LEC2) gene network [46].
We reinforced this regulation hypothesis for the CcGH3.15 by comparing through the Spearman’s correlation method the gene expression data of CaGH3s members with previously published data from our group for the CaBBM [18] gene. A strong correlation value was found (ρ = 0.971) only for CaGH3.15 while the maximum correlation value observed for the other GH3 genes was ρ = 0,50, meaning that only CaGH3.15 and CaBBM are co-expressed in these conditions (Fig. 8).
Expression profile of CaBBM and its regulatory relationships with CaGH3.9, CaGH3.13, CaGH3.15 and CaGH3.16. a Expression pattern of CaBBM in non-embryogenic cells (NEC), embryogenic cells (EC) and embryogenic cell suspension (ECS) measured by RT-qPCR; b Co-expression analysis of CaBBM gene with CaGH3.9, CaGH3.13, CaGH3.15 and CaGH3.16, inferred by Spearman’s method; dark blue means significant positive Spearman’s correlation, light blue non-significant and light red non-significant negative Spearman’s correlation
We suggest that the CaGH3.15, one of the members of GH3 gene family in C. arabica, could be regulated by CaBBM, and it possibly influences auxin homeostasis during somatic embryogenesis in coffee. This is consistent with the higher IAA-Glu conjugate concentration in comparison to free auxin and IAA-Ala detected during somatic embryogenesis induction in C. canephora, which reflects the requirement of decreasing auxin content during somatic embryogenesis process [47].
In C. canephora, the somatic embryogenesis happens directly in the explant, without an embryogenic calli formation step. The auxin content decreases during the induction phase, what may also be due to its transport to explant extremities, where the embryos need this phytohormone supply to develop [48]. The process in C. arabica is through indirect embryogenesis, with the need of embryogenic calli induction before somatic embryo regeneration, and two types of auxins were included in the medium [40]. Embryogenic and non-embryogenic calli derive from the same explant (leaf cuts, Fig. 6b), but those without embryogenic competence appear earlier, sometimes with more than two months of difference. Therefore, auxin homeostasis through amino acid conjugation, which we suggest that can be an effect of CaGH3.15 upregulation demonstrated in this work, might be a differentiation factor between these two types of calli and necessary for embryogenic competence achievement of the coffee cells.
Somatic embryogenesis is a bottleneck process for C. arabica clonal propagation and genetic transformation. Although many breakthroughs were achieved recently to scale this technology to commercial level, the improvements are mostly due to empirical research and key points of process yet remain unclear, with the need of wider cellular and molecular comprehension [49]. Here, we identified a GH3 gene member potentially associated to early somatic embryogenesis process in C. arabica, which may have influence on auxin degradation, due to conjugation with acidic amino acids. Further analysis needs to be performed to unravel the many aspects related to SE at cellular and molecular level, to deepen the understanding of this process and establishing an efficient clonal propagation protocol based on totipotent cell regeneration, which is essential in the pursuit of this crop ideotypes in genome editing era [50].
Conclusions
Here we report that the GH3 family is conserved in C. canephora, with some of the members clustered by similarity at amino acid sequence level with proteins already known to be associated to auxin conjugation. Four of these coffee GH3 members have most of the conserved amino acid sets related to acidic amino acid conjugation to auxin. Besides structural properties, one out of all CcGH3 members, the CcGH3.15, have binding sites in its promoter region for some transcription factors that might be associated to embryogenic features.
The homolog of CcGH3.15 in C. arabica, named CaGH3.15, has the transcriptional profile most correlated with embryogenic competence, addressed here by using coffee cells with contrasting embryogenic potential, and correlation with the expression profile of CaBBM, the closest coffee homolog to BBM transcription factor, a major plant embryogenesis regulator. All these findings together indicates that GH3 genes may have influence on the internal fine tuning of auxin content in the cell, necessary for totipotency acquirement and maintenance in the embryogenic competent cells. As the coffee market mostly relies on this species cultivation, all knowledge and the available tools need to be explored to ensure a sustainable coffee production chain.
Methods
Identification, phylogenetic and structural analyses of GH3 members in C. canephora
The GH3 amino acid sequences of A. thaliana, Z. mays and O. sativa were used as query for blastp analysis against C. canephora proteome in Coffee Genome Hub Database19, with e-value threshold of 1.0 × 10− 5. The resulting sequences were then submitted to NCBI Conserved Domains Database (NCBI-CDD) aiming to verify the presence of conserved domains presented in proteins of this family, PLN02247 and pfam03321. The sequences that satisfied this requisite were considered as putative GH3 members.
The genomic sequences of these putative GH3 members were submitted to MEGA 7.0 software [51] to perform an alignment by ClustalW algorithm [52] and phylogenetic analyses by Neighbor-joining [53] and p-distance [54] methods, with 1000 bootstrap replications. The resulting file with phylogenetic relationships, together with coding and genomic sequences of GH3 genes were used to design a graphical representation of gene structure with phylogenetic relationships in Gene Structure Display Server (GSDS 2.0, http://gsds.cbi.pku.edu.cn/) [55]. A second phylogenetic tree, constructed with same software and by same methods, was based on amino acid sequences of putative C. canephora GH3s and those from A. thaliana, Z. mays and O. sativa, aiming to speculate CcGH3s phylogenetic relationship. The proteins AtGH3.20 and CcGH3.10 were excluded from the phylogenetic tree because the pairwise distances of some pairs that included these proteins were not calculated by the program (MEGA7) with the parameter we have set.
The putative C. canephora proteins were also aligned for analysis of conserved sets of amino acids related to substrate specificity [11]. Finally, the four selected putative CcGH3 proteins were submitted to Swiss-model software (http://swissmodel.expasy.org/) [56], in which CcGH3.9, CcGH3.13 and CcGH3.15 tridimensional structures were modeled based on GH3.5 of A. thaliana (5kod.2.A template) and CcGH3.16 based on GH3 protein from Vitis vinifera, all built with ProMod3 version 1.1.0.
Motif-bind network construction
The upstream regions (3 kb before ATG) from C. canephora GH3 genes CcGH3.9, CcGH3.13, CcGH3.15 and CcGH3.16 were downloaded from Coffee Genome Hub database (CGH) and were submitted to regulation prediction on Plant Transcription Factor Database 4.0 (http://planttfdb.cbi.pku.edu.cn/) [23] to predict which transcription factors can bind to each GH3 promoter. The detected genes were then used as source nodes, the GH3 genes as target nodes and the number of motifs found in each GH3 promoter for each gene was used as a score to construct a network. With the locus id of each detected gene, their amino-acid sequences were downloaded from CGH, submitted to TF prediction server and this classification in transcription factor family was used for the network enrichment. Finally, the transcription factors were also discriminated by the number of possible interactions with CcGH3s to enrich the network.
Achievement and histology of embryogenic, non-embryogenic and suspension cells
Leaves of greenhouse grown C. arabica cv. Catuaí Amarelo IAC 62 plants were used as explants for embryogenic, non-embryogenic calli and embryogenic cell suspension achievement, accordingly to an already established protocol [57]. The embryogenic cell suspension was cultivated in Erlenmeyer flasks with T3 medium [58], with initial density of 10 g L− 1 in absence of light at 25 °C and 100 rpm.
Samples of the plant materials were fixed with FAA70 (10% formaldehyde + 5% acetic acid + 70% ethanol, v/v) for 48 h at room temperature, dehydrated in a graded series of 60, 70, 80, 90 and 100% ethanol. Then the samples were embedded in epoxy resin (Historesin® Leica) according to manufacturer’s protocol. The blocks were sectioned into 2 μm slices using manual rotary microtome (Easypath EP-31-20,091), stained with 0.05% toluidine blue and analyzed with a light microscope (Zeiss, Axio Scope). Images were analyzed using AxioVision 4.8 capture system.
RT-qPCR analysis of putative GH3 genes and correlation with CaBBM
Total RNA was extracted from non-embryogenic calli, embryogenic calli and embryogenic cell suspension (3 months) using Invitrogen (Life Technologies, Carlsbad, CA, USA) Concert™ Plant RNA reagent. RNA extracts were treated with Ambion (Life Technologies) Turbo DNA-free kit reagents in order to remove any genomic DNA. The quantity and purity of total RNA was assessed with an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, NC, USA). The synthesis of cDNA from 1,000 ng aliquots of RNA was carried out using Applied Biosystems (Life Technologies) High-Capacity cDNA Reverse Transcription kit according to the recommendations of the company.
RT-qPCR analyses were performed using an Applied Biosystems 7500 Real-Time PCR system with a reaction mix containing 5.0 μL of SYBR® Green PCR Master Mix (Applied Biosystems), 2 ng of cDNA, 0.4 μM of primers and RNase-free water to a total volume of 10 μL. Amplification conditions were initial activation at 95 °C for 10 min and 40 cycles of denaturation at 95 °C for 15 s and combined annealing and extension at 60 °C for 1 min. The primers design were suitable to RT-qPCR technique and the amplification of both homeologs in C. arabica genome, derived from C. canephora and C. eugenioides, was assured through analysis of the sequences relative to CcGH3 genes in Phytozome database. (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Carabica_er) using blastn tool and analysis of multiple alignments performed by ClustalW [52] algorithm and visualized by GeneDoc software [59]. Expression data were normalized with reference genes 24S and RPL39 and relatively quantified by applying Pfaffl method [60]. Shapiro-Wilk test was performed to verify the normality of data distribution. Data that did not fit into test’s prerequisites were transformed with the method [log10(x + 0.5)]. Statistical data analysis was performed by two-way ANOVA upon three biological replicates (n = 3) per sample followed by a comparison among gene expressions for each GH3 gene member at the cell types using the Tukey’s test at 5% significance (P < 0.05).
The co-expression analysis was made by using the expression profile of CaGH3.9, CaGH3.13, CaGH3.15, CaGH3.16 genes together with CaBBM [18], with all the data obtained in the same samples, with the same reference genes and equipment. The correlation values were generated by Spearman’s correlation analysis using R software.
Availability of data and materials
All amino acid sequences used into the network analysis are available on the Supplementary data 2. Embryogenic and non-embryogenic calli herein used were obtained under in vitro culture conditions previously described in the materials and methods. The primer sequences for amplification of the GH3 family genes are also available at the Supplementary files. In case of any information required for reproduction of the experimental data is not present elsewhere within publication content, please directly contact the corresponding authors on reasonable request.
Abbreviations
- AMP:
-
Adenosine monophosphate
- ANOVA:
-
Analysis of Variance
- AP2:
-
Transcription factor with Apetala2 domain
- ARF:
-
Auxin response factor
- BBM:
-
Baby Boom
- BBR-BPC:
-
BBR-Basic pentacysteine
- Blastp:
-
Basic Local Alignment Search Tool (proteins)
- C2H2:
-
Trancription factor encoding C2H2 zinc finger domains
- CDD:
-
Conserved domain database
- cDNA:
-
Complement DNA
- CGH:
-
Coffee Genome Hub
- Dof:
-
DNA binding with one finger
- EC:
-
Embryogenic cells
- ECS:
-
Embryogenic cell suspension
- ERF:
-
Ethylene Responsive Factor
- FAA:
-
Formaldehyde,Acetic acid, Alcohol (ethanol)
- G2-like:
-
Golden 2-like
- GH3:
-
Gretchen Hagen 3
- GSDS:
-
Gene Structure Display Server
- HD-ZIP:
-
Homeodomain and leucine zipper containing transcription factors
- HSF:
-
Heat stress transcription factors
- IAA:
-
Indole-3-acetic acid
- IAC:
-
1H-indol-3-Yacetic acid
- KNOX:
-
Knotted-like domain containing transcription factors
- LAFL:
-
(LEC1, ABI3, FUS3, LEC2)
- LEC2:
-
LEAFY COTYLEDON 2
- MIKC-MADS:
-
MIKC-type MADS box
- NCBI:
-
National Center for Biotechnology Information
- NEC:
-
Non-embryogenic cells
- PlantTFDB:
-
Plant Transcription Factor Database
- RT-qPCR:
-
Quantitative reverse transcription Polymerase Chain Reaction
- SBP:
-
Squamosa promoter binding proteins
- SD:
-
Standard deviation
- SE:
-
Somatic embryogenesis
- TALE:
-
Transcription factor containing from theThree-amino-acid-loop-extension class
- TF:
-
Transcription factor
References
International Coffee Organization: Total production by all exporting countries. http://www.ico.org/prices/po-production.pdf (2018). Accessed 25 Feb 2019.
Doepker C, Lieberman HR, Smith AP, Peck JD, El-Sohemy A, Welsh BT. Caffeine: friend or foe? Ann Rev Food Sci T. 2016;7:117–37. https://doi.org/10.1146/annurev-food-041715-033243.
Koehnlein EA, Bracht A, Nishida VS, Peralta RM. Total antioxidant capacity and phenolic content of the Brazilian diet: a real scenario. Int J Food Sci Nutr. 2014;65:293–8. https://doi.org/10.3109/09637486.2013.879285.
Georget F, Courtel P, Garcia EM, Hidalgo M, Alpizar E, Breitler JC, Bertrand B, Etienne H. Somatic embryogenesis-derived coffee plantlets can be efficiently propagated by horticultural rooted mini-cuttings: a boost for somatic embryogenesis. Sci Hort. 2017;216:177–85. https://doi.org/10.1016/j.scienta.2016.12.017.
Etienne H, Bertrand B, Dechamp E, Maurel P, Geroget F, Guyot R, Breitler JC. Are genetics and epigenetic instabilities of plant embryogenic cells a fatality? The experience of coffee somatic embryogenesis. Human Genet Embryol. 2016;6:136. https://doi.org/10.4172/2161-0436.1000136.
Campos NA, Panis B, Carpentier SC. Somatic embryogenesis in coffee: the evolution of biotechnology and the integration of omics technologies offer great opportunities. Front Plant Sci. 2017;8:1–12. https://doi.org/10.3389/fpls.2017.01460.
Ikeuchi M, Favero DS, Sakamoto Y, Iwase A, Coleman D, Rymen B, Sugimoto K. Molecular mechanisms of plant regeneration. Annu Rev Plant Biol. 2019;70:377–406. https://doi.org/10.1146/annurev-arplant-050718-100434.
Shin J, Seo PJ. Varying Auxin levels induce distinct pluripotent states in callus cells. Front Plant Sci. 2018;9:1653. https://doi.org/10.3389/fpls.2018.01653.
Nowak K, Gaj MD. Transcription factors in the regulation of somatic embryogenesis. In: Loyola-Vargas VM, Ochoa-Alejo N, editors. Somatic embryogenesis: fundamental aspects and applications. New York: Springer; 2016. p. 53–79.
Zheng Z, et al. Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat Plants. 2016;2:1–19. https://doi.org/10.1038/nplants.2016.25.
Westfall CS, et al. Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science. 2012;336:1708–11. https://doi.org/10.1126/science.1221863.
Peat TS, et al. Crystal structure of an indole-3-acetic acid amido synthetase from grapevine involved in auxin homeostasis. Plant Cell. 2012;24:4525–38. https://doi.org/10.1105/tpc.112.102921.
Okrent RA, Wildermuth MC. Evolutionary history of the GH3 family of acyl adenylases in rosids. Plant Mol. Biol. 2011;76:489–505. https://doi.org/10.1007/s11103-011-9776-y.
Kumar R, Agarwal P, Tyagi AK, Sharma AK. Genome-wide investigation and expression analysis suggest diverse roles of auxin-responsive GH3 genes during development and response to different stimuli in tomato (Solanum lycopersicum). Mol. Genet. Genomics. 2012;287:221–35. https://doi.org/10.1007/s00438-011-0672-6.
Yuan H, et al. Genome-wide analysis of the GH3 family in apple (Malus × domestica). BMC Genomics. 2013;14:297. https://doi.org/10.1186/1471-2164-14-297.
Yang Y, et al. Genome-wide identification, expression analysis of GH3 family genes in Medicago truncatula under stress-related hormones and Sinorhizobium meliloti infection. Appl. Microbiol. Biot. 2015;99:841–54. https://doi.org/10.1007/s00253-014-6311-5.
Westfall CS, et al. Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. P. Natl. Acad. Sci. USA. 2016;113:13917–22. https://doi.org/10.1073/pnas.1612635113.
Freitas NC, et al. Validation of reference genes for qPCR analysis of Coffea arabica L. somatic embryogenesis-related tissues. Plant Cell Tiss. Org. 2017;128:663–78. https://doi.org/10.1007/s11240-016-1147-6.
Silva AT, Barduche D, Livramento KG, Ligterink W, Paiva LV. Characterization of a putative Serk-like ortholog in embryogenic cell suspension cultures of Coffea arabica L. Plant Mol Biol Rep. 2014;32:176–84. https://doi.org/10.1007/s11105-013-0632-x.
Berger N, Dubreucq B, Roudier F, Dubos C, Lepiniec L. Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant Cell. 2011;23:4065–78. https://doi.org/10.1105/tpc.111.087866.
Denoeud F, et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 2014;345:1181–4. https://doi.org/10.1126/science.1255274.
Biasini M, et al. SWISS-MODEL: modeling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–8. https://doi.org/10.1093/nar/gku340.
Jin J, et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45:D1040–5. https://doi.org/10.1093/nar/gkw982.
Liu J, et al. The Hsp70 gene family in Solanum tuberosum: genome-wide identification, phylogeny, and expression patterns. Sci Rep. 2018;8:1–11. https://doi.org/10.1038/s41598-018-34878-7.
Lu Y, et al. Genome-wide identification and expression analysis of glycine-rich RNA-binding protein family in sweet potato wild relative Ipomoea trifida. Gene. 2019;686:177–86. https://doi.org/10.1016/j.gene.2018.11.044.
Xu X, et al. Genome-wide identification and characterization of laccase gene family in Citrus sinensis. Gene. 2019;689:114–23. https://doi.org/10.1016/j.gene.2018.12.015.
Colina F, et al. Genome-wide identification and characterization of CKIN/SnRK gene family in Chlamydomonas reinhardtii. Sci Rep. 2019;9:1–16. https://doi.org/10.1038/s41598-018-35625-8.
Wang R, Li M, Wu X, Wang J. The gene structure and expression level changes of the GH3 gene family in Brassica napus relative to its diploid ancestors. Genes. 2019;10:1–25. https://doi.org/10.3390/genes10010058.
Bernales M, et al. Expression of two indole-3-acetic acid (IAA)-amido synthetase (GH3) genes during fruit development of raspberry (Rubus idaeus heritage). Sci Hortic. 2019;246:168–75. https://doi.org/10.1016/j.scienta.2018.09.077.
Staswick PE, et al. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17:616–27. https://doi.org/10.1105/tpc.104.026690.
Khan S, Stone J. Arabidopsis thaliana GH3.9 in auxin and jasmonate cross talk. Plant Signal Behav. 2007;2:483–7. https://doi.org/10.4161/psb.2.6.4498.
Wójcikowska B, Gaj MD. Expression profiling of AUXIN RESPONSE FACTOR genes during somatic embryogenesis induction in Arabidopsis. Plant Cell Rep. 2017;36:843–58. https://doi.org/10.1007/s00299-017-2114-3.
Cao A, et al. Comparative transcriptome analysis of SE initial dedifferentiation in cotton of different SE capability. Sci Rep. 2017;7:1–13. https://doi.org/10.1038/s41598-017-08763-8.
Ge F, et al. Metabolomic and proteomic analysis of maize embryonic callus induced from immature embryo. Sci Rep. 2017;7:1–16. https://doi.org/10.1038/s41598-017-01280-8.
Hay A, Tsiantis M. KNOX genes: versatile regulators of plant development and diversity. Development. 2010;137:3153–65. https://doi.org/10.1242/dev.030049.
Tsuda K, Hake S. Diverse functions of KNOX transcription factors in the diploid body plan of plants. Curr Opin Plant Biol. 2015;27:91–6. https://doi.org/10.1016/j.pbi.2015.06.015.
Indoliya Y, et al. Decoding regulatory landscape of somatic embryogenesis reveals differential regulatory networks between japonica and indica rice subspecies. Sci Rep. 2016;6:1–15. https://doi.org/10.1038/srep23050.
Ribas AF, et al. Agrobacterium-mediated genetic transformation of Coffea arabica (L.) is greatly enhanced by using established embryogenic callus cultures. BMC Plant Biol. 2011;11:1–15. https://doi.org/10.1186/1471-2229-11-92.
Pádua MS, et al. Morphological characteristics and cell viability of coffee plants calli. Cienc Rural. 2014;44:660–5. https://doi.org/10.1590/S0103-84782014000400014.
Freitas NC, Barreto HG, Torres LF, Freire LL, Rodrigues LA, Diniz LE, Beijo LA, Paiva LV. In silico and in vivo analysis of ABI3 and VAL2 genes during somatic embryogenesis of Coffea arabica: competence acquisition and developmental marker genes. Plant Cell Tissue Organ Culture (PCTOC). 2019;137(3):599–611. https://doi.org/10.1007/s11240-019-01594-7.
Spartz AK, et al. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabdopsis. Plant Cell. 2014;26:2129–42. https://doi.org/10.1105/tpc.114.126037.
Rensing SA. Gene duplication as a driver of plant morphogenetic evolution. Curr Opin Plant Biol. 2014;17:43–8. https://doi.org/10.1016/j.pbi.2013.11.002.
Zhao T, et al. Phylogenomic synteny network analysis of MADS-box transcription factor genes reveals lineage-specific transpositions, ancient tandem duplications, and deep positional conservation. Plant Cell. 2017;29:1278–92. https://doi.org/10.1105/tpc.17.00312.
Zheng Q, Perry SE. Alterations in the transcriptome of soybean in response to enhanced somatic embryogenesis promoted by orthologs of AGAMOUS-Like15 and AGAMOUS-Like18. Plant Physiol. 2014;164:1365–77. https://doi.org/10.1104/pp.113.234062.
Simonini S, Kater MM. Class I BASIC PENTACYSTEINE factors regulate HOMEOBOX genes involved in meristem size maintenance. J Exp Bot. 2014;65:1455–65. https://doi.org/10.1093/jxb/eru003.
Horstman A, et al. The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 2017;175:848–57. https://doi.org/10.1104/pp.17.00232.
Ayil-Gutiérrez B, Galaz-Ávalos RM, Peña-Cabrera E, Loyola-Vargas VM. Dynamics of the concentration of IAA and some of its conjugates during the induction of somatic embryogenesis in Coffea canephora. Plant Signal. Behav. 2013;8:e26998–1-10. https://doi.org/10.4161/psb.26998.
Márquez-López RE, Pérez-Hernández C, Ku-González Á, Galaz-Ávalos RM, Loyola-Vargas VM. Localization and transport of indole-3-acetic acid during somatic embryogenesis in Coffea canephora. Protoplasma. 2018;255:695–708. https://doi.org/10.1007/s00709-017-1181-1.
Etienne H, et al. Coffee somatic embryogenesis: how did research, experience gained and innovations promote the commercial propagation of elite clones from the two cultivated species? Front Plant Sci. 2018;9:1–21. https://doi.org/10.3389/fpls.2018.01630.
Breitler JC, et al. CRISPR/Cas9-mediated efficient targeted mutagenesis has the potential to accelerate the domestication of Coffea canephora. Plant Cell Tiss Org. 2018;134:383–94. https://doi.org/10.1007/s11240-018-1429-2.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–4. https://doi.org/10.1093/molbev/msw054.
Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensivity of progressive multiple sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25 https://doi.org/10.1093/oxfordjournals.molbev.a040454.
Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford: Oxford University Press; 2000.
Hu B, et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–7. https://doi.org/10.1093/bioinformatics/btu817.
Bordoli L, et al. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. https://doi.org/10.1038/nprot.2008.197.
Teixeira JB, et al. Multiplicação clonal de café (Coffea arabica L.) via embriogênese somática. 2004. https://www.embrapa.br/busca-de-publicacoes/-/publicacao/185448/multiplicacao-clonal-de-cafe-coffea-ararabica-l-via-embriogencese-somatica. Accessed 20 Feb 2019.
Boxtel JV, Berthouly M. High frequency somatic embryogenesis from coffee leaves. Factors influencing embryogenesis and subsequent proliferation and regeneration in liquid medium. Plant cell Tiss. Org. 1996;44:7–17. https://doi.org/10.1007/BF00045907.
Nicholas KB, Nicholas HB: GeneDoc: a tool for editing and annotating multiple sequence alignments. http://www.psc.edu/biomed/genedoc (1997). Accessed 10 Jan 2019.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2011;29:2002–7. https://doi.org/10.1093/nar/29.9.e45.
Acknowledgements
We would like to thank the National Council for Scientific and Technological Development (CNPq) from Ministry of Science and Technology, Brazil, and the Foundation of Support Research of the State of Minas Gerais (FAPEMIG) for the financial support provided to this study. The Coordination for the Improvement of Higher Education Personnel (CAPES) from Ministry of Education, for the grants awarded to the authors. We also thank the Federal University of Lavras (UFLA) for providing the infrastructure and equipment to accomplish this research, including real time PCR machine, computers and servers for bioinformatics analyses, which is supported by Ministry of Science and Technology and Ministry of Education.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
RTP performed the bioinformatics analysis, interpreted the results and wrote the manuscript; WPFM constructed RT-qPCR graphs, interpreted the results, revised the manuscript and formatted the manuscript; NCF conducted the in vitro culture of cell types and performed RT-qPCR experiments; TBC performed the bioinformatics analysis and revised the manuscript; DOP performed statistical analysis, reviewed and formatted the manuscript; LVP acquired the funding, supervised the entire study, and reviewed the manuscript. All authors significantly contributed to produce and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1:
Data S1. Amino acids sequences related to GH3 proteins in C. canephora available at Coffee Genome Hub website. Three (Cc00_g28970, Cc02_g39040 and Cc00_g04550) out of the twenty amino acid sequences lacked the domains commonly shared by GH3 proteins (PLN02247 and pfam03321); therefore these sequences were not selected for analysis.
Additional file 2:
Data S2. Complete set of amino acid sequences used into the network analysis.
Additional file 3: Figure S1.
Gene distribution of CcGH3 members on C. canephora chromosomes using the option “locus search” in the Coffe Genome Hub website. The colors within each chromosome indicate ancestral blocks corresponding to the 7 core eudicot chromosomes (DENOUED et al., 2014). Red lines stand for the position of each CcGH3 over chromosomes. The most CcGH3 members are not presented in any chromosome, so they were allocated in chromosome 0 (not available). The genes are distributed as follow into each chromosome (chr): chr1: CcGH3.9; chr2: CcGH3.10, CcGH3.11 and CcGH3.12; chr5: CcGH3.13, CcGH3.14 and CcGH3.15; chr7: CcGH3.16; chr10: CcGH3.17.
Additional file 4:
Figure S2 Tridimensional structure model for the four GH3 proteins selected in Coffea canephora.
Additional file 5:
Figure S3 RNA integrity assessment by agarose gel electrophoresis. This figure was cropped from the original picture to exhibit the RNAs extracted from three biological replicates referred to each cell type used for RT-qPCR analysis. The bands clearly separated and visible represent two rRNA subunits point to the good RNA quality. From the left to the right are the samples: Embryogenic cell suspension, biological replicates 1, 2 and 3; Embryogenic calli, biological replicates 1, 2 and 3; Non-embryogenic calli, biological replicates 1, 2 and 3.
Additional file 6:
Table S1 Number of binding possibilities for each transcription factor in selected GH3 promoter regions.
Additional file 7:
Table S1 Appendix. Description of binding possibilities for each transcription factor in selected GH3 genes promoter regions.
Additional file 8:
Table S2 Concentration and quality assessment of RNA samples used for RT-qPCR analysis.
Additional file 9:
Table S3 Primers used in RT-qPCR experiments.
Additional file 10:
: Table S4 Endogenous gene expression stability analyzed by RefFinder software (http://fulxie.0fees.us/).
Additional file 11:
Table S4 Appendix. Primer pairs used into analysis of expression stability for endogenous genes (Freitas et al. 2017).
Additional file 12:
Table S5 Gene expression profile correlations between CaGH3s and CaBBM. Scores were obtained by Spearman’s correlation.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Pinto, R.T., Freitas, N.C., Máximo, W.P.F. et al. Genome-wide analysis, transcription factor network approach and gene expression profile of GH3 genes over early somatic embryogenesis in Coffea spp. BMC Genomics 20, 812 (2019). https://doi.org/10.1186/s12864-019-6176-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-6176-1