Abstract
Background
Recent studies show that galling Hymenoptera and Diptera are able to synthesize the plant hormone indole-3-acetic acid (auxin) from tryptophan and that plant response to insect-produced auxin is implicated in gall formation. We examined the leaf transcriptome of galled and ungalled leaves of individuals of the Hawaiian endemic plant Metrosideros polymorpha (Myrtaceae) subject to infestation by psyllid (Hemiptera) gall-makers in the genus Trioza (Triozidae).
Results
Transcript libraries were sequenced using Illumina technology and the reads assembled de novo into contigs. Functional identification of contigs followed a two-step procedure, first identifying contigs by comparison to the completely sequenced genome of the related Eucalyptus, followed by identifying the equivalent Arabidopsis gene using a pre-computed mapping between Eucalyptus and Arabidopsis genes. This allowed us to use the rich functional annotation of the Arabidopsis genome to assess the transcriptional landscape of galling in Metrosideros. Comparing galled and ungalled leaves, we find a highly significant enrichment of expressed genes with a gene ontology (GO) annotation to auxin response in the former. One gene consistently expressed in all galled trees examined but not detected in any libraries from ungalled leaves was the Metrosideros version of SMALL AUXIN UPREGULATED (SAUR) 67 which appears to be a marker for leaf-galling in Metrosideros.
Conclusions
We conclude that an auxin response is involved in galling by Metrosideros psyllids. The possibility should therefore be considered that psyllids (like other insects examined) are able to synthesize auxin.
Similar content being viewed by others
Background
The biology of galls - what is known?
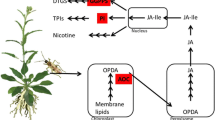
A gall is a plant structure resulting from the alteration of plant developmental processes by a galling organism, and which increases the fitness of the galler by providing a nutrient rich, protected environment [1]. It can be considered an extension of the phenotype of the galling organism [2]. Galls are caused by a variety of organisms but notably bacteria, nematodes, insects, mites and fungi. Most gallers are parasites, but a few, like the pollinating wasps of figs (Ficus) are beneficial to the host. Many of the parasitic galling organisms, such as the gall midge pests of cereals [3, 4], are of considerable economic importance as serious agricultural threats. Despite this importance remarkably little is known about the mechanisms by which the developmental processes of the plant host are perturbed by the parasite. However it has been established that at least some insect gallers directly synthesize the plant growth regulator auxin, specifically the major auxin indole-3-acetic acid (IAA) [5–7]. Auxin induces expression of two major classes of auxin-responsive genes: the GH3 family and the small auxin upregulated (SAUR) family [8]. A study of galling by the Asian rice gall midge (Rawat et al. 2012) found induction of both GH3 and SAUR genes [9].
A diverse array of insects form galls, particularly in the Hymenoptera, Diptera and Hemiptera [10]. In the Hemiptera, gall-making is particularly abundant in the aphids [11, 12], scale insects [13] and the psyllids [14–17]. The subject of this paper is a radiation of psyllid gall-makers on the common native Hawaiian tree, Metrosideros polymorpha.
Metrosideros in Hawaii – its morphology and its galls
Metrosideros is a tree and shrub genus of the eucalyptus and guava family, Myrtaceae, with around 50 species distributed across the Pacific region and a small radiation in the Hawaiian Islands [18–20]. One species, M. polymorpha (local name: ‘ōhi’ lehua) is an abundant, variable and ecologically important species in the Hawaiian Islands [21, 22]. It is host to a group of galling psyllids, which are often so abundant that they disfigure the leaves throughout the plant [23]. M. polymorpha has glabrous (hairless) biotypes as well as hairy-leaved biotypes and the hairy-leaved biotypes are noticeably less heavily galled than the hairless. No complete genome of Metrosideros has so far been released but good genomic resources are available for Eucalyptus [24–27], which, like Metrosideros, is in the family Myrtaceae.
The gall-makers – a radiation of species and gall types
Psyllids, or jumping plant lice, form the superfamily Psylloidea (Hemiptera, suborder Sternorhyncha). They are plant phloem feeders, usually highly host-specific and several are important agricultural pests such as the potato psyllid (Bactericera cockerelli) and the Asian citrus psyllid (Diaphorina citri) [28]. The immature stages may be free-living or may form galls of various types from simple pits to more elaborate structures. Because of the economic importance of some psyllids, genomic resources are emerging rapidly [29–32]. A number of endemic psyllids are present on the Hawaiian Islands [23, 33] including a radiation of the genus Trioza feeding exclusively on Metrosideros. [34]. These psyllid species include stem galling, leaf galling (flat and cone galls), pit galling and free-living types. A modern revision of Hawaiian psyllids, including descriptions of the new species referred to here, is currently being undertaken by one of us (DMP).
On the island of Hawai’i, two of the leaf gallers (producing flat and cone galls) are common and often gall the same individual leaves. In the early 1990s, a common garden was established on the island of Hawai’i to grow and study different morphotypes of M. polymorpha [35, 36], and over time these leaf galling psyllids have colonized many of the plants in the common garden. Both psyllids are native locally and can be found in adjacent forest areas. Cone and flat leaf gallers produce recognizably different gall phenotypes. These galls also dehisce by different mechanisms: flat leaf galls will usually dehisce on the underside of the leaf by irregular fissures, and cone leaf galls dehisce on the upper surface by a circular fissure that gives the appearance of a trap door (Fig. 1). A closely related species on the island of O’ahu, which also produces flat leaf galls, Trioza ohiacola, lays its eggs on the lower leaf surface. When the first instar nymph hatches and starts feeding, it is initially exposed on the surface of the leaf; the leaf tissue, which often shows a reddish discolouration at the site of feeding, then encloses the nymph and eclosion to the second instar occurs in a completely closed gall. All instars up to the last (fifth) instar remain in the closed gall (Fig. 1). A similar progression is likely to occur in both psyllid species studied here.
Metrosideros polymorpha. Figure 1. a View of plant. b-d Left to right, glabrous, intermediate and pubescent morphs. e Cone galls (inset: opening of cone gall on upper surface of leaf). f Flat galls (inset: opening of flat gall on lower leaf surface). g Psyllid nymphs (Trioza), above: first instar before gall formation; below: 5th instar removed from gall
In order to study the perturbation of leaf developmental processes by the leaf-galling species we analyse the transcriptome of galled and ungalled leaves.
Results
Insect contigs from galled and ungalled leaves
Contigs from plant 816 (leaf samples with and without galls) were mapped against a transcriptome of an adult Hawaiian psyllid (flat leaf galler, Trioza nov. sp. 1). As expected, numerous putative insect loci (9419 independent sequences) were retrieved from the galled sample. Fewer putative insect contigs were retrieved from the ungalled sample (671). Even samples that appear to be ungalled may have undetected eggs or young nymphs that have not yet formed visible galls, despite efforts to exclude psyllid presence from ungalled samples. The presence of insect contigs in ungalled samples may therefore reflect this undetected psyllid presence. The strong representation of insect contigs in leaf transcriptomes indicates the potential ease of co-analyzing insect and plant gene expression. When the insect contigs were blasted against the pea aphid genome a large number of putative insect gene orthologues were identified. The insect contigs are not considered further in this paper.
Presence/absence and differential expression of plant contigs from galled and ungalled leaf of the same plant and their Arabidopsis gene orthologues
The primary analysis used here is the comparison of galled and ungalled leaves in carefully matched samples of the same individual tree (816). Because galling causes a massive disruption to the phenotype of the leaf with implied disruption to physiology and development we were particularly interested in genes present under galling but not detectable in normal leaves, i.e. genes switched on de novo as a result of the galling trauma. We found 666 such Arabidopsis gene orthologues in the genotype 816 comparison (Fig. 2). This dataset we refer to as the tree 816 (galled only) dataset. We examined the tree 816 (galled only) dataset for auxin related genes and found a surprisingly large number of genes that are known to be expressed in response to exogenous application of auxin (Table 2), notably genes in the SAUR and GH3 gene families. We also found genes annotated to auxin response to be enriched in a GO analysis of the tree 816 (galled only) dataset (see below).
a Genes (Arabidopsis orthologues) present in galled and ungalled samples of Metrosideros genotype 816. 666 genes are detected in the galled sample but not in the ungalled. b Differential expression. Of the 12958 genes detected in both samples, 898 show substantially greater expression in the galled sample
In addition to the auxin responsive genes present only in the galled sample, there are some auxin response genes that, while present in the ungalled dataset, are significantly upregulated in the galled sample. These include 6 SAUR family genes (AT3G12955.1, AT1G14000.1, AT1G10210.1, AT4G36800.1, AT5G63310.1, AT3G61900.1) and genes involved in auxin signalling, including the auxin response transcription factor ETTIN (ARF3: AT2G33860.1) and VH1-INTERACTING KINASE (VIK) which is involved in auxin and brassinosteroid signalling.
Gall-faithful gene transcripts not present in ungalled leaves
The previous analyses (above) focused on genotype 816. Next, in order to test the generality of the results and to find consistent expression indicators of galling, we analysed the dataset for genes expressed in all of the diverse group of galled samples and absent in all of the ungalled samples (Table 1). Two genes were consistently recovered as transcripts from all individuals with galls yet undetectable in all samples without galls (Fig. 3). These gall-faithful and gall-exclusive genes are potentially interesting as putative markers of the galling response. One gene corresponds to one of the auxin responsive genes, SMALL AUXIN UPREGULATED 67 (SAUR67: AT1G29510), discussed above. It should be noted that SAUR67 (so named in TAIR version 10) has previously been referred to as SAUR68 [37]. Expression of several genes of the SAUR family appear to be associated with the galling response in Metrosideros but this member appears to be particularly consistent, not occurring in any ungalled leaves. The protein alignment of the Metrosideros sequence (GenBank: KT884616) with Eucalyptus (Eucgr.I01491) and Arabidopsis putative orthologs is shown in Fig. 4.
Venn diagram of genes present in all galled samples and genes present in at least one ungalled sample. Only two genes (Arabidopsis orthologues) have evidence of expression in all galled samples yet are absent in all ungalled samples. These genes are discussed in Results
The other gene corresponds to the Arabidopsis gene HARDY (HRD). HRD (AT2G36450) is of considerable interest as it encodes a drought resistance gene [38] as a member of the dehydration responsive element binding (DREB) subfamily A-4 of the ERF/AP2 transcription factor family [39]. Arabidopsis HARDY has three putative paralogues in Eucalyptus (Eucgr.K02071.1, Eucgr.K00220.1, Eucgr.A01537.1) consistent with the expansion of DREB genes in that genus [40]. The Metrosideros sequence has highest similarity to Eucgr.K02071.1.
Expression of this gene in galled leaves may possibly be indicative of reduced water potential. Several other drought adaptation genes of the late embryogenesis abundant (LEA) gene family are also found in the galled leaf (816.3) but not in ungalled (816.1): AT2G18340, AT5G06760, AT2G40170, AT1G52690. LEA proteins generally accumulate in response to reduced water availability. A study of galling of the invasive plant Parthenium hysterophorus by Epiblema strenuana (Lepidoptera: Tortricidae) showed a number of physiological effects leading to reduced water potential and drought stress [41].
Gene ontology over-representation analyses
For gene ontology enrichment analyses we used the Arabidopsis mappings, as the Arabidopsis genome has a rich functional annotation. For the genes expressed only in the galled leaf (tree 816), among the comprehensive set of Biological Process GO terms tested, 34 showed evidence of enrichment with a p-value <0.05. However only 7 of these survived false discovery rate (FDR) correction for multiple testing (Table 3). Of particular interest is “response to auxin stimulus” as auxin has been implicated in many other gall systems [5, 6]. The expression of a greater-than-expected number of auxin responsive genes under galling supports the conclusion that an auxin response is involved in the Metrosideros system.
In contrast, the genes present in the ungalled leaf but not in the galled leaf (222) had no significant over-representation of any biological process when FDR corrected. The genes present in both galled and ungalled leaves (12958) had a very large number of biological process GO categories significantly enriched even when corrected for multiple testing (242): as expected these were categories relating to photosynthesis and general leaf development and physiology.
To assess whether the over-representation of auxin response genes is general in galled leaves, we examined pairwise comparisons of all galled samples vs all ungalled samples using all genotypes (Table 4). Genes expressed in galled leaves but not in ungalled, were enriched in GO term “response to auxin” in seven of the pairwise comparisons. In this test we are only testing enrichment for auxin response genes so multiple-test correction is not strictly necessary. However, we note that in six of these seven cases the reslts would also be significant if all GO categories were tested. Importantly, by contrast, none of the ungalled samples showed enrichment for this GO term in any pairwise comparisons (Table 4). Although only 7 out of 12 pairwise comparisons showed significant GO enrichment for auxin respnse in galled leaves, it should be noted that all the galled samples express auxin response genes not found in ungalled leaves (e.g. SAUR67 above).
Discussion
An auxin response is associated with gall formation
The significant enrichment of auxin-response genes associated with gall-formation in this system is strongly indicative of phytohormone involvement in gall development. Our results show that expression of auxin response genes is general in all galled samples. This upregulation, in many samples, involves a particularly large number of auxin response genes, sufficient for a significant enrichment to be detected, in galled leaves only, in over half the pairwise comparisons between galled and ungalled leaves.
This is consistent with many previous studies, which have also shown auxin involvement [5, 6, 10, 12, 42] and auxin response gene expression [9]. By contrast we observe no obvious transcriptional activation or upregulation of auxin synthesis genes, which implies that the auxin may be exogenous, in which case it is presumably supplied by the immature insect within the gall. This accords with recent studies that have shown that diverse groups of insects can synthesize auxin [5, 7]. No study has yet determined whether psyllids synthesize auxin, but such a finding would not be surprising, and could be investigated. More puzzling is how very similar species of psyllids (such as the flat galler, Trioza nov. sp. 1, and the cone-galler, Trioza nov. sp. 2) produce such different types of gall. Not only are the gall shapes different but the galls dehisce by different mechanisms on different sides of the leaf. Auxin alone is unlikely to be the sole agent as it is difficult to see how differences in auxin production alone could produce such developmentally different outcomes in terms of gall morphology.
The potential for co-sequencing of insect and plant transcriptomes and cross species network analyses
One potential avenue for further progress is to examine the differences in gene expression in the immature insect over different gall developmental stages, looking for (for instance) differences in gene expression between cone-gallers and flat-gallers. This would be particularly powerful if done in the context of correlated changes in plant gene expression profiles.
We have shown how insect and plant transcriptomes can be co-sequenced in the same sample and the two RNA populations separated in silico. The main problem is the lack of a well-annotated hemipteran genome. The pea aphid (Acyrthosiphon pisum) is the closest completely sequenced genome [43] but annotation is still progressing. Very well-annotated genomes such as the Drosophila genome are too phylogenetically distant to be of much direct use.
By contrast it is possible, as we have shown here, to use the Eucalyptus genome to characterize the Metrosideros transcriptome; and Arabidopsis, which has the best annotated plant genome, is not too distant to be cross-mapped gene-by-gene with the Eucalyptus genome and Metrosideros transcriptome. Whole genome sequencing projects are currently in progress for Metrosideros and there is a good prospect for the release of a Metrosideros whole genome assembly in the near future. There are also whole genome sequencing projects for two psyllids: the potato psyllid (Bactericera cockerelli) and the Asian citrus psyllid (Diaphorina citri). Release of these genomes, particularly if they can be well cross-annotated with the pea aphid Acyrthosiphon pisum genome, may facilitate the elucidation of plant-insect co-expression networks during psyllid gall formation. What has been called “the power of paired genomes” [44] could potentially lead to the elucidation of a detailed plant-insect interactome and an understanding of how two separate developmental systems co-evolve.
The Hawaiian Metrosideros gall radiation as a system for gall biology
On the face of it, an endemic insect-plant system on remote islands in the Pacific may not seem a promising model for understanding a fundamental biological phenomenon. However, the Hawaian Metrosideros-feeding psyllids have some interesting features: they comprise a number of very closely related insects that have all evolved on the same host yet have come to interact with that host very differently. Some have reverted to a free-living lifestyle without galling, while the galling species variously produce stem-galls, flat galls and cone galls. They all manipulate the genome of the same host species to do so. It is therefore not only possible to discover how galling mechanisms work, but also how they evolve.
Methods
Sampling
Metrosideros leaf tissue was collected from five plants in a Metrosideros common garden situated near Volcano Village (N 19.475594, W −155.260161, 1265 m), on the island of Hawai’i (Table 1). The sampling strategy was two part. First: carefully matched leaf pairs (galled and ungalled) from the same individual plant (816) were sampled at the same time and height in the canopy (breast height: ~1.5 m above ground level). Secondly, to test the generality of the results, a further four individual plants were selected to represent a range of leaf morphologies (glabrous, intermediate, pubescent) and gall types (flat leaf and cone leaf galls) (Table 1). All samples were collected on the same day (11th March 2014) between 11 am and 3 pm. All leaves were selected from the 2nd or 3rd node below the terminal leaf bud. Leaf tissue was flash frozen in liquid nitrogen in the field and transported in a dry shipper to the University of British Columbia, Vancouver, for sequencing.
RNA extraction and sequencing
Leaf samples were stored at −80 C before RNA extraction. RNA was processed as previously described [45]. Briefly, RNA was extracted using Pure LinkTM Plant RNA Reagent (Invitrogen). RNA was quantitated using a QubitR 2.0 (Life Technologies) and quality-checked using an Agilent 2100 Bioanalyzer. Only samples with a RIN value of 7 or above were used for library construction. Sequencing libraries were prepared and barcoded using an Illumina Library Preparation kit. All seven libraries (Table 1) were sequenced in one lane of an Illumina Hi-Seq 2000 at the sequencing facility at the Biodiversity Research Centre, University of British Columbia (UBC), generating 100 bp paired end reads. Reads (Fastq files) for each sample are deposited in the European Nucleotide Archive (ENA).
Processing of reads, assembly of contigs and identification of contigs to genes
The raw reads were assembled to contigs in Trinity r20140717 [46] using recommended parameters. The contigs were blasted against the complete CDS library of Eucalyptus grandis downloaded from Phytozome [47] using blastn [48] and an expectation value of 1e-6. Only their top high scoring Eucalyptus hit was then used for further analysis of the contigs. A second blast search was performed, with similar parameters, against a previously assembled transcriptome of an adult Trioza nov. sp. 1 psyllid, assembled in Trinity (Michael Brewer pers. comm.) in order to identify the psyllid component present in the sequenced material.
Once contigs had been matched to Eucalyptus genes, they were identified as their putative Arabidopsis orthologs by means of a pre-computed mapping table as available from Phytozome version 9: http://phytozome.jgi.doe.gov [47]. This table was used to identify orthologs between the Eucalyptus grandis (version 1.0) gene annotation to the Arabidopsis thaliana (version 10) gene annotation. Lists of transcripts (as Eucalyptus gene orthologs) present in each sample were constructed for comparing galled and ungalled samples (presence/absence analysis).
Expression analysis
The main analyses in this paper were conducted with data on the presence/absence of contigs in different samples. However, quantitative differential expression analysis was also performed using Trinity [46] on specimen 816 by comparing galled (816.3) and ungalled (816.1) leaves. In order to compare read count, the raw sequencing data from both samples were pooled together. The reads were then aligned and estimated for abundance using the toolset provided in the Trinity package. From these results a minimum FPKM threshold was estimated and a measure of abundance of specific contigs per sample can then be made. Using Trinity's provided expression analysis tools, differential expression was calculated between the two leaf samples implementing the ‘edgeR’ method [46, 49]. Using the initial contig count and FPKM estimation, true FPKM values were generated and normalised. Finally, contigs were defined into associated clusters per sample in R 3.2.0 [50]. Contigs identified as being present in both samples and differentially expressed were then blasted (as per above) in order to identify Eucalyptus gene orthologues and ultimately Arabidopsis orthologues (again as above).
Analytical methods
Sequential analysis was performed (see sampling above), first using genotype 816 (matched galled/ungalled libraries: Tables 2 and 3). Finally the generality of the results were tested using all libraries (Table 4). Genes were analyzed for gene ontology term enrichment using the AmiGO Term Enrichment Service http://amigo.geneontology.org/rte and agriGO http://bioinfo.cau.edu.cn/agriGO/ [51].
Availability of supporting data
All short read data sets used for the analyses in this article are deposited in EBI’s European Nucleotide Archive (ENA), under study number PRJEB11301, accessible at www.ebi.ac.uk/ena/data/view/PRJEB11301. The sequence of Metrosideros SAUR67 (full length cds) is deposited in GenBank (NCBI), accession number: KT884616 (MpSAUR67).
Abbreviations
- AP2:
-
APETALA2 [gene]
- BLASTn:
-
Basic local alignment search tool (nucleotide)
- bp:
-
Base pairs
- CDS:
-
Coding DNA sequence
- DREB:
-
Dehydration responsive element binding [gene family]
- EBI:
-
European Bioinformatics Institute
- ENA:
-
European Nucleotide Archive
- ERF:
-
Ethylene-responsive element binding factor [gene family]
- FDR:
-
False discovery rate
- FPKM:
-
Fragments Per Kilobase of transcript per Million mapped reads
- GH3:
-
Gretchen Hagen3 [gene family]
- GO:
-
Gene ontology
- HRD:
-
HARDY [gene]
- IAA:
-
Indole-3-acetic acid
- LEA:
-
Late embryogenesis abundant [gene family]
- M. polymorpha :
-
Metrosideros polymorpha
- NCBI:
-
National Center for Biotechnology Information
- nov.sp.:
-
New species
- RIN:
-
RNA integrity number
- RNA:
-
Ribonucleic acid
- SAUR:
-
Small auxin upregulated [gene family]
- UBC:
-
University of British Columbia
References
Blanche R. Life in a Gall: The Biology and Ecology of Insects that Live in Plant Galls. Victoria, Australia: CSIRO Publishing; 2012.
Stone GN, Cook JM. The structure of cynipid oak galls: patterns in the evolution of an extended phenotype. Proc R Soc B Biol Sci. 1998;265(1400):979–88.
Harris MO, Stuart JJ, Mohan M, Nair S, Lamb RJ, Rohfritsch O. Grasses and Gall midges: plant defense and insect adaptation. Annu Rev Entomol. 2003;48:549–77.
Zhao CY, Escalante LN, Chen H, Benatti TR, Qu JX, Chellapilla S, et al. A massive expansion of effector genes underlies gall-formation in the wheat pest Mayetiola destructor. Curr Biol. 2015;25(5):613–20.
Yamaguchi H, Tanaka H, Hasegawa M, Tokuda M, Asami T, Suzuki Y. Phytohormones and willow gall induction by a gall-inducing sawfly. New Phytol. 2012;196(2):586–95.
Tooker JF, Helms AM. Phytohormone dynamics associated with gall insects, and their potential role in the evolution of the gall-inducing habit. J Chem Ecol. 2014;40(7):742–53.
Suzuki H, Yokokura J, Ito T, Arai R, Yokoyama C, Toshima H, et al. Biosynthetic pathway of the phytohormone auxin in insects and screening of its inhibitors. Insect Biochem Mol Biol. 2014;53:66–72.
Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot. 2005;95(5):707–35.
Rawat N, Neeraja CN, Nair S, Bentur JS. Differential gene expression in gall midge susceptible rice genotypes revealed by suppressive subtraction hybridization (SSH) cDNA libraries and microarray analysis. Rice. 2012;5:8.
Raman A. Gall induction by hemipteroid insects. J Plant Interact. 2012;7(1):29–44.
Wool D. Galling aphids: specialization, biological complexity, and variation. Annu Rev Entomol. 2004;49:175–92.
Takei M, Yoshida S, Kawai T, Hasegawa M, Suzuki Y. Adaptive significance of gall formation for a gall-inducing aphids on Japanese elm trees. J Insect Physiol. 2015;72:43–51.
Gullan PJ, Miller DR, Cook LG. Gall-inducing scale insects (Hemiptera: Sternorrhyncha: Coccoidea). In: Raman A, Schaefer CW, Withers TM, editors. Biology, Ecology, and Evolution of Gall-Inducing Arthropods. Enfield, USA, Plymouth, UK: Science Publishers; 2005. p. 159–229.
Hodkinson I. Life cycle variation and adaptation in jumping plant lice (Insecta: Hemiptera: Psylloidea): a global synthesis. J Nat Hist. 2009;43(1–2):65–179.
Hodkinson ID. The biology and ecology of the gall-forming Psylloidea. In: Ananthakrishnan R, editor. The biology of gall forming insects. London: Edward Arnold; 1984. p. 59–77.
Burckhardt D. Biology, ecology, and evolution of gall-inducing psyllids (Hemiptera: Psylloidea). In: Raman A, Schaefer CW, Withers TM, editors. Biology, Ecology, and Evolution of Gall-Inducing Arthropods. Enfield, Plymouth: Science Publishers; 2005. p. 143–57.
Yang MM, Raman A. Diversity, richness, and patterns of radiation among gall-inducing psyllids (Hemiptera : Psylloidea) in the orient and eastern Palearctic. Orient Insects. 2007;41:55–65.
Percy DM, Garver AM, Wagner WL, James HF, Cunningham CW, Miller SE, et al. Progressive island colonization and ancient origin of Hawaiian Metrosideros (Myrtaceae). Proc R Soc B Biol Sci. 2008;275(1642):1479–90.
Harbaugh DT, Wagner WL, Percy DM, James HF, Fleischer RC. Genetic Structure of the Polymorphic Metrosideros (Myrtaceae) Complex in the Hawaiian Islands Using Nuclear Microsatellite Data. Plos One. 2009;4(3):e4698.
Stacy EA, Johansen JB, Sakishima T, Price DK, Pillon Y. Incipient radiation within the dominant Hawaiian tree Metrosideros polymorpha. Heredity. 2014;113:334–42.
Treseder KK, Vitousek PM. Potential ecosystem-level effects of genetic variation among populations of Metrosideros polymorpha from a soil fertility gradient in Hawaii. Oecologia. 2001;126(2):266–75.
Morrison KR, Stacy EA. Intraspecific divergence and evolution of a life-history trade-off along a successional gradient in Hawaii’s Metrosideros polymorpha. J Evol Biol. 2014;27(6):1192–204.
Zimmerman EC. Insects of Hawaii. Volume 5. Homoptera: Sternorhyncha. Superfamily Psylloidea. Honolulu: University of Hawaii Press; 1948.
Mizrachi E, Hefer CA, Ranik M, Joubert F, Myburg AA: De novo assembled expressed gene catalog of a fast-growing Eucalyptus tree produced by Illumina mRNA-Seq. Bmc Genomics. 2010;11.
Grattapaglia D, Vaillancourt RE, Shepherd M, Thumma BR, Foley W, Kulheim C, et al. Progress in Myrtaceae genetics and genomics: Eucalyptus as the pivotal genus. Tree Genetics & Genomes. 2012;8(3):463–508.
Myburg AA, Grattapaglia D, Tuskan GA, Hellsten U, Hayes RD, Grimwood J, et al. The genome of Eucalyptus grandis. Nature. 2014;510(7505):356.
Hefer CA, Mizrachi E, Myburg AA, Douglas CJ, Mansfield SD. Comparative interrogation of the developing xylem transcriptomes of two wood‐forming species: Populus trichocarpa and Eucalyptus grandis. New Phytol. 2015;206:1391–405.
Ouvrard D, Chalise P, Percy DM. Host-plant leaps versus host-plant shuffle: a global survey reveals contrasting patterns in an oligophagous insect group (Hemiptera, Psylloidea). Systematics and Biodiversity. 2015. in press.
Hail D, Hunter WB, Dowd SE, Bextine BR. Expressed Sequence Tag (EST) survey of life stages of the potato psyllid, Bactericera cockerelli, using 454 pyrosequencing. Southwestern Entomologist. 2010;35(3):463–6.
Nachappa P, Levy J, Tamborindeguy C. Transcriptome analyses of Bactericera cockerelli adults in response to “Candidatus Liberibacter solanacearum” infection. Mol Genet Genomics. 2012;287(10):803–17.
Reese JC, Christenson MK, Leng N, Saha S, Cantarel B, Lindeberg M, et al. Characterization of the Asian citrus psyllid transcriptome. J Genomics. 2014;2:54.
Hunter WB, Reese J, Consortium IPG. The Asian Citrus Psyllid Genome (Diaphorina citri, Hemiptera). J Citrus Pathology. 2014;1:1.
Hodkinson ID. The psyllids (Homoptera, Psylloidea) of the Austro-oriental, Pacific and Hawaiian zoogeographical realms - an annotated check list. J Nat Hist. 1983;17(3):341–77.
Nishida T, Haramoto FH, Nakahara LM. Altitudinal distribution of endemic psyllids (Homoptera, Psyllidae) in the Metrosideros ecosystem. Proc Hawaiian Entomol Soc. 1980;23(2):255–62.
Cordell S, Goldstein G, Mueller-Dombois D, Webb D, Vitousek PM. Physiological and morphological variation in Metrosideros polymorpha, a dominant Hawaiian tree species, along an altitudinal gradient: the role of phenotypic plasticity. Oecologia. 1998;113:188–96.
Fisher JB, Goldstein G, Jones TJ, Cordell S. Wood vessel diameter is related to elevation and genotype in the Hawaiian tree Metrosideros polymorpha (Myrtaceae). Am J Bot. 2007;94:709–15.
Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portoles S, Rodriguez-Conception M, et al. Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J. 2007;26(22):4756–67.
Karaba A, Dixit S, Greco R, Aharoni A, Trijatmiko KR, Marsch-Martinez N, et al. Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proc Natl Acad Sci U S A. 2007;104(39):15270–5.
Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms. 2012;1819(2):86–96.
Cao PB, Azar S, SanClemente H, Mounet F, Dunand C, Marque G, Marque C, Teulieres C. Genome-Wide Analysis of the AP2/ERF Family in Eucalyptus grandis: An Intriguing Over-Representation of Stress-Responsive DREB1/CBF Genes. Plos One. 2015;10(4):e0121041.
Florentine SK, Raman A, Dhileepan K. Effects of gall induction by Epiblema strenuana on gas exchange, nutrients, and energetics in Parthenium hysterophorus. Biocontrol. 2005;50(5):787–801.
Bartlett L, Connor EF. Exogenous phytohormones and the induction of plant galls by insects. Arthropod Plant Interact. 2014;8(4):339–48.
Richards S, Gibbs RA, Gerardo NM, Moran N, Nakabachi A, Stern D, Tagu D, Wilson ACC, Muzny D, Kovar C et al. Genome Sequence of the Pea Aphid Acyrthosiphon pisum. Plos Biology. 2010;8(2):e1000313.
Gerardo NM, Wilson ACC. The power of paired genomes. Mol Ecol. 2011;20(10):2038–40.
Ojeda DI, Santos-Guerra A, Oliva-Tejera F, Valido A, Jaén-Molina R, Xinxin X, et al. Bird-pollinated Macaronesian Lotus (Leguminosae) evolved within a group of entomophilous ancestors with post-anthesis flower colour change. Perspect Plant Ecol Evol Syst. 2013;15:193–204.
Grabherr M, Haas B, Yassour M, Levin J, Thompson D, Amit I, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644–52.
Goodstein DM, Shu SQ, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(D1):D1178–86.
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402.
Robinson MD, McCarthy D, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
R CoreTeam. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2015.
Du Z, Zhou X, Ling Y, Zhang ZH, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–70.
Acknowledgements
Funding for this study came from NSERC (Canada) Discovery grant to QC and Natural History Museum (NHM) funding to DMP. We also acknowledge use of the NHM ctag server. Financial support that assisted in fieldwork and the Trioza nov. sp. 1 transcriptome sequencing is gratefully acknowledged from the University of California, Berkeley, and NSF Dimensions of Biodiversity award (DEB 1241253). We thank Dario Ojeda Alayon (UBC) for kindly performing the RNA extractions, We also thank Debbie Adam, Michael Brewer, Kari Goodman, and Anastasia Kuzmin for further assistance with lab work, library preparation and sequencing. We would also like to thank Brian Haas for his assistance with the expression analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SB analysed the data, contributed analytical tools and contributed to the manuscript. DMP conducted the fieldwork. DMP and QC designed the study, obtained funding for the study, contributed to the analysis and wrote the manuscript. CH contributed analytical tools and assisted the analysis. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Bailey, S., Percy, D.M., Hefer, C.A. et al. The transcriptional landscape of insect galls: psyllid (Hemiptera) gall formation in Hawaiian Metrosideros polymorpha (Myrtaceae). BMC Genomics 16, 943 (2015). https://doi.org/10.1186/s12864-015-2109-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-015-2109-9