Abstract
Background
Provision of long-chain polyunsaturated fatty acids (LC-PUFA) in vertebrates occurs through the diet or via endogenous production from C18 precursors through consecutive elongations and desaturations. It has been postulated that the abundance of LC-PUFA in the marine environment has remarkably modulated the gene complement and function of Fads in marine teleosts. In vertebrates two fatty acyl desaturases, namely Fads1 and Fads2, encode ∆5 and ∆6 desaturases, respectively. To fully clarify the evolutionary history of LC-PUFA biosynthesis in vertebrates, we investigated the gene repertoire and function of Fads from species placed at key evolutionary nodes.
Results
We demonstrate that functional Fads1Δ5 and Fads2∆6 arose from a tandem gene duplication in the ancestor of vertebrates, since they are present in the Arctic lamprey. Additionally, we show that a similar condition was retained in ray-finned fish such as the Senegal bichir and spotted gar, with the identification of fads1 genes in these lineages. Functional characterisation of the isolated desaturases reveals the first case of a Fads1 enzyme with ∆5 desaturase activity in the Teleostei lineage, the Elopomorpha. In contrast, in Osteoglossomorpha genomes, while no fads1 was identified, two separate fads2 duplicates with ∆6 and ∆5 desaturase activities respectively were uncovered.
Conclusions
We conclude that, while the essential genetic components involved LC-PUFA biosynthesis evolved in the vertebrate ancestor, the full completion of the LC-PUFA biosynthesis pathway arose uniquely in gnathostomes.
Similar content being viewed by others
Background
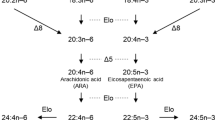
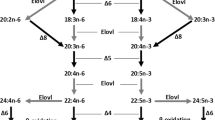
Long-chain (≥C20) polyunsaturated fatty acids (LC-PUFA) and their derivatives are biologically active molecules that are involved in neural function, signalling and regulation of lipid metabolism, inflammation and cell division [1]. Among LC-PUFA, arachidonic acid (ARA, 20:4n-6), eicosapentaenoic acid (EPA, 20:5n-3) and docosahexaenoic acid (DHA, 22:6n-3) play particularly important roles in the abovementioned physiological processes [2, 3]. In vertebrates, biosynthesis of LC-PUFA such as ARA, EPA and DHA is achieved by sequential reactions towards the dietary essential fatty acids (EFA) linoleic acid (LA, 18:2n-6) and α-linolenic acid (ALA, 18:3n-3), which are catalysed by fatty acyl desaturase (Fads) and elongation of very long-chain fatty acid (Elovl) enzymes [3]. Briefly, ARA and EPA are synthesised from LA and ALA, respectively, by two distinct pathways called the “Δ6 pathway” (Δ6 desaturation – elongation – Δ5 desaturation) or the “Δ8 pathway” (elongation – Δ8 desaturation – Δ5 desaturation). Moreover, DHA biosynthesis generally proceeds through the so-called “Sprecher pathway”, comprising two consecutive elongation steps from EPA to produce 24:5n-3, which is then Δ6 desaturated to 24:6n-3 prior to being converted to DHA by partial β-oxidation in peroxisomes [4, 5]. Interestingly, an alternative pathway involving direct Δ4 desaturation of 22:5n-3 to DHA has been described in teleosts [6, 7] and, more recently, found to operate in mammalian cells [8].
In mammals, Δ5 and Δ6 desaturation reactions are specifically catalysed by FADS1 and FADS2 enzymes, respectively [2]. Orthologues of both fads1Δ5 and fads2Δ6 were previously identified in the cartilaginous fish Scyliorhinus canicula, an indication that they emerged before gnathostome origin [9]. Furthermore, while mammals, birds, reptiles and amphibians also possess fads1 and fads2 genes, an orthologue of fads1 has not be identified to date in Teleostei [3, 9]. In addition, the repertoire of Teleostei fads2 varies significantly among lineages, with some species possessing one (e.g. Danio rerio), two (e.g. Monopterus albus), three (e.g. Oreochromis niloticus) or four (e.g. Salmo salar) fads2 paralogues, whereas others lack completely fads genes in their genomes (e.g. Tetraodon nigroviridis) [3]. With regards to function, many Teleostei Fads2 retain the Δ6 desaturase phenotype but, interestingly, they also exhibit a more varied spectrum of activities including bifunctional Δ6Δ5 desaturase [10,11,12,13] and Δ4 desaturase [7] as a result of a functionalisation process hypothesised to have occurred in response to dietary availability in natural prey [9].
Although the gene complement and functions of fads are well understood in Chondrichthyes, numerous Teleostei species and Tetrapoda [2, 3], the lack of information in lineages such as Cyclostomata, Polypteriformes, Holostei and post 3R lineages (e.g. Elopomorpha and Osteoglossomorpha) hampers the full comprehension of FADS function in vertebrates. Hence, to fully clarify the history of LC-PUFA biosynthesis in vertebrates, we isolated and functionally characterised the fads complement from species placed at key phylogenetic nodes, i.e. the cyclostome Lethenteron camtschaticum (Arctic lamprey), representative of the most ancient lineage of extant vertebrates, and four species of ray-finned fish. Among the latter, we investigated two species that diverged before the teleost specific whole genome duplication (3R WGD), namely the Polypteriforme, Polypterus senegalus (Senegal bichir) and the Lepisosteiforme, Lepisosteus oculatus (spotted gar) [14], and two others that diverged after the teleost specific 3R whole genome duplication (WGD), namely the Elopomorpha, Anguilla japonica (Japanese eel) and the Osteoglossomorpha, Pantodon buchholzi (African butterfly fish) [15, 16]. Our findings provide a definitive understanding of the evolutionary history of key components of essential LC-PUFA biosynthesis and demonstrate that functional fads1Δ5 and fads2Δ6 emerged in the vertebrate ancestor.
Methods
Sequence collection and phylogenetic analysis
The initial sequence collection for phylogenetic analysis was performed through blastp and blastn searches in NCBI, using as query Homo sapiens FADS1 (NP_037534.3) and FADS2 (NP_004256.1) sequences. Additionally, to ensure a full collection of fads sequences in Teleostei a second search was performed in NCBI targeting specifically the nucleotide collection and non-redundant protein database. From these results a set of Fads amino acid sequences representative of the major vertebrate clades was collected for phylogenetic analysis (Accession numbers provided in Fig. 1). Searches revealed that the Scleropages formosus presented a 3’partial Fads-like sequence (XP_018598908.1); this sequence was completed by performing blastn searches in S. formosus transcriptome SRA reads (NCBI accession: SRX1668426 to 32). Additionally, Gnathonemus petersii and Osteoglossum bicirrhosum fads-like genes were assembled from their genomic SRA (SRX2235994 and 95) with Geneious V 7.1.9 using as reference the previously curated S. formosus fads. The 86 sequences from the databases and 9 sequences functionally characterised in the present study were aligned with MAFFT v7.306 [17]. The best alignment method was determined automatically resulting in L-INS-i method [18]. Columns containing 90% gaps were stripped from sequence alignment leaving a total of 451 sites for phylogenetic analysis. Sequence alignment was then submitted to PhyML v3.0 server [19] for Maximum Likelihood (ML) phylogenetic evolutionary model and was automatically selected by smart model selection SMS resulting in LG + G + I, and branch support was calculated using Abayes [20]. The resulting tree was visualised using FigTree v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/) and rooted with invertebrate Fads sequences.
Maximum likelihood phylogenetic analysis of FADS1 and FADS2 amino acid sequences. Values at nodes indicate posterior probabilities, * indicates FADS isolated and functionally analysed in this work. Black arrow (3R WGD) approximates the timing of the teleost duplication. Accession numbers are indicated
Isolation of fads for functional characterisation in yeast
Selected Fads from species within major pre and post 3R WGD lineages were functionally characterised by expressing their ORF in yeast S. cerevisiae. In order to isolate the corresponding fads ORF sequences, initial tblastn searches using S. canicula fads1 (AEY94454) and fads2 (AEY94455) as queries were carried out in the genome assembly of L. camtschaticum (https://www.ncbi.nlm.nih.gov/assembly/GCA_000466285.1) and A. japonica (GCA_000470695), and transcriptome SRA from P. senegalus (SRX796491, SRX732875) and P. buchholzi (SRX666400). The resulting hits were assembled into predicted full-length ORF using the corresponding bait sequences as references. With L. oculatus, although annotations of fads1- and fads2-like genes were available at GenBank (XM_015338726 for fads1) and Ensembl (ENSLOCG00000007048 and ENSLOCG00000007031 for fads1 and fads2, respectively), these gene predictions were poor due to the genome coverage. Therefore, we performed manual mapping of the genomic sequence (NC_023205) using transcript sequences of fads1- and fads2-like genes retrieved from their transcriptome SRA (SRX661022). The obtained transcripts were then assembled into full-length ORF sequences referring to the genomic sequence. All final assembled/predicted fads ORF sequences were then used as reference to design primers for further cloning into the yeast expression vector pYES2 (Thermo Fisher Scientific, Waltham, MA, USA). Briefly, cDNA prepared from total RNA extracted from L. camtschaticum, P. senegalus, L. oculatus, P. buchholzi and A. japonica was used to amplify the corresponding fads ORF by PCR using primers containing restriction sites at both the start and stop codon (see Additional file 1 for primer details and PCR conditions). Each PCR product was digested with the corresponding restriction enzymes and ligated into similarly restricted pYES2 [6, 10]. All pYES2 clones were confirmed by sequencing (GATC Biotech Constance, Germany) prior to being used in functional assays in yeast.
Yeast expression assays
Yeast transformation with pYES2 and the culture of yeast S. cerevisiae were carried out as described previously [10, 21]. The resulting transgenic yeast expressing each fads were grown in the presence of PUFA including Δ6 (18:3n-3 and 18:2n-6), Δ8 (20:2n-6 and 20:3n-3), Δ5 (20:4n-3 and 20:3n-6) and Δ4 (22:5n-3 and 22:4n-6) desaturase substrates. The PUFA substrates were exogenously supplemented at final concentrations of 0.5 mM (C18), 0.75 mM (C20) and 1.0 mM (C22) to compensate for decreased efficiency of uptake with increased chain length [21]. All FA substrates (98–99% pure), except for 18:4n-3 and 20:4n-3, were purchased from Nu-Chek Prep, Inc. (Elysian, MN, USA). Moreover, 18:4n-3 and 20:4n-3 were obtained from Sigma-Aldrich (St Louis, MO, USA) and Cayman Chemicals (Ann Arbor, MI, USA), respectively. After 48 h incubation, yeast cells were collected, washed twice in distilled water and kept at − 20 °C until further analysis.
Fatty acid analysis of yeast
Total lipid was extracted from yeast and used to prepare fatty acid methyl esters (FAME) as described in detail previously [10]. FAME extraction, purification and analysis were performed as described by Li et al. [6]. Substrate fatty acid conversions from exogenously added PUFA substrates were calculated by the proportion of substrate fatty acid converted to a desaturated product as [product area/(product area + substrate area)] × 100 [21]. When appropriate, GC-MS was used to confirm the identity of the desaturation products [6].
Results
Gene orthologues of fads1 and fads2 emerged in the ancestor of vertebrates
To address the orthology of the identified Fads-like sequences an ML phylogenetic analysis was conducted with a total of 86 amino acid (aa) sequences, including species from cyclostomes (e.g. Arctic lamprey L. camtschaticum), tetrapods (e.g. amphibians, birds, mammals), Chondrichthyes (e.g. elephant shark Callorhinchus milii), ray-finned fishes (e.g. P. senegalus, L. oculatus, A. japonica and P. buchholzi), as well as Fads sequences from several protostomes (Capitella teleta, Lingula anatina and Lottia gigantea) and the invertebrate deuterostome Saccoglossus kowalevskii. The resulting phylogenetic tree displays two well-supported monophyletic clades, each containing vertebrate Fads1 and Fads2 sequences, respectively, with the invertebrate Fads appearing as an independent clade from both vertebrate Fads (Fig. 1). In the Fads1 group, in addition to the described gene orthologues from various lobe-finned fish (e.g. Latimeria chalumnae) and Chondrichthyes species (C. milii and S. canicula) [9], we find a putative Fads1 from the Arctic lamprey (L. camtschaticum), the Senegal bichir (P. senegalus), spotted gar (L. oculatus) and the teleost Japanese eel (A. japonica), but not in the Osteoglossomorpha or any other teleost examined (Fig. 1). A similar genetic distribution is observed in the Fads2 clade, with an orthologue of fads2 found in the Arctic lamprey (Fig. 1). Moreover, Fads2 were also present in the pre-3R whole genome duplication (WGD) lineages such as Polypteriformes (P. senegalus) and Lepisosteiformes (L. oculatus) located at the ray-finned fish Fads2 clade. With the exception of Arapaima gigas, all Osteoglossomorpha species examined were found to possess two Fads2 (termed “a” and “b”), which are distributed among two well supported groups (0.9). Although, the branching tree pattern of the Osteoglossomorpha fads2 gene duplicates could be indicative of an origin related with the teleost-specific 3R WGD, the absence of synteny precludes further analysis.
Newly cloned fads1 and fads2 exhibit conserved Δ5 and Δ6 signature residues
To further characterise the newly identified Fads sequences, we cloned the full open reading frame (ORF) of each gene and performed Pfam searches of the deduced amino acid (aa) sequences. All the sequences presented the characteristic signature motifs of Fads, namely the heme binding motif (HPGG) and three histidine boxes HXXXH, HXXHH and QXXHH [22, 23] (Fig. 2, brown boxes). Next, we searched for critical aa residues that have been recently demonstrated to account for Δ5 (NP_445897.2) and Δ6 (NP_112634.1) desaturase activities in rat FADS enzymes [24]. Regarding Fads1, the analysed fish species preserved an overall Δ5 pattern with a conserved methionine (M) to leucine (L) substitution (Fig. 2, black box 3), possibly presenting no impact on the substrate specificity given the comparable biochemical properties of these residues. An additional valine (V) residue (Fig. 2, black box 5), also suggested to determine substrate selectivity in rat Fads1 [24], was not conserved in the analysed fish sequences, nor in the human sequence. Curiously, the fads2b sequences from the Osteoglossomorpha P. buchholzi and S. formosus presented key residues for Δ5 function, conserved with rat Fads1, serine (S) (Fig. 2, red box 2) and M/L (Fig. 2, red box 3). In contrast, the fads2a of P. buchholzi and S. formosus showed typical Δ6 residues (Fig. 2).
Sequence alignment of FADS1 and FADS2 amino acid sequences. Orange boxes correspond to the conserved histidine boxes, the yellow box indicates residues proposed to be involved in substrate specificity [27], and blue boxes indicate residues replaced in rat FADS2 Δ6 desaturase to obtain Δ5 activity [27]. The heme binding motif HPGG is also shown
Functional analysis indicates conserved and derived fads activities in various vertebrate species
Functional analysis of the newly isolated Fads was carried out by the heterologous expression of the ORF in the yeast S. cerevisiae grown in the presence of potential exogenously added polyunsaturated fatty acid (PUFA) substrates. Inactivity towards PUFA in control (untransformed) yeast was confirmed by fatty acid profiles characterised by major endogenous yeast fatty acids (16:0, 16:1n-7, 18:0 and 18:1n-9) and unmetabolised exogenously added PUFA (data not shown) [25]. Transgenic yeast expressing fads1 from all species examined (L. camtschaticum, P. senegalus, L. ocelatus and A. japonica) showed Δ5 desaturase activity towards 20:4n-3 and 20:3n-6 since they were able to produce 20:5n-3 and 20:4n-6, respectively (Table 1 and Additional file 2). Moreover, yeast expressing fads2 from L. camtschaticum, P. senegalus, L. oculatus and P. buchholzi (Fads2a) showed ∆6 activity, desaturating 18:3n-3 to 18:4n-3 and 18:2n-6 to 18:3n-6 (Table 1 and Additional file 2). In the vast majority of cases, Fads2 exhibited Δ8 desaturase capability as they were able to desaturate 20:2n-6 and 20:3n-3 to 20:3n-6 and 20:4n-3, respectively (Table 1 and Additional file 2), a feature previously confirmed in Fads2 of A. japonica [26]. Interestingly, transgenic yeast expressing fads2b of the African butterfly fish, P. buchholzi, showed neither Δ6 nor Δ8 desaturase activity but, as described above for Fads1 enzymes, a clear Δ5 desaturase profile, biosynthesising 20:4n-6 (ARA) and 20:5n-3 (EPA) from 20:3n-6 and 20:4n-3, respectively. No Δ4 desaturase activity was detected in any of the assayed Fads, which is in agreement with none of the sequences possessing the key aa residues responsible for Δ4 function (Fig. 2, yellow box) [27].
Discussion
Gene duplication has long been recognised as a decisive contributor in the shaping vertebrate genomes, providing spare genetic material for adaptive evolution, mutation, and genetic drift [28,29,30,31]. Yet, degeneration and loss are the most common fates encountered with duplicate genes [3], with significant gene loss occurring shortly after WGD episodes [28, 29, 32,33,34,35,36]. To evaluate the impact of gene duplication and loss on LC-PUFA biosynthesis in vertebrates, we investigated the genetic repertoire and function of fads genes in species placed at key phylogenetic points, namely the transition from jawless to jawed vertebrates, and the pre/post 3R period in the evolution of the ray-finned fish.
Sequence and phylogenetic data revealed that fads1 and fads2 orthologues are present in the genome of the Arctic lamprey (L. camtschaticum), supporting the hypothesis that fads1 and fads2 originated most likely thorough a tandem gene duplication in the vertebrate ancestor (Fig. 3) [9]. Additionally, we found that fads1 and fads2 are also retained in ray-finned fish such as the Senegal bichir (P. senegalus) and the spotted gar (L. oculatus), which diverged prior to the Teleostei specific 3R WGD (Fig. 3). Unexpectedly, a fads1 orthologue was also identified in species such as the Japanese eel (A. japonica) and the Indo-Pacific tarpon (Megalops cyprinoides) belonging to Elopomorpha, a group that diverged after the 3R WGD [15, 16]. These results indicate that fads1, previously hypothesised to be lost in Teleostei [3, 9, 11], is actually retained in some Teleostei such as the Elopomorpha (Fig. 3). We were unable to recover a fads1 orthologue in P. buchholzi nor in any of the other Osteoglossomorpha species analysed, indicating that this gene is likely lost in this lineage. Further, in silico search for fads1 in relevant genome databases of representatives of Euteleosteomorpha or Otomorpha lineages suggest an absence of fads1 in Clupeocephala, which further supports Elomorpha as the only Teleostei retaining a functional fads1. Yet, all Fads1 desaturases characterised in the present work (L. camtschaticum, P. senegalus, L. ocelatus and A. japonica) exhibit ∆5 desaturase activity in agreement with the previous functional assessments in Sarcopterygii such as H. sapiens FADS1 [37] and chondrichthyan Fads1 [9].
Evolutionary history and distribution of Fads1 and Fads2 in vertebrates, combined with the corresponding desaturase activities. Squares correspond to fads1 circle to fads2, black arrows indicate the approximate timing of whole genome duplications in the ancestral vertebrate (2R) and the teleost specific duplication (3R), grey square highlights lineages lacking fads1
In contrast to the lack of fads1 within most Teleostei, phylogenetic analysis confirmed that fads2 is ubiquitously present across the entire vertebrate clade, with exceptions represented by teleosts lacking fads-like orthologues in their genomes, namely Takifugu rubripes and T. nigroviridis [3, 38]. Thus, vertebrate Fads2 includes not only those previously reported from numerous species within Teleostei, Sarcopterygii and Chondrichthyes [3], but also the presently functionally characterised Fads2 from Cyclostomata (Arctic lamprey), Polypteriformes (Senegal bichir), Lepisosteiformes (spotted gar), Elopomorpha (Japanese eel) and Osteoglossomorpha (African butterfly fish). Interestingly, we found that Osteoglossomorpha species presented two copies of fads2 that clustered into two separate groups in the phylogenetic analysis and therefore suggesting a potential origin dating to the 3R WGD. With regards to their functional characterisation, the African butterfly fish fads2b demonstrated that this gene did not encode a ∆6 desaturase, but rather a desaturase that showed ∆5 activity (Table 1). A similar functionalisation scenario among teleost Fads2 was also suggested in some salmonids such as Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss), with the acquisition of ∆5 activity occurring in one of the several fads2 copies that arose from tandem gene duplication [3, 39, 40]. In fact, fads gene duplication appears to be a frequent event in vertebrate evolution, with most mammals possessing an additional gene, Fads3 [41,42,43], and the well-established fads2 duplication occurred in Teleostei [6, 10, 11, 39, 44]. Gene duplication is often followed by low purifying selection; thus, the presence of Δ5 functionalised Fads2b in Teleostei, such as osteoglossomorpha, is consistent with subfunctionalisation and/or neofunctionalisation processes occurring rapidly after gene duplication [35, 45].
The conversion of a fads2Δ6 ancestor, as deduced from the data from the Senegal bichir and the spotted gar, into a fads2Δ5 in African butterflyfish (fads2b) may be viewed as a mechanism to overcome the bottleneck generated by the loss of fads1Δ5 in most Teleostei that would otherwise be unable to biosynthesise essential fatty acids such as EPA and ARA. Yet, such a constraint triggered distinct evolutionary routes. In addition to the abovementioned case in salmonids, whereby acquisition of Δ5 desaturase occurred in one of the several Fads2 copies, in other species such as D. rerio [10] with one fads2, the acquisition of Δ5 desaturase activity has been accompanied by retention of Δ6 activity, thus resulting in desaturases with dual (or bifunctional) Δ6Δ5 activities (Fig. 3), which have also been found in other Teleostei [11, 13, 46, 47]. Further functionalisation cases amongst teleost Fads2 include the presence of Δ4 desaturases, often co-existing with Δ5 desaturase activity within the same enzyme [6, 11, 12]. Still, the majority of Teleostei Fads2 are primarily Δ6 desaturases without Δ5 or Δ4 desaturase activities [3]. Interestingly, the functional characterisation data confirm that, with the exception of the African butterfly fish Fads2b (Δ5 desaturase), all the presently analysed Fads2 showed capability for Δ8 desaturation, an intrinsic enzymatic ability within vertebrate Fads2 and hence not regarded as a functionalisation case [42, 44].
The LC-PUFA biosynthetic pathway is punctuated by alternate steps of fatty acid elongation and desaturation therefore, a comprehensive interpretation of the evolution of this key metabolic pathway in vertebrates can be only fully attained if the repertoire and function of Elovl enzymes is also considered. Similarly to Fads1 and Fads2, Elovl2 and Elovl5, major elongation enzymes involved in LC-PUFA biosynthesis [48], emerged in vertebrate ancestry possibly as a consequence of genome duplications, and as demonstrated by the existence of both orthologues in Arctic lamprey and elephant shark [49]. Therefore, the ability to biosynthesise EPA and ARA was present in vertebrates such as lamprey. For DHA biosynthesis, two consecutive elongation reactions from EPA are required to produce 24:5n-3, which is then Δ6 desaturated to 24:6n-3 before it is partly β-oxidised to DHA (22:6n-3) [4]. While the Δ6 desaturation activity towards 24:5n-3 has been shown to be an intrinsic characteristic of non-Δ4 Fads2 desaturases and it is widely distributed from basal gnathostomes to recently emerged Teleostei [7], the elongation capacity towards C22 substrates such as 22:5n-3 hinders the “Sprecher pathway” in cyclostomes since the Arctic lamprey Elovl2 could not elongate 22:5n-3 [49]. In pre 3R WGD Teleostei, along with Fads1Δ5 and Fads2Δ6 reported here, it is likely that Elovl2 and Elovl5 (present in at least spotted gar, Elovl2- XP_015210453.1 and Elovl5- XP_006638754.1) constitute a complete LC-PUFA biosynthesis cascade, as demonstrated in chondrichthyans [49], a scenario that can be also postulated for post 3R WGD Teleostei such as Elopomorpha. Yet, other 3R WGD lineages including cyprinids, siluriformes and salmonids have Elovl2 and Elovl5 [13, 25, 50,51,52] and, while lacking Fads1 gene orthologues, complete the LC-PUFA biosynthetic cascade with functionalised Fads2Δ5 desaturase (e.g. D. rerio). Consequently, a key event in Teleostei evolution included also that of Elovl2, crucial for elongation reactions involved in DHA biosynthesis, although Elovl4 has been seen to partly compensate such gene loss in Teleostei [53]. The parallel, and contingent, evolution of two separate gene families, Fads and Elovl, in vertebrate ancestry, allowed for the combination of pre-existing metabolic islands into an integrated and functional enzymatic cascade, which was indispensable for adaptive paths with respect to LC-PUFA habitat availability.
Conclusion
In the present study, we established that the fatty acid desaturation dependent of fads1Δ5 and fads2Δ6 orthologues arose before vertebrate radiation, as deduced from the Arctic lamprey data. Additionally, fads1 was retained in pre 3R WGD lineages such as Polypteriformes (Senegal bichir) and Lepisosteiformes (spotted gar) and post 3R WGD Elopomorpha (Japanese eel) but was probably lost in the Osteoglossomorpha and Clupeocephala. Moreover, in Osteoglossomorpha the two reported fads2 genes exhibited the expected Δ6 desaturase activity, while the second fads2 paralogue (fads2b) had Δ5 activity. This observation supports the existence of alternative evolutionary routes that mitigate the loss of the canonical fads1Δ5 in Teleostei and puts forward Osteoglossomorpha Fads2 as, possibly, the most ancient representative of Fads2 plastic functionalisation.
Abbreviations
- 2R WGD:
-
Vertebrate whole genome duplication, or two rounds of whole genome duplication
- 3R WGD:
-
Teleost specific whole genome duplication
- ALA:
-
α-linolenic acid
- ARA:
-
Arachidonic acid
- DHA:
-
Docosahexaenoic acid
- EFA:
-
Essential fatty acid
- Elovl:
-
Fatty acid elongase
- EPA:
-
Eicosapentaenoic acid
- Fads:
-
Fatty acid desaturase
- FAME:
-
Fatty acid methyl esters
- GC-MS:
-
Gas chromatography–mass spectrometry
- LA:
-
Linoleic acid
- LC-PUFA:
-
Long-chain polyunsaturated fatty acids
- ORF:
-
Open reading frame
- PCR:
-
Polymerase chain reaction
- PUFA:
-
Polyunsaturated fatty acids
- SRA:
-
Sequence read archive
- WGD:
-
Whole genome duplication
References
Schmitz G, Ecker J. The opposing effects of n−3 and n−6 fatty acids. Prog Lipid Res. 2008;47(2):147–55.
Guillou H, Zadravec D, Martin PG, Jacobsson A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: insights from transgenic mice. Prog Lipid Res. 2010;49(2):186–99.
Castro LFC, Tocher DR, Monroig O. Long-chain polyunsaturated fatty acid biosynthesis in chordates: insights into the evolution of fads and Elovl gene repertoire. Prog Lipid Res. 2016;62:25–40.
Sprecher H. Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochim Biophys Acta. 2000;1486(2–3):219–31.
Ferdinandusse S, Ebberink MS, Vaz FM, Waterham HR, Wanders RJA. The important role of biochemical and functional studies in the diagnostics of peroxisomal disorders. J Inherit Metab Dis. 2016;39:531–43.
Li Y, Monroig O, Zhang L, Wang S, Zheng X, Dick JR, You C, Tocher DR. Vertebrate fatty acyl desaturase with Δ4 activity. Proc Natl Acad Sci. 2010;107(39):16840–5.
Oboh A, Kabeya N, Carmona-Antoñanzas G, Castro LFC, Dick JR, Tocher DR, Monroig O. Two alternative pathways for docosahexaenoic acid (DHA, 22:6n-3) biosynthesis are widespread among teleost fish. Sci Rep. 2017;7(1):3889.
Park HG, Park WJ, Kothapalli KSD, Brenna JT. The fatty acid desaturase 2 (FADS2) gene product catalyzes Δ4 desaturation to yield n-3 docosahexaenoic acid and n-6 docosapentaenoic acid in human cells. FASEB J. 2015;29(9):3911–9.
Castro LFC, Monroig Ó, Leaver MJ, Wilson J, Cunha I, Tocher DR. Functional desaturase Fads1 (Δ5) and Fads2 (Δ6) orthologues evolved before the origin of jawed vertebrates. PLoS One. 2012;7(2):e31950.
Hastings N, Agaba M, Tocher DR, Leaver MJ, Dick JR, Sargent JR, Teale AJ. A vertebrate fatty acid desaturase with Δ5 and Δ6 activities. Proc Natl Acad Sci. 2001;98(25):14304–9.
Fonseca-Madrigal J, Navarro JC, Hontoria F, Tocher DR, Martínez-Palacios CA, Monroig Ó. Diversification of substrate specificities in teleostei Fads2: characterization of Δ4 and Δ6Δ5 desaturases of Chirostoma estor. J Lipid Res. 2014;55(7):1408–19.
Kuah MK, Jaya-Ram A, Shu-Chien AC. The capacity for long-chain polyunsaturated fatty acid synthesis in a carnivorous vertebrate: functional characterisation and nutritional regulation of a Fads2 fatty acyl desaturase with Delta4 activity and an Elovl5 elongase in striped snakehead (Channa striata). Biochim Biophys Acta. 2014;1851(3):248–60.
Oboh A, Betancor MB, Tocher DR, Monroig O. Biosynthesis of long-chain polyunsaturated fatty acids in the African catfish Clarias gariepinus: molecular cloning and functional characterisation of fatty acyl desaturase (fads2) and elongase (elovl2) cDNAs7. Aquaculture. 2016;462:70–9.
Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics. 2011;188(4):799–808.
Betancur-R R, Wiley EO, Arratia G, Acero A, Bailly N, Miya M, Lecointre G, Ortí G. Phylogenetic classification of bony fishes. BMC Evol Biol. 2017;17(1):162.
Hughes LC, Orti G, Huang Y, Sun Y, Baldwin CC, Thompson AW, Arcila D, Betancur RR, Li C, Becker L, et al. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc Natl Acad Sci U S A. 2018;115(24):6249–54.
Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9(4):286–98.
Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33(2):511–8.
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–21.
Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol. 2011;60(5):685–99.
Lopes-Marques M, Ozório R, Amaral R, Tocher DR, Monroig Ó, Castro LFC. Molecular and functional characterization of a fads2 orthologue in the Amazonian teleost, Arapaima gigas. Comp Biochem Physiol B Biochem Mol Biol. 2017;203:84–91.
Pereira SL, Leonard AE, Mukerji P. Recent advances in the study of fatty acid desaturases from animals and lower eukaryotes. Prostaglandins Leukot Essent Fatty Acids. 2003;68(2):97–106.
Los DA, Murata N. Structure and expression of fatty acid desaturases. Biochim Biophys Acta Lipids Lipid Metab. 1998;1394(1):3–15.
Watanabe K, Ohno M, Taguchi M, Kawamoto S, Ono K, Aki T. Identification of amino acid residues that determine the substrate specificity of mammalian membrane-bound front-end fatty acid desaturases. J Lipid Res. 2016;57(1):89–99.
Agaba MK, Tocher DR, Zheng X, Dickson CA, Dick JR, Teale AJ. Cloning and functional characterisation of polyunsaturated fatty acid elongases of marine and freshwater teleost fish. Comp Biochem Physiol B Biochem Mol Biol. 2005;142(3):342–52.
Wang S, Monroig Ó, Tang G, Zhang L, You C, Tocher DR, Li Y. Investigating long-chain polyunsaturated fatty acid biosynthesis in teleost fish: functional characterization of fatty acyl desaturase (Fads2) and Elovl5 elongase in the catadromous species, Japanese eel Anguilla japonica. Aquaculture. 2014;434:57–65.
Lim Z, Senger T, Vrinten P. Four amino acid residues influence the substrate chain-length and regioselectivity of Siganus canaliculatus Δ4 and Δ5/6 desaturases. Lipids. 2014;49(4):357–67.
Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–5.
Wagner A. The fate of duplicated genes: loss or new function? BioEssays. 1998;20(10):785–8.
Holland PW. More genes in vertebrates? J Struct Funct Genom. 2003;3:75–84.
Schonfeld P, Reiser G. Why does brain metabolism not favor burning of fatty acids to provide energy? Reflections on disadvantages of the use of free fatty acids as fuel for brain. J Cereb Blood Flow Metab. 2013;33(10):1493–9.
Louis EJ. Evolutionary genetics: making the most of redundancy. Nature. 2007;449(7163):673–4.
Lynch M, Katju V. The altered evolutionary trajectories of gene duplicates. Trends Genet. 2004;20(11):544–9.
Kondrashov FA, Kondrashov AS. Role of selection in fixation of gene duplications. J Theor Biol. 2006;239:141–51.
Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7(5):R43.
Albalat R, Canestro C. Evolution by gene loss. Nat Rev Genet. 2016;17(7):379–91.
Cho HP, Nakamura M, Clarke SD. Cloning, expression, and fatty acid regulation of the human Δ-5 desaturase. J Biol Chem. 1999;274(52):37335–9.
Leaver MJ, Bautista JM, Björnsson BT, Jönsson E, Krey G, Tocher DR, Torstensen BE. Towards fish lipid nutrigenomics: current state and prospects for fin-fish aquaculture. Rev Fish Sci. 2008;16(sup1):73–94.
Hastings N, Agaba MK, Tocher DR, Zheng X, Dickson CA, Dick JR, Teale AJ. Molecular cloning and functional characterization of fatty acyl desaturase and Elongase cDNAs involved in the production of Eicosapentaenoic and docosahexaenoic acids from α-linolenic acid in Atlantic Salmon (Salmo salar). Mar Biotechnol. 2004;6(5):463–74.
Abdul Hamid NK, Carmona-Antoñanzas G, Monroig Ó, Tocher DR, Turchini GM, Donald JA. Isolation and functional characterisation of a fads2 in rainbow trout (Oncorhynchus mykiss) with Δ5 desaturase activity. PLoS One. 2016;11(3):e0150770.
Marquardt A, Stöhr H, White K, Weber BHF. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66(2):175–83.
Park WJ, Kothapalli KSD, Reardon HT, Kim LY, Brenna JT. Novel fatty acid desaturase 3 (FADS3) transcripts generated by alternative splicing. Gene. 2009;446(1):28–34.
Blanchard H, Legrand P, Pédrono F. Fatty acid desaturase 3 (Fads3) is a singular member of the fads cluster. Biochimie. 2011;93(1):87–90.
Monroig Ó, Li Y, Tocher DR. Delta-8 desaturation activity varies among fatty acyl desaturases of teleost fish: high activity in delta-6 desaturases of marine species. Comp Biochem Physiol B Biochem Mol Biol. 2011;159(4):206–13.
He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial Neofunctionalization in duplicate gene evolution. Genetics. 2005;169(2):1157–64.
Tanomman S, Ketudat-Cairns M, Jangprai A, Boonanuntanasarn S. Characterization of fatty acid delta-6 desaturase gene in Nile tilapia and heterogenous expression in Saccharomyces cerevisiae. Comp Biochem Physiol B Biochem Mol Biol. 2013;166(2):148–56.
Kuah M-K, Jaya-Ram A, Shu-Chien AC. A fatty acyl desaturase (fads2) with dual Δ6 and Δ5 activities from the freshwater carnivorous striped snakehead Channa striata. Comp Biochem Physiol A Mol Integr Physiol. 2016;201(Supplement C):146–55.
Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res. 2006;45(3):237–49.
Monroig Ó, Lopes-Marques M, Navarro JC, Hontoria F, Ruivo R, Santos MM, Venkatesh B, Tocher DR, Castro LFC. Evolutionary functional elaboration of the Elovl2/5 gene family in chordates. Sci Rep. 2016;6:20510.
Monroig Ó, Rotllant J, Sanchez E, Cerda-Reverter JM, Tocher DR. Expression of long-chain polyunsaturated fatty acid (LC-PUFA) biosynthesis genes during zebrafish Danio rerio early embryogenesis. Biochim Biophys Acta. 2009;1791(11):1093–101.
Morais S, Monroig O, Zheng X, Leaver MJ, Tocher DR. Highly unsaturated fatty acid synthesis in Atlantic salmon: characterization of ELOVL5- and ELOVL2-like elongases. Mar Biotechnol (NY). 2009;11(5):627–39.
Gregory MK, James MJ. Rainbow trout (Oncorhynchus mykiss) Elovl5 and Elovl2 differ in selectivity for elongation of omega-3 docosapentaenoic acid. Biochim Biophys Acta. 2014;1841(12):1656–60.
Monroig Ó, Wang S, Zhang L, You C, Tocher DR, Li Y. Elongation of long-chain fatty acids in rabbitfish Siganus canaliculatus: cloning, functional characterisation and tissue distribution of Elovl5- and Elovl4-like elongases. Aquaculture. 2012;350–353:63–70.
Acknowledgements
The authors would like to thank Wataru Kawamura in the Tokyo University of Marine Science and Technology for sampling of Japanese eel and Jonathan Wilson for Senegal bichir, spotted gar and African butterfly fish samples.
Funding
We acknowledge Fundação para a Ciência e a Tecnologia for the support to M.L.-M (SFRH/BD/84238/2012). LFCC research is supported by Norte2020 and FEDER (Coral - Sustainable Ocean Exploitation - Norte-01-0145-FEDER-000036) a project supported by the North Portugal Regional Operational Program (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF). The funding bodies played no role in the design of the study and collection, analysis, interpretation of data, or in writing the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article and corresponding supplementary information files (Additional files 1 and 2).
Author information
Authors and Affiliations
Contributions
The original idea for this study was conceived by LFCC and OM. NK, BV, YQ and MLM performed all the experimental analysis; RR, MS and DRT participated in the discussion regarding lipid metabolism and physiology. The manuscript was drafted by MLM, LFCC and OM, and edited by all other co-authors. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
All animal tissue sampling procedures were evaluated and approved by CIIMAR Managing Animal Welfare Body (ORBEA) ethics committee, according to the European Union Directive 2010/63/EU.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Primer sets, corresponding PCR conditions. (DOCX 23 kb)
Additional file 2:
Figure S1. GC-MS Chromatograms of Fads functional characterisation. (PDF 1338 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lopes-Marques, M., Kabeya, N., Qian, Y. et al. Retention of fatty acyl desaturase 1 (fads1) in Elopomorpha and Cyclostomata provides novel insights into the evolution of long-chain polyunsaturated fatty acid biosynthesis in vertebrates. BMC Evol Biol 18, 157 (2018). https://doi.org/10.1186/s12862-018-1271-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-018-1271-5