Abstract
Background
The molecular bases explaining the diversity of dental tissue mineralization across gnathostomes are still poorly understood. Odontodes, such as teeth and body denticles, are serial structures that develop through deployment of a gene regulatory network shared between all gnathostomes. Dentin, the inner odontode mineralized tissue, is produced by odontoblasts and appears well-conserved through evolution. In contrast, the odontode hypermineralized external layer (enamel or enameloid) produced by ameloblasts of epithelial origin, shows extensive structural variations. As EMP (Enamel Matrix Protein) genes are as yet only found in osteichthyans where they play a major role in the mineralization of teeth and others skeletal organs, our understanding of the molecular mechanisms leading to the mineralized odontode matrices in chondrichthyans remains virtually unknown.
Results
We undertook a phylogenetic analysis of the SPARC/SPARC-L gene family, from which the EMPs are supposed to have arisen, and examined the expression patterns of its members and of major fibrillar collagens in the spotted catshark Scyliorhinus canicula, the thornback ray Raja clavata, and the clawed frog Xenopus tropicalis. Our phylogenetic analyses reveal that the single chondrichthyan SPARC-L gene is co-orthologous to the osteichthyan SPARC-L1 and SPARC-L2 paralogues. In all three species, odontoblasts co-express SPARC and collagens. In contrast, ameloblasts do not strongly express collagen genes but exhibit strikingly similar SPARC-L and EMP expression patterns at their maturation stage, in the examined chondrichthyan and osteichthyan species, respectively.
Conclusions
A well-conserved odontoblastic collagen/SPARC module across gnathostomes further confirms dentin homology. Members of the SPARC-L clade evolved faster than their SPARC paralogues, both in terms of protein sequence and gene duplication. We uncover an osteichthyan-specific duplication that produced SPARC-L1 (subsequently lost in pipidae frogs) and SPARC-L2 (independently lost in teleosts and tetrapods).Our results suggest the ameloblastic expression of the single chondrichthyan SPARC-L gene at the maturation stage reflects the ancestral gnathostome situation, and provide new evidence in favor of the homology of enamel and enameloids in all gnathostomes.
Similar content being viewed by others
Background
Dentin and enamel are found in serially developing skeletal organs called odontodes which include oral and pharyngeal teeth, dermal scales and denticles [1,2,3]. Stem-gnathostome fossils suggest that initially odontodes covered the body and were subsequently recruited to the mouth region [1, 2, 4, 5]. The early morphogenesis of all odontodes is initiated by similar epithelial-mesenchymal interactions and relies on a well-conserved genetic cascade in extant gnathostomes [6,7,8,9,10,11,12,13,14]. Dentine, the inner odontode matrix, is evolutionarily conserved in terms of tissue structure, while structural variation has been described for the odontode outer mineralized region, mostly known as enamel or enameloid. Enamel was originally described as a sarcopterygian-specific tissue exclusively produced by ameloblasts, devoid of collagen fibers, and clearly demarcated from the underlying dentin produced by odontoblasts [6, 15]. By contrast, enameloid is produced by odontoblasts (with or without an ameloblastic contribution), is continuous with dentin, and is characterized by variable degrees of collagen contents [6, 15, 16]. Furthermore, in caudate amphibian and teleost species harboring enameloid-covered odontodes, ameloblasts express type I fibrillar collagens [17,18,19,20].
A body of literature has led to two incompatible views of odontode evolution. On the one hand, it has been proposed that the osteichthyan last common ancestor harbored enameloid-covered odontodes [2, 21]. This is because (i) enameloid is present at the surface of stem gnathostome odontodes, chondrichthyan teeth and dermal denticles, actinopterygian oral and pharyngeal teeth, and caudate amphibian larval teeth [22,23,24,25] and (ii) teleost and chondrichthyan enameloids were proposed to be homologous [2, 21]. On the other hand, it has been inferred that the osteichthyan last common ancestor harbored enamel-covered odontodes [2, 26,27,28,29]. Indeed, evidence showing that (i) EMP (Enamel Matrix Proteins) genes are specifically expressed in ameloblasts during mammalian enamel matrix secretion and maturation, [30] and (ii) EMPs are present in the gar genome [4], suggest that the surface tissue of tooth and ganoid scales of non-teleost actinopterygians (polypterids and lepisosteids, [26, 27]) are homologous to the sarcopterygian enamel [4]. These data led to the prediction that type I fibrillary collagens will be expressed in chondrichthyan ameloblasts [2], together with EMP-related genes belonging to the SCPP (secretory calcium-binding phosphoproteins) gene family.
The SPARC (secreted protein acidic and rich in cysteine), SPARC-L1 (SPARC-Like1) and SCPP proteins are crucial extracellular matrix components involved in several major vertebrate innovations [17, 20, 31,32,33]. The classical evolutionary scenario posits that the SPARC-L1 and SPARC paralogues arose from two rounds of vertebrate-specific genome duplications [34, 35]. The gar and coelacanth SPARC-L1 genomic loci were recently shown to contain a SPARC-L2 gene of unknown evolutionary history [5, 36]. The SCPP gene family has as yet only been found in osteichthyan vertebrates, and is thought to have originated through a series of tandem duplications of the SPARC-Like1 (SPARC-L1) gene [5, 32, 37,38,39,40]. The SCPP family includes P/Q-rich protein genes (such as the EMPs) and acidic, bone and dentin protein genes also known as small integrin-binding ligand, N-linked glycoproteins (SIBLING) genes [5, 20, 34, 41, 42]. SPARC-L1 and SPARC are extracellular proteins harboring collagen-interacting and calcium-binding domains, thereby contributing to matrix deposition and mineralization [35, 43,44,45]. Accordingly, both SPARC and type I fibrillar collagen genes are highly expressed in osteichthyan odontoblasts [20, 43, 46,47,48]. All examined osteichthyan EMP and SIBLING members are expressed by odontoblasts and/or ameloblasts [17, 20, 49, 50], and code for extracellular regulators of dentin (e.g. Dmp-1, a SIBLING member) or enamel (e.g. Amtn and Enam, two EMP members) mineralization [51,52,53]. A high rate of gene gain, loss and divergence has dramatically modified the SCPP repertoire in distinct osteichthyan lineages, blurring orthology relationships [5, 17, 20, 32,33,34, 36, 54]. Nevertheless, a few conserved EMP genes can unambiguously be identified in sarcopterygians and actinopterygians, as is the case of Enam and Ambn [5, 36, 41]. Changes in the SCPP gene content led to proposals that this causes odontode matrix structural variations in osteichthyans [17, 20, 34]. However, no SCPP gene has been detected in the elephant shark genome so far, which only seems to harbor putative SPARC and SPARC-L1 orthologues [55].
In this study, we sought to improve understanding of jawed vertebrate odontode evolution by (i) clarifying the evolutionary relationships between SPARC, SPARC-L1 and SPARC-L2, (ii) determining whether or not members of the SPARC and SPARC-L gene families are specifically expressed by ameloblasts and/or by odontoblasts in chondrichthyans, using embryos from the spotted catshark Scyliorhinus canicula (S.c.) and the thornback ray Raja clavata (R.c.), (iii) evaluating whether type I fibrillar collagens are expressed in ameloblasts in S.c., R.c. and in the osteichthyan Xenopus tropicalis (X.t.), and (iv) examining SPARC gene expression in odontodes of a species lacking both SPARC-L1 and SPARC-L2, an extremely rare situation which has so far only been reported in the Xenopus genus.
Results
Reconstruction of SPARC/SPARC-L1/−L2 gene phylogeny
We performed phylogenetic analyses of the SPARC and SPARC-L1/−L2 homologous protein sequences obtained in public databases, as well as the 13 novel sequences identified in jaw transcriptomes [56], including two isolated transcripts from the lesser spotted catshark and the thornback ray (Additional file 1). As outgroups sequences, we used the orthologous protein from two amphioxus species, and the lamprey SPARC-A and SPARC-B sequences (Additional file 1). Our alignment allowed phylogenetic reconstructions based on 235 amino-acid positions (Fig. 1 and Additional file 2: a-c). Branch supports were higher when the amphioxus sequences were excluded from the analyses, both for Bayesian (compare Fig. 1 and Additional file 2: a) and Maximum Likelihood reconstructions (compare Additional file 2: b and c). All phylogenetic reconstructions led to a maximal support of a SPARC clade (Fig. 1 and Additional file 2: a-c) including both the chondrichthyan and the osteichthyan SPARC sequences. A lesser resolution within the osteichthyan SPARC sequences was probably due to a lack of phylogenetic signal at this level of the tree.
Phylogenetic relationships of the vertebrate SPARC and SPARC-L gene families. Bayesian inference was run with Phylobayes and the site-heterogeneous CAT+Γ4 model of sequence evolution. The tree was rooted with the lamprey SPARC-A and SPARC-B sequences. Posterior probabilities are indicated on the branches
The evolutionary history of the gnathostome SPARC-L1/−L2 clade was more difficult to decipher from these analyses (Fig. 1). Indeed, in addition to SPARC-L1, both the spotted gar and coelacanth harbored a second paralogue which had previously been named “SPARCL2”, “SPARCL1-like”, “SPARCL1L1” or “SPARCL1A” [5, 32, 35, 36]. In agreement with Qu et al. [5] and based on the phylogenetic arguments described below, we referred to these gar and coelacanth long-branch copies as “SPARC-L2”, in opposition to the osteichthyan SPARC-L1 sequences which were found in more species and comparatively evolved at a slower rate (Fig. 1). Here we identified a new SPARC-L2 sequence from the bichir (Polypterus senegalus) jaw transcriptomic data (Fig. 1). The monophyly of the osteichthyan SPARC-L2 group was robust in all analyses (Fig. 1 and Additional file 2: a-c). This SPARC-L2 clade consistently clustered with the osteichthyan SPARC-L1 group in Bayesian reconstructions, albeit with medium robustness (posterior probability 0,79, see Fig. 1, also Additional file 2: a). Both ML trees were congruent with this clade, but with weak support (Additional file 2: b-c). In all analyses, the catshark, thornback ray, whale shark and elephant shark homologues clustered together as the sister group to the osteichthyan SPARC-L1/SPARC-L2 clade, although with medium to weak support (Fig. 1 and Additional file 2: a-c). This topology strongly supports the idea that an osteichthyan-specific gene duplication led to the SPARC-L1 and SPARC-L2 copies, which directly implies that the use of the SPARC-L1 name should be avoided for the chondrichthyan genes. We therefore respectively named Sc-SPARC-L and Rc-SPARC-L the spotted catshark and thornback ray genes related to the osteichthyan clade that groups the SPARC-L1 and SPARC-L2 genes together.
Expression patterns of SPARC and SPARC-L genes in developing teeth and dermal denticles
SPARC and SPARC-L gene expression was studied in the lesser spotted catshark (S.c.) and thornback ray (R.c.) odontodes (Fig. 2). SPARC and Enam (an EMP gene) expression was studied in the frog (X.t., Fig. 3).
Histology and gene expression in the developing odontodes of Scyliorhinus canicula and Raja clavata. Lower jaw longitudinal sections (anterior, left; dorsal, up) of a 20 cm long S.c. juvenile (a), a 9 cm long S.c. embryo (b-f), and a 9 cm long R.c embryos (m-r), revealing the presence of tooth series at the maturation stage as well as less developed secretory stage tooth bud harboring columnar ameloblasts (black arrowhead). Thoracic transverse sections are shown for of 6 cm (g-l) and 7 cm (g’-l’) long S.c. embryos, focusing on developing primary dorsal dermal denticles at late morphogenesis and late maturation stage, respectively (dorsal to the top). Sections were stained with HES (a, g, g’, m) or in situ hybridized against SPARC (b, h, h’, n), SPARC-L (c, i, i’, o), Col1a1 (d, j, j’, p), Col1a2 (e, k, k’, q) and Col2a1-L (f, l, l’, r). The asterisks locate the mineralized matrix in teeth and denticles at the late mineralization stage, separating the ameloblasts (Am, located by the dashed lines) from the odontoblasts (Od, delineated by the orange line). MC: Meckel. The scale bar in (a) represents 100 μm in (a-f, m-r), and the scale bar in (g) represents 50 μm in (g-l’)
Histology and gene expression in developing teeth of Xenopus tropicalis. Longitudinal sections of the X.t. upper jaw at the NF57 developmental stages were stained with HES (a, g) or processed by in situ hybridization for the indicated genes (b-f and h-l). Ameloblasts and odontoblasts are delineated by black dotted lines or by an orange line, respectively. The asterisks locate the mineralized matrix. The scale bars in (a) and (g) respectively represent 20 μm in (a-f) and (g-l)
In the catshark, the early stage of dentin and enameloid matrix synthesis could be identified by the presence of columnar ameloblasts at the tooth and denticle surface (secretory stage: Fig. 2a and g), while later tooth and denticle buds were covered with cubic ameloblasts facing significant amount of mineralized matrix (maturation stage: Fig. 2a and g’). At all developmental stages, the Sc-SPARC expression was restricted to odontoblasts in both teeth and dermal denticles (Fig. 2b, h, h’). In contrast, Sc-SPARC-L was expressed only in the ameloblasts in both developing teeth and dermal denticles at the maturation stages (Fig. 2c and i’). At the earlier secretory stage, ameloblasts did not display any detectable expression of the SPARC-L transcripts (Fig. 2c and i).
In the thornback ray, most dermal denticles develop as massive dermal thorns, making it technically challenging to obtain denticle sections at different stages (but see ref. [57]). We therefore focused on tooth development only. Longitudinal sections of the thornback ray lower jaw showed developing teeth at the secretory and maturation stages, similar to the catshark observations (compare Fig. 2a and m). We detected a faint Rc-SPARC expression in the tooth bud mesenchyme (Fig. 2n). In contrast, Rc-SPARC-L was clearly expressed in the inner epithelial cell-layer (ameloblasts), exclusively at the maturation stage when secretion was well advanced and ameloblasts have lost their secretory morphology (Fig. 2o).
To date, only the Xenopus genus (X. laevis and X. tropicalis) has been reported to have lost both the SPARC-L1 and SPARC-L2 paralogues, a unique situation amongst osteichthyans [20]. In Xenopus tropicalis, SPARC expression has previously been described during embryogenesis and osteogenesis, but not during odontogenesis [35, 58]. To evaluate whether the SPARC-L1 and SPARC-L2 loss was accompanied by compensatory mechanisms that might have involved changes in the expression domain of other related genes, we examined SPARC expression in developing teeth of Xenopus tropicalis. We find that, at stages NF57 and NF59, Xt-SPARC expression was restricted to the odontoblasts (Fig. 3a, b, g, h). For the sake of comparison with chondrichthyan SPARC-L expression, we chose to also examine the expression pattern of Enam, an EMP gene, whose expression had not been reported in Xenopus so far. We showed that Xt-Enam is expressed specifically in the ameloblasts at both developmental stages (Fig. 3c, i).
Expression patterns of type I and II collagen genes in developing teeth and dermal denticles
A phylogeny of the type I and II fibrillar collagen families including chondrichthyan sequences was previously published, leading to the unambiguous identification of the Col1a1, Col1a2 and Col2a1 paralogues used in the present study [59]. In the catshark, Sc-Col1a1, Sc-Col1a2 transcripts were robustly expressed in secretory odontoblasts, both in developing teeth (Fig. 2d-f) and in dermal denticles at the late morphogenesis stage (Fig. 2j-l’). Sc-Col2a1 expression was weak in mineralizing tooth buds and undetectable in denticle buds (Fig. 2f, l, l’). Sc-Col2a1 was also faintly expressed in early tooth buds ameloblasts (Fig. 2f, arrowhead).
In the thornback ray, Rc-Col1a1 and Rc-Col1a2 were strongly expressed in putative odontoblasts (Fig. 2p-q). Rc-Col2a1 was expressed at much lower levels in secretory ameloblasts at early developmental stage of tooth bud development (Fig. 2q, arrowhead).
The absence of expression of type I fibrillar collagen genes in the ameloblasts of these two chondrichthyans stands in sharp contrast with the situation observed in other species with enameloid-covered teeth, such as teleosts, and salamander larvae [17, 18, 20]. Mammalian ameloblasts synthesize an enamel matrix (not enameloid) and are known to be devoid of fibrillar collagen expression. In order to polarize evolutionary change, we examined Xenopus tropicalis also with enamel-producing ameloblasts, and found that both Xt-Col1a1 and Xt-Col1a2 transcripts were strongly expressed by odontoblasts, yet clearly excluded from ameloblasts, at two distinct stages of tooth development (Fig. 3d, e, j, h). No expression could be detected in developing tooth buds for Xt-Col2a1 (Fig. 3f, l), while a signal was observed in the head cartilage present on the same section and used as an internal positive control (not shown).
Discussion
Evolution of the SPARC-L family
Our phylogenetic analyses suggest a new evolutionary scenario for the expansion of the SPARC-L gene family in gnathostomes. Recent genomic data have shown that SPARC-L1 and SPARC-L2 are located close to each other in the gar genome, and that the synteny around both genes is conserved between gar and coelacanth [5, 32, 36]. Here we identified SPARC-L2 in the bichir jaw transcriptome, but this gene is absent in either tetrapod or teleost genomes. Therefore, SPARC-L2 has probably been independently lost early in the teleost and tetrapod lineages. We could not identify any SPARC-L2 sequence in the transcriptome of Lepidosiren paradoxa or Protopterus aethiopicus, suggesting that this copy was lost in the last common ancestor of Dipnoi and Tetrapods. Except in pipidae frogs, SPARC-L1 is identified in all available osteichthyan genomes. This suggests osteichthyans ancestrally exhibited a repertoire of at least three SPARC/SPARC-L paralogues: SPARC which is present in all species examined to date, and SPARC-L1 and SPARC-L2 which were independently lost in distinct lineages (illustrated in Fig. 4). In chondrichthyans, we identified only two genes in the available transcriptomics and genomics data: the SPARC orthologue and one paralogue which we refer here as SPARC-L (formerly coined SPARC-L1 [60]). In the elephant shark genome, SPARC-L is located in a region of conserved synteny compared to the osteichthyan SPARC-L1/SPARC-L2 locus [5, 36, 61]. Our phylogenetic analyses support the notion that the chondrichthyan SPARC-L clade is the sister-group to the osteichthyan SPARC-L1/SPARC-L2 clade (Fig. 1), implying that SPARC-L1 and SPARC-L2 arose through an osteichthyan-specific tandem duplication of an ancestral gnathostome SPARC-L gene. The chondrichthyan SPARC-L is therefore co-orthologous to the osteichthyan SPARC-L1 and SPARC-L2 genes. As a consequence, the chondrichthyan gene is referred to as “SPARC-L” forthwith, as this nomenclature best reflects the evolutionary history of the family (Figs. 1 and 4). In this scenario, the SPARC and SPARC-L paralogues arose during the two rounds of vertebrate-specific whole genome duplications.
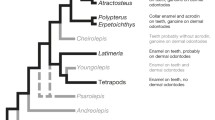
Favored model for the evolution of gene content and ameloblastic expression in gnathostomes. A cladogram shows the classically accepted phylogenetic relationships of Scyliorhinus canicula (S.c.), Raja clavata (R.c.), Lepisosteus oculatus (L.o.), Danio rerio (D.r.), Takifugu rubripes (T.r.), Latimeria chalumnae (L.c.), Xenopus tropicalis (X.t.) and Mus musculus (M.m.), as well as putative ancestral situations and polarized evolutionary changes. Pentagons summarize the gene content identified in each species and inferred to have existed in their last common ancestor. Ameloblastic gene expression status is summarized as transcribed (blue), undetected (white) or unknown (grey). Dotted arrows represent unresolved ambiguities with respect to the origin of the SCPP members. See text for details and references
Longer branches reveal that the SPARC-L/SPARC-L1/SPARC-L2 proteins evolve faster than the SPARC sequences, potentially interfering with phylogeny reconstruction and leading to weaker node support (Fig. 1 and Additional file 2). Hence, an alternative hypothesis to the aforementioned scenario is that SPARC-L is a highly derived SPARC-L1 or SPARC-L2 copy, which duplication was followed by the loss of the other duplicate in all chondrichthyan lineages. Nevertheless, the tree topology, the synteny data, and the presence of a single SPARC-L gene in all chondrichthyan transcriptomes and genomes examined to date provide sound arguments in favor of SPARC-L1 and SPARC-L2 being of an osteichthyan origin. Interestingly, an accelerated rate of evolution is a shared feature of the SPARC-L/SPARC-L1/SPARC-L2 and SCPP genes [17, 34, 42]. We therefore propose the term “SPARC-L gene family” is used to refer to the full repertoire of gnathostome SPARC-L, SPARC-L1, SPARC-L2 and SCPPs, since these are all derived from a single ancestral gnathostome SPARC-L gene. We can conclude that duplicates in the SPARC-L family: (1) are found at a single locus of conserved synteny between all examined gnathostome genomes, (2) encode rapidly diverging protein sequences (when compared to their SPARC orthologues), and (3) have been prone to frequent independent gene gains and losses in osteichthyans.
A conserved collagen/SPARC module
We show that odontoblasts co-express SPARC, Col1a1, Col1a2 and Col2a1 in catshark teeth and dermal denticles, and in thornback ray teeth, thereby supporting the consistency of this co-expression in selachians and the serial nature of chondrichthyan odontodes [9,10,11]. SPARC, Col1a1 and Col1a2 are also co-expressed in X.t. odontoblasts (this study), as previously reported in teleost and mammalian representatives [17, 20, 47]. These observations confirm the classical hypothesis that the dentin is homologous across jawed vertebrates, and we therefore propose that SPARC and type I collagen genes are final targets of the odontoblastic regulatory network active in teeth and dermal denticles [9, 14].
In contrast, while the absence of any detectable ameloblastic expression of the collagen/SPARC module in S.c., R.c. and X.t. is strikingly similar to the situation reported in mammals [47], it is markedly different from the fugu, zebrafish and salamander expression patterns [17, 18, 20]. Based on this, we propose that, in the gnathostome last common ancestor, the ameloblasts did not express the collagen/SPARC module, and that this character (absence of expression) was subsequently modified in distinct lineages. One possible scenario is that the collagen/SPARC ameloblastic expression originated in the osteichthyan last common ancestor, and that at least two secondary losses occurred, in mammals and in the clawed frog lineage after it separated from salamanders (three evolutionary events). However, in this study, we favor a more parsimonious scenario (Fig. 4) according to which the collagen/SPARC module was recruited twice in odontoblasts, in teleosts and salamander [17, 18, 20].
SPARC-L expression supports the homology of the odontode outer layer
In mice, SPARC-L1 is broadly expressed and dispensable for development [62,63,64], and to the best of our knowledge, no detailed SPARC-L1 and/or SPARC-L2 expression patterns have been reported in the odontodes of any osteichthyan species. Therefore, the ameloblast-specific expression of the SPARC-L gene in elasmobranch teeth and dermal denticles is remarkable and strikingly similar to some of the osteichthyan SCPP members. Indeed, we find that in X.t., in which both SPARC-L1 and SPARC-L2 have been lost, the EMP gene Enam is exclusively expressed in ameloblasts, which is comparable to the previously reported expression of Amel and Odam in Xenopus [20] and of Amel and Amtn in salamander [18, 65]. There are at least two hypotheses that might account for these ameloblast-specific expression patterns: (i) that they follow two convergent acquisitions in chondrichthyans (for SPARC-L) and osteichthyans (for SCPPs), or (ii) that the chondrichthyan SPARC-L and the osteichyan SCPP genes have inherited their ameloblatic-specific expression from the ancestral SPARC-L gene. The latter hypothesis (shown in Fig. 4) is well supported if we assume that the osteichthyan SCPP genes (particularly the EMP members) have largely taken over the function ancestrally fulfilled by SPARC-L. Hence, the SCPP emergence may have rendered the SPARC-L1/−L2 proteins dispensable for enamel mineralization and other skeletal tissues. This strong redundancy might have allowed the occurrence of independent gene loss and of decreased expression levels that affected the SPARC-L1/−L2 genes in distinct osteichthyan lineages.
SPARC-L is expressed during the enameloid late maturation stage in both examined chondrichthyans, which is similar to the timing of Amtn, Odam and Scppq1 expression (all being SCPP genes) in various tetrapods ameloblasts [20, 65,66,67,68,69]. Hence, the ancestral gnathostome SPARC-L gene might first have been involved in the maturation of the outer odontode layer, without necessarily contributing to its protein composition per se. In this respect, the role of EMPs as the major architectural proteins of the enamel matrix, best exemplified by the more recent emergence of the Amel gene, may represent a sarcopterygian-specific evolutionary innovation [41]. Based on our results, we propose that all odontode external layers are homologous, and that secondary modifications (e.g. shifts in the expression timing of EMP genes in sarcopterygians, recruitment of the collagen/SPARC module in teleost and salamander ameloblasts) have led to a variety of derived structures known today as enamel, enameloid, ganoine or acrodin. In the future, describing SPARC-L1 and SPARC-L2 expression patterns in odontodes of non-tetrapod species, and unravelling the localization and function of chondrichthyan SPARC-L proteins during the mineralization process, will represent a major breakthrough in our understanding of vertebrate odontode evolution.
Conclusions
We show here that the SPARC gene is conserved in all gnathostomes examined to date and that the chondrichthyan SPARC-L gene is co-orthologous to the osteichthyan SPARC-L1 and SPARC-L2 genes. We introduce the “SPARC-L gene family” nomenclature to describe the full repertoire of gnathostome SPARC-L, SPARC-L1, SPARC-L2 and SCPP members, which are all derived from an ancestral gnathostome SPARC-L gene.
A highly conserved odontoblastic collagen/SPARC module confirms gnathostome dentin homology. The exclusion of the collagen/SPARC module from ameloblasts is a shared feature of the amphibian and the two elasmobranch species examined here, and of mammals. In addition, the ameloblastic expression of the single SPARC-L gene in chondrichthyans, similar to the EMPs in osteichthyans, probably best reflects the ancestral gnathostome situation. Taken together, our results provide new evidence in favor of a more general definition of the odontode external layer and support its homology in all gnathostomes.
Methods
Transcriptomic data
Sequences belonging to the SPARC and SPARC-L1/−L2 gene families were obtained by screening jaw transcriptomes (Illumina sequencing, see [56] for taxon sampling details and reference to SRA IDs). These available transcriptomic data were assembled at ISEM-Montpellier (France) and subsequently screened using BLAST on the Montpellier Bioinformatics Biodiversity (MBB) platform (http://mbb.univ-montp2.fr/MBB//index.php). The species chosen in these transcriptomic data were: the catshark Scyliorhinus canicula (late embryonic stage 33, 9 cm total length (TL) and 20 cm TL juvenile); the thornback ray Raja clavata (late embryonic stage 33, courtesy of the Centre de découverte de la pêche en Mer (Maréis), Etaples sur Mer, France); the salamander Pleurodeles waltl (6 month-old, reared in J-Y S. lab facility); two lungfishes: Lepidosiren paradoxa (juvenile, 10.0 cm TL, pet shop, Challet Herault) and Protopterus aethiopicus (juvenile, 12.0 cm TL, pet shop Challet Herault); the bichir Polypterus senegalus (2 year-old, reared in J-Y S. lab facility); the bowfin Amia calva (juvenile, 10.0 cm TL, pet shop, Challet Herault); the sturgeon Acipenser baerii (larvae, Sturgeon SCEA farm, France) and two gymnophionans Typhlonectes compressicauda and Rhinatrema bivittatum (juveniles, courtesy of Philippe Gaucher, CNRS French Guiana).
Reconstruction of the SPARC/SPARC-L1/−L2 phylogeny
Sequences of the SPARC and SPARC-L gene families were obtained as first hits after similarity analyses (BLAST) using SPARC and SPARC-L1 sequences of the mouse transcripts (NCBI references: BC004638.1 and NM_010097.4 respectively) against all available transcriptomes. New nucleotide sequences of the SPARC and SPARC-L gene families were deposited in Genbank under the IDs: MH206590 (Lepidosiren paradoxa SPARC), MH206591 (Pleurodeles waltl SPARCL1), MH206592 (Pleurodeles waltl SPARC), MH206593 (Protopterus aethiopicus SPARC), MH206594 (Polypterus senegalus SPARCL2), MH206595 (Polypterus senegalus SPARC), MH206596 (Typhlonectes compressicauda SPARC), MH206597 (Rhinatrema bivittatum SPARC), MH206598 (Scyliorhinus canicula SPARCL), MH206599 (Raja clavata SPARC), MH206600 (Raja clavata SPARCL), MH206601 (Acipenser baerii SPARCL1), and MH206602 (Amia calva SPARCL1). Phylogenetic analyses were performed to infer the evolutionary history of these two gene families and provide orthology assignments. Protein sequences coming from public databases and the jaw transcriptomic data (see Additional file 1) were aligned using MAFFT [70]. Members of the SCPP gene family were not included as they only share their 5′ sequence with SPARC-L1, which includes a small coding region. Regions of ambiguous sequence homology were removed with GBlocks [71] (−b1 21 -b2 21 -b3 16 -b4 5 -b5 half), generating a 235 amino-acid alignment in the complete set of sequences (Additional file 3) that has been analyzed without (Fig. 1 and Additional file 2: b) or with (Additional file 2: a, c) the amphioxus sequences. Both datasets were used to reconstruct gene phylogenies with either Bayesian or Maximum Likelihood approaches (Additional file 2). Bayesian inferences were run using Phylobayes 4,1 [72] with the site-heterogeneous CAT+Γ4 model of sequence evolution. Two independent chains were run for 10,000 cycles, and convergence was checked using pbcomp and tracecomp programs. A burnin of 50% was used to obtain the consensus tree (Fig. 1, Additional file 2: a). Maximum Likelihood analyses were run using IQ-TREE [73] with site-heterogeneous C20 + R4 + F model of sequence evolution. 100 bootstraps were performed and then mapped onto the best Maximum Likelihood tree (Additional file 2: b, c). Both models take into account the site-heterogeneity of substitution processes, which is particularly important given the peculiar characteristics of SPARC sequences (with highly conserved cysteine positions). Reconstructions were rooted with the amphioxus sequences when included (Additional file 2: a, c) or with the lamprey sequences (Fig. 1 and Additional file 2: b).
Specimens, cryosections and histological sections
Lesser spotted catshark (Scyliorhinus canicula) embryos were obtained in Montpellier from the Station Méditerrannéenne de l’Environnement Littoral, Sète, France. Thornback ray (Raja clavata) embryos were kindly provided by the Centre de découverte de la pêche en Mer (Maréis), Etaples sur Mer, France. Embryos were raised in a smell tank at 18 °C until they reached the proper stages of tooth development (stage 33 as described in [74, 75]). Embryos were taken off the eggshell and euthanized following all European animal-care specifications, by overdose of MS-222 (Sigma), then fixed 48 h in PFA 4% in PBS 1× at 4 °C. They were finally transferred and stored in ethanol at − 20 °C.
Adult Xenopus tropicalis are maintained at the University of Concepcion, following standard protocols established for this species. Embryos and tadpoles were raised after natural mating and staged according to the Nieuwkoop and Faber developmental table [76]. Anesthesia of tadpoles was performed with a MS-222 (Sigma) solution at 200 mg/mL and specimen were subsequently decapitated in agreement with international bioethical recommendations [77, 78].
Catshark, thornback ray, and frog specimens were embedded in wax. 7 μm-thick histological sections were cut and then stained with standard protocols (eosin, hematoxylin and safran reaction for catshark and thornback ray sections (RHEM platform at IRCM, Montpellier); hematoxyline and chromotrope 2R (C3143 Sigma) for frog sections). Clawed frog in situ hybridizations were made on 7 μm thick paraffin sections of the upper jaw oriented longitudinally. Spotted catshark and thornback ray in situ hybridizations were performed on 14 μm thick cryostat sections (para-sagitally in lower jaws and transversally in the body trunk). Parts of the specimens that were not used for this study were conserved in ethanol at − 20 °C for further studies on gene expression.
DIG-labeled riboprobe synthesis
Identified thornback ray Col1a1, Col1a2, Col2a1, SPARC and SPARC-L1 sequences were used to design primers and amplify selected sequences from a cDNA extract obtained from the jaw of a stage 33 embryo (primer sequence given in Additional file 4). PCR products were ligated into the pGEM-Teasy vector using the TA cloning kit (Promega). Identified catshark SPARC and SPARC-L sequences were used to screen a cDNA library of catshark embryo RNA extracts [79] and one clone for each gene was isolated. Xt-Col1a1, Xt-Col1a2, Xt-Col2a1, and Xt-SPARC (GenBank NM_001011005.1; NM_001079250.1; NM_203889 and AY575077, respectively) were amplified from stage NF60 calvaria cDNA. Xt-Enam (GenBank NM_001145743) was amplified from stage NF60 upper jaws. All PCR products were blunt-cloned into the pBluescript vector (see Additional file 4 for primers). All inserts were amplified using primers present in the vector backbone or in the insert (see Additional file 4 for internal primers), and PCR products were used as a template to synthesize antisense DIG riboprobes in a 3 μl reaction containing 100–200 ng PCR product and using the DIG RNA labeling mix (Roche) and T7 or Sp6 RNA polymerase (Promega) following manufacturer’s instructions. DIG-labelled riboprobes were purified on MicroSpin G50 column (GE Healthcare).
In situ hybridization and image processing
In situ hybridization of xenopus, catshark and thornback ray jaw sections were performed as described previously [59]. Images of the catshark in situ hybridization and histological staining were taken under a Hamamatsu NanoZoomer 2.0-HT Slide Scanner (Montpellier RIO Imaging facility, INM Optique) for the analysis of gene expression patterns. Slides were first scanned with a 20× objective and selected sections were scanned again with a 40× objective when necessary.
Abbreviations
- EMP:
-
Enamel Matrix Proteins
- R.c. :
-
Raja clavata
- S.c. :
-
Scyliorhinus canicula
- SCPP:
-
Secretory Calcium-binding PhosphoProtein
- SIBLING:
-
Small Integrin-Binding LIgand, N-linked Glycoproteins
- SPARC:
-
Secreted Protein Acidic and Rich in Cysteine
- X.t. :
-
Xenopus tropicalis
References
Donoghue PCJ, Rucklin M. The ins and outs of the evolutionary origin of teeth. Evol. Dev. 2016;18:19–30.
Sire J-Y, Donoghue PCJ, Vickaryous MK. Origin and evolution of the integumentary skeleton in non-tetrapod vertebrates. J Anat. 2009;214:409–40.
Sire JY. Teeth outside the mouth in teleost fishes: how to benefit from a developmental accident. Evol. Dev. 2001;3:104–8.
Qu Q, Sanchez S, Blom H, Tafforeau P, Ahlberg PE. Scales and tooth whorls of ancient fishes challenge distinction between external and oral “teeth”. PLoS One. 2013;8:e71890.
Qu Q, Haitina T, Zhu M, Ahlberg PE. New genomic and fossil data illuminate the origin of enamel. Nature. 2015;526:108–11.
Ørvig T. Phylogeny of tooth tissue: evolution of some calcified tissues in early vertebrates. In: Miles AEW, editor. Struct. Chem. Organ. Teeth. New-York and London: Academic Press; 1967. p. 45–110.
Reif W-E. Evolution of dermal skeleton and dentitions in vertebrates: the odontode-regulation theory. Evol Biol. 1982;15:287–368.
Borday-Birraux V, Van der Heyden C, Debiais-Thibaud M, Verreijdt L, Stock DW, Huysseune A, et al. Expression of Dlx genes during the development of the zebrafish pharyngeal dentition: evolutionary implications. Evol. Dev. 2006;8:130–41.
Debiais-Thibaud M, Oulion S, Bourrat F, Laurenti P, Casane D, Borday-Birraux V. The homology of odontodes in gnathostomes: insights from dlx gene expression in the dogfish, Scyliorhinus canicula. BMC Evol Biol. 2011;11:307.
Debiais-Thibaud M, Chiori R, Enault S, Oulion S, Germon I, Martinand-Mari C, et al. Tooth and scale morphogenesis in shark: an alternative process to the mammalian enamel knot system. BMC Evol Biol BMC Evolutionary Biology. 2015;15:292.
Debiais-Thibaud M, Germon I, Laurenti P, Casane D, Borday-Birraux V. Low divergence in Dlx gene expression between dentitions of the medaka (Oryzias latipes) versus high level of expression shuffling in osteichtyans. Evol. Dev. 2008;10:464–76.
Hecht J, Stricker S, Wiecha U, Stiege A, Panopoulou G, Podsiadlowski L, et al. Evolution of a core gene network for skeletogenesis in chordates. PLoS Genet. 2008;4:e1000025.
Smith MM, Johanson Z, Butts T, Ericsson R, Modrell M, Tulenko FJ, et al. Making teeth to order: conserved genes reveal an ancient molecular pattern in paddlefish (Actinopterygii). Proc R Soc B Biol Sci. 2015;282
Gillis JA, Alsema EC, Criswell KE. Trunk neural crest origin of dermal denticles in a cartilaginous fish. Proc Natl Acad Sci. 2017;114:201713827.
Poole DFG. Poole1967.pdf. In: Miles EW, editor. Struct Chem Organ teeth. New-York and London: Academic press; 1967. p. 111–49.
Robinson C, Brookes SJ, Shore RC, Kirkham J. The developping enamel matrix: nature and function. Eur J Oral Sci. 1998;106:282–91.
Sire J-Y, Géraudie J, Zylberberg L, Meunier FJ, Zylberberg L. On the origin of ganoine: histological and ultrastructural data on the experimental regeneration of the scales of Calamoichthys calabaricus (Osteichthyes, Brachyopterygii, Polypteridae). Am J Anat. 1987;180:391–402.
Owen R. Odontography or a treatise on the comparative anatomy of the teeth, their physiological relations, mode of development, and microscopic structure in the vertebrate animals. London: Hippolyte Bailliere; 1845.
Zivile Z, Richter M, Karatajute-Talimaa V, Smith MM. Tissue diversity and evolutionary trends of the dermal skeleton of Silurian thelodonts. Hist Biol An Int J Paleobiol. 2013;25:143–54.
Assaraf-Weill N, Gasse B, Silvent J, Bardet C, Sire J-Y, Davit-Béal T. Ameloblasts express type I collagen during amelogenesis. J Dent Res. 2014;93:502–7.
Davit-Béal T, Chisaka H, Delgado S, Sire J-Y. Amphibian teeth: current knowledge, unanswered questions, and some directions for future research. Biol Rev. 2007;82:49–81.
Sire J.-Y. Light and TEM study of nonregenerated and experimentally regenerated scales of Lepisosteus oculatus (Holostei) with particular attention to ganoine formation. Anat Rec. 1994;240:189–207.
Ru M, Donoghue PCJ, Ru M. Romundina and the evolutionary origin of teeth. Biol Lett. 2015;11:1–5.
Sire J-Y. Ganoine formation in the scales of primitive actinopterygian fishes, lepisosteids and polypterids. Connect Tissue Res. 1995;33:213–22.
Donoghue PCJ, Sansom IJ. Origin and early evolution of vertebrate skeletonization. Microsc Res Tech. 2002;59:352–72.
Kawasaki K. The SCPP gene repertoire in bony vertebrates and graded differences in mineralized tissues. Dev Genes Evol. 2009;219:147–57.
Kawasaki K, Suzuki T, Weiss KM. Phenogenetic drift in evolution: the changing genetic basis of vertebrate teeth. Proc Natl Acad Sci U S A. 2005;102:18063–8.
Huysseune A, Takle H, Soenens M, Taerwe K, Witten PE. Unique and shared gene expression patterns in Atlantic salmon (Salmo salar) tooth development. Dev Genes Evol. 2008;218:427–37.
Sasagawa I, Oka S, Mikami M, Yokosuka H, Ishiyama M, Imai A, et al. Immunohistochemical and western blotting analyses of Ganoine in the ganoid scales of Lepisosteus oculatus : an Actinopterygian fish. J. Exp. Zool. Part B Mol. Dev. Evol. 2016;326:193–209.
Gillis AJ, Donoghue PCJ. The homology and phylogeny of Chondrichthyan tooth Enameloid. J Morphol. 2007;268:33–49.
Kawasaki K, Buchanan AV, Weiss KM. Biomineralization in humans: making the hard choices in life. Annu Rev Genet. 2009;43:119–42.
Kawasaki K, Mikami M, Nakatomi M, Braasch I, Batzel P, Postlethwait JH, et al. SCPP genes and their relatives in gar: rapid expansion of mineralization genes in Osteichthyans. J. Exp. Zool. Part B Mol. Dev. Evol. 2017;328:645–65.
Kawasaki K, Lafont A-G, Sire J-Y. The evolution of milk casein genes from tooth genes before the origin of mammals. Mol Biol Evol. 2011;28:2053–61.
Kawasaki K, Buchanan AV, Weiss KM. Gene duplication and the evolution of vertebrate skeletal mineralization. Cells Tissues Organs. 2007;186:7–24.
Bertrand S, Fuentealba J, Aze A, Hudson C, Yasuo H, Torrejon M, et al. A dynamic history of gene duplications and losses characterizes the evolution of the SPARC family in eumetazoans. Proc R Soc B Biol Sci. 2013;280:20122963.
Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. 2016;48(4):427–37.
Ryll B, Sanchez S, Haitina T, Tafforeau P, Ahlberg PE. The genome of Callorhinchus and the fossil record: a new perspective on SCPP gene evolution in gnathostomes. Evol. Dev. 2014;16:123–4.
Rücklin M, Donoghue PCJ, Cunningham JA, Marone F, Stampanoni M. Developmental paleobiology of the vertebrate skeleton. J Paleontol. 2014;88:676–83.
Donoghue PCJ, Sansom IJ, Downs JP. Early evolution of vertebrate skeletal tissues and cellular interactions, and the canalization of skeletal development. J Exp Zool B Mol Dev Evol. 2006;306B:278–94.
Venkatesh B, Lee AP, Ravi V, Maurya AK, Korzh V, Lim ZW, et al. On the origin of SCPP genes. Evol Dev. 2014;16:125–6.
Kawasaki K, Amemiya CT. SCPP genes in the coelacanth: tissue mineralization genes shared by sarcopterygians. J. Exp. Zool. Part B Mol. Dev. Evol. 2014;322:390–402.
Kawasaki K. Evolutionary genetics of vertebrate tissue Mineralization : the origin and evolution of the secretory calcium-binding phosphoprotein family. J Exp Zool Part B Mol Dev Evol. 2006;306B:295–316.
Brekken RA, Sage EH. SPARC, a matricellular protein: at the crossroads of cell–matrix communication. Matrix Biol. 2001;19:815–27.
Martinek N, Shahab J, Saathoff M, Ringuette M. Haemocyte-derived SPARC is required for collagen-IV-dependent stability of basal laminae in Drosophila embryos. J Cell Sci. 2011;124:670.
Sullivan MM, Barker TH, Funk SE, Karchin A, Seo NS, Hook M, et al. Matricellular Hevin regulates Decorin production and collagen assembly. J Biol Chem. 2006;281:27621–32.
Bleicher F, Couble ML, Farges JC, Couble P, Magloire H. Sequential expression of matrix protein genes in developing rat teeth. Matrix Biol. 1999;18:133–43.
Liao H, Brandsten C, Lundmark C, Christersson C, Wurtz T. Osteonectin RNA and collagen a1(I) RNA in the developping rat maxilla. Eur J Oral Sci. 1998;106:418–23.
Sommer B, Bickel M, Hofstetter W, Wetterwald A. Expression of matrix proteins during the development mineralized tissues. Bone. 1996;19:371–80.
Iwasaki K, Bajenova E, Somogyi-Ganss E, Miller M, Nguyen V, Nourkeyhani H, et al. Amelotin - a novel secreted, ameloblast-specific protein. J Dent Res. 2005;84:1127–32.
Park JC, Park JT, Son HH, Kim HJ, Jeong MJ, Lee CS, et al. The amyloid protein APin is highly expressed during enamel mineralization and maturation in rat incisors. Eur J Oral Sci. 2007;115:153–60.
Hu JCC, Hu Y, Smith CE, McKee MD, Wright JT, Yamakoshi Y, et al. Enamel defects and ameloblast-specific expression in Enam knock-out/lacZ knock-in mice. J Biol Chem. 2008;283:10858–71.
Ye L, MacDougall M, Zhang S, Xie Y, Zhang J, Li Z, et al. Deletion of dentin matrix Protein-1 leads to a partial failure of maturation of Predentin into dentin, Hypomineralization, and expanded cavities of pulp and root canal during postnatal tooth development. J Biol Chem. 2004;279:19141–8.
Abbarin N, San Miguel S, Holcroft J, Iwasaki K, Ganss B. The enamel protein amelotin is a promoter of hydroxyapatite mineralization. J Bone Miner Res. 2015;30:775–85.
Lin Q, Fan S, Zhang Y, Xu M, Zhang H, Yang Y, et al. The seahorse genome and the evolution of its specialized morphology. Nature Nature Publishing Group. 2016;540:395–9.
Ravi V, Lam K, Tay B-H, Tay A, Brenner S, Venkatesh B. Elephant shark (Callorhinchus milii) provides insights into the evolution of Hox gene clusters in gnathostomes. Proc Natl Acad Sci U S A. 2009;106:16327–32.
Irisarri I, Baurain D, Brinkmann H, Delsuc F, Sire J-Y, Kupfer A, et al. Phylotranscriptomic consolidation of the jawed vertebrate timetree. Nat Ecol Evol. 2017;1(9):1370–8.
Miyake T, Vaglia JL, Taylor LH, Hall BK. Development of dermal denticles in skates (Chondrichthyes, Batoidea): patterning and cellular differentiation. J Morphol. 1999;241:61–81.
Espinoza J, Sanchez M, Sanchez A, Hanna P, Torrejon M, Buisine N, et al. Two families of Xenopus tropicalis skeletal genes display well-conserved expression patterns with mammals in spite of their highly divergent regulatory regions. Evol. Dev. 2010;12:541–51.
Enault S, Muñoz DN, Silva WTAF, Borday-birraux V, Bonade M, Oulion S, et al. Molecular footprinting of skeletal tissues in the catshark Scyliorhinus canicula and the clawed frog Xenopus tropicalis identifies conserved and derived features of vertebrate calcification. Front Genet. 2015;6:1–14.
Yu W-P, Rajasegaran V, Yew K, Loh W-L, Tay B-H, Amemiya CT, et al. Elephant shark sequence reveals unique insights into the evolutionary history of vertebrate genes: a comparative analysis of the protocadherin cluster. Proc Natl Acad Sci U S A. 2008;105:3819–24.
Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505:174–9.
Soderling JA, Reed MJ, Corsa A, Sage EH. Cloning and expression of murine SC1, a gene product homologous to SPARC. J Histochem Cytochem. 1997;45:823–35.
McKinnon PJ, McLaughlin SK, Kapsetaki M, Margolskee RF. Extracellular matrix-associated protein Sc1 is not essential for mouse development. Mol Cell Biol. 2000;20:656–60.
Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582.
Gasse B, Liu X, Corre E, Sire JY. Amelotin gene structure and expression during enamel formation in the opossum monodelphis domestica. PLoS One. 2015;10:0–9.
Gasse B, Sire JY. Comparative expression of the four enamel matrix protein genes, amelogenin, ameloblastin, enamelin and amelotin during amelogenesis in the lizard Anolis carolinensis. Evodevo BioMed Central. 2015;6:1–9.
Lee HK, Lee DS, Ryoo HM, Park JT, Park SJ, Bae HS, et al. The odontogenic ameloblast-associated protein (ODAM) cooperates with RUNX2 and modulates enamel mineralization via regulation of MMP-20. J Cell Biochem. 2010;111:755–67.
Dos Santos Neves J, Wazen RM, Kuroda S, Francis Zalzal S, Moffatt P, Nanci A. Odontogenic ameloblast-associated and amelotin are novel basal lamina components. Histochem Cell Biol. 2012;137:329–38.
Moffatt P, Wazen RM, Dos Santos Neves J, Nanci A. Characterisation of secretory calcium-binding phosphoprotein-proline-glutamine-rich 1: a novel basal lamina component expressed at cell-tooth interfaces. Cell Tissue Res. 2014;358:843–55.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80.
Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–52.
Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–8.
Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74.
Ballard WW, Mellinger J, Lechenault H. A series of stages for development of Scyliorhinus canicula the lesser spotted dogfish (Chondrichthyes: Scyliorhinidae). J Exp Zool. 1993;267:1–43.
Maxwell EE, Fröbisch NB, Heppleston AC. Variability and conservation in late chondrichthyan development: ontogeny of the winter skate (Leucoraja ocellata). Anat Rec (Hoboken). 2008;291:1079–87.
Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin). Amsterdam: North-Holland Publishing Company; 1967.
Ramlochansingh C, Branoner F, Chagnaud BP, Straka H. Efficacy of tricaine methanesulfonate (MS-222) as an anesthetic agent for blocking sensory-motor responses in Xenopus laevis tadpoles. PLoS One. 2014;9:1–11.
Close B, Banister K, Baumans V, Bernoth EM, Bromage N, Bunyan J, et al. Recommendations for euthanasia of experimental animals: part 1. DGXI of the European Commission Lab Anim. 1996;30:293–316.
Oulion S, Borday-Birraux V, Debiais-Thibaud M, Mazan S, Laurenti P, Casane D. Evolution of repeated structures along the body axis of jawed vertebrates, insights from the Scyliorhinus canicula Hox code. Evol. Dev. 2011;13:247–59.
Acknowledgements
We thank Marie-Ka Tilak and Claire Dupont for their help with molecular biology; Solenn Soriano for catshark handling; Frédéric Delsuc and the MBB (Montpellier Bioinformatics Biodiversity) platform for help with the transcriptomic analyses; the RHEM (Réseau d’Histologie Expérimentale de Montpellier at the Institute of Cancer Research of Montpellier (IRCM – U1194)) and MRI (Montpellier RIO Imaging, member of the national infrastructure France-BioImaging supported by the French National Research Agency (ANR-10-INBS-04, «Investments for the future»)) platforms for histological staining and imaging; the genotyping and sequencing (GenSeq) facilities of ISEM (Institut des Sciences de l’Evolution-Montpellier) and Labex Centre Méditerranéen Environnement Biodiversité. This is manuscript #ISEM 2018-108.
Funding
FONDECYT REGULAR grant 1151196 to SM; CNRS PEPS ExoMod to MDT; ANR Jaws project 12-BSV7–0020 to J-Y S. Funding bodies had no role in the design of the study, nor in the collection, analysis, interpretation of data, nor in writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files. All sequence data analyzed in this study are available through public databases, with reference IDs found in Additional file 1.
Author information
Authors and Affiliations
Contributions
MDT and SM designed the study. SE, DM and SV produced and analyzed the molecular, histological and in situ hybridization data. PS carried out the phylogenetic analyses. J-Y S carried out the BLASTs and gene identification. MDT, SM and J-Y S drafted the manuscript. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Collection and handling of catshark and ray embryos were carried out in full compliance of institutional, national and international guidelines [French Décret n° 2013–118 du 1er février 2013; European Communities Council Directive of 22 September 2010 (2010/63/UE)] and did not require further approval by an ethics committee.
The Ethics Committee of the University of Concepcion (Concepcion, Chile) approved all experimental procedures carried out during this study with Xenopus embryonic stages, which were performed following the guidelines outlined in the Biosafety and Bioethics Manual of the National Commission of Scientific and Technological Research (CONICYT, Chilean Government).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
SPARC and SPARC-L sequences used in the phylogenetic reconstruction. (XLSX 10 kb)
Additional file 2:
Bayesian (a) and Maximum Likelihood (b, c) trees. Amphioxus sequences are included in a and c, excluded in b. (PDF 722 kb)
Additional file 3:
Protein sequence alignment – 235 amino-acids. (PHY 15 kb)
Additional file 4:
List of primers used in this study. (DOCX 17 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Enault, S., Muñoz, D., Simion, P. et al. Evolution of dental tissue mineralization: an analysis of the jawed vertebrate SPARC and SPARC-L families. BMC Evol Biol 18, 127 (2018). https://doi.org/10.1186/s12862-018-1241-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-018-1241-y