Abstract
Various secretory calcium-binding phosphoprotein (SCPP) genes are involved in the formation of the bone, dentin, enamel, and enameloid in bony vertebrates. By contrast, no SCPP gene is found in cartilaginous vertebrates. In order to explain this difference, I investigated the origin and early evolution of SCPP genes. First, I examined the phylogeny of SPARC-family genes that include evolutionary precursors of SCPP genes. Then, I analyzed the genomic arrangement of the SCPP genes and three SPARC-family genes, SPARCL1, SPARCL1L1, and SPARCR1. The results are consistent with our previous hypothesis that an SCPP gene-like structure arose in the 5′ half of SPARCL1L1 in a common ancestor of jawed vertebrates, at about the same time as the origin of mineralized skeleton. It is possible that cartilaginous vertebrates secondarily lost early SCPP genes, while bony vertebrates gained various new SCPP genes. Some of these new SCPP genes appear to have specifically involved in scale formation; however, these scale genes were lost in tetrapods.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- SCPP genes
- SPARC gene family
- Mineralized skeleton
- Bony vertebrates
- Cartilaginous vertebrates
- Jawed vertebrates
- Scale formation
- Gene duplication
- Genome duplication
- Vertebrate evolution
1 Introduction
Among cardinal traits evolved in vertebrates is mineralized skeleton, which arose in a common ancestor of jawed vertebrates after the divergence of the lineage leading to modern jawless vertebrates (Fig. 17.1) (Donoghue and Sansom 2002). The bone, dentin, enamel, and enameloid are principal mineralized skeletal tissues (Donoghue and Sansom 2002). Among these tissues, the bone was secondarily lost in cartilaginous vertebrates (Eames et al. 2007), and enamel is thought to have originated in bony vertebrates (Schultze 2016).
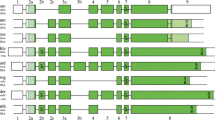
The arrangement of SCPP genes, SPARC family genes, USO1, G3BP2, and RCHY1 in the human, coelacanth, gar, zebrafish, elephant shark, and lamprey genomes. The phylogeny of these vertebrates and the origin of mineralized skeleton are shown on the top. Vertical bars represent chromosomes or contigs (names on the top). Regions separated by >200 kilobases are shown by double slashes. Horizontal bars represent P/Q-rich SCPP genes (red), acidic SCPP genes (blue), SPARC-family genes (yellow, circled), and other genes (green, circled). Genes with different transcriptional directions are shown on different sides (right, plus strand; left, minus strand). Orthologs are connected with a dashed line (a question mark represents unconfirmed orthologs). See Kawasaki et al. (2017) for more details, including lamprey spock3
In bony vertebrates, formation of mineralized tissues involves various secretory calcium-binding phosphoprotein (SCPP) genes, which arose by gene duplication and form gene clusters (Fig. 17.1) (Kawasaki and Weiss 2003). Two types of SCPP genes are known; one encodes acidic SCPPs and the other Pro and Glu (P/Q)-rich SCPPs. Most acidic SCPP genes are employed in the formation of the bone and/or dentin, whereas many P/Q-rich SCPP genes are expressed during the enamel and/or enameloid formation (Kawasaki 2011). In contrast to bony vertebrates, no SCPP gene is found in the genomes of cartilaginous vertebrates (Venkatesh et al. 2014). In order to explain this difference, I investigated the origin and early evolution of SCPP genes.
SCPP genes evolve rapidly, but all SCPP genes retain a characteristic exon-intron structure, which allows us to identify SCPP genes without relying on sequence similarities (Kawasaki and Weiss 2003). This characteristic exon-intron structure is also found in the 5′ half of the SPARC-like 1 (SPARCL1) and SPARCL1-like 1 (SPARCL1L1), both located adjacent to SCPP genes in the genomes of coelacanth and gar (Fig. 17.1) (Kawasaki et al. 2004, 2017). Moreover, the 5′ half of elephant shark sparcl1l1 is highly similar to SPP1 and other acidic SCPP genes, encoding an extremely acidic sequence, a cluster of Ser-Xaa-Glu (Xaa represents any amino acids, and the Ser residue is thought to be phosphorylated) near the N-terminus, and one or more Arg-Gly-Asp (RGD) integrin-binding sequences (Fig. 17.2). Based on these findings, we proposed that SCPP genes arose from the 5′ half of SPARCL1L1 (Kawasaki et al. 2017).
The 3′ half of SPARCL1 and SPARCL1L1 encodes evolutionarily conserved amino acid sequences, known as the follistatin-like (FS) domain and the extracellular calcium-binding EF-hand (EC) motif (Bradshaw 2012). Genes encoding the FS domain and the EC motif constitute the SPARC gene family, which includes SPARC, SPARCL1, SPARCL1L1, and two SPARC-related genes, SPARCR1 and SPARCR2 (Kawasaki et al. 2017). Previous studies suggested the duplication history of these genes (Fig. 17.2) (Bertrand et al. 2013; Torres-Núñez et al. 2015; Kawasaki et al. 2017). A tandem duplication split the SPARC and SPARCR lineages before the divergence of protostomes and deuterostomes. In the SPARC lineage, SPARC and the common ancestor of SPARCL1 and SPARCL1L1 arose in two vertebrate genome duplications (VGD1/VGD2) (Ohno 1970), thought to have occurred in the common ancestor of jawless vertebrates and jawed vertebrates (Kuraku et al. 2009). In the SPARCR lineage, SPARCR1 and SPARCR2 originated also in VGD1/VGD2. SPARCL1 and SPARCL1L1 arose subsequently by tandem duplication in the common ancestor of cartilaginous vertebrates and bony vertebrates (Fig. 17.2).
Duplicated SPARC family genes appear to have differentiated asymmetrically; while one duplicate maintained ancient characteristics, the other duplicate obtained new characteristics. The new characteristics, encoded by the differentiated genes, include a small N-terminal acidic domain arisen early in the SPARC lineage, a large N-terminal acidic domain in the common ancestor of SPARCL1 and SPARCL1L1, and a cluster of phospho-Ser (pSer) residues in the N-terminus and an RGD integrin-binding sequence in SPARCL1L1 (Fig. 17.2) (Kawasaki et al. 2017). We hypothesized that the most derived characteristics arose in SPARCL1L1 evolved into SPP1 and other acidic SCPP genes (Fig. 17.2). This hypothesis infers that an SPP1-like structure originated in the common ancestor of jawed vertebrates, at about the same time as the origin of skeletal mineralization (Fig. 17.2) (Kawasaki et al. 2017). In the present study, I reexamined this hypothesis and analyzed the genomic arrangement of SCPP genes and their adjacent genes.
2 Materials and Methods
In the present study, four genes were added to our previous analysis (Kawasaki et al. 2017). These genes are sparcl1 in the whale shark and Asian arowana (GenBank XM_020510428.1 and XM_018735764.1, respectively) and sparc in the Pacific hagfish and gray bichir (reconstructed from GenBank SRX2541845 and SRX796491, respectively). Amino acid sequences of the FS domain and the EC motif, deduced from the nucleotide sequences, were used to construct a maximum likelihood (ML) tree and a Bayesian inference (BI) tree, as described previously (Kawasaki et al. 2017). I considered 95% or higher bootstrap values in the ML tree and 95% or higher posterior probabilities in the BI tree as statistically significant.
3 Results and Discussion
3.1 Phylogenetic Analysis
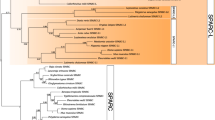
Among the newly analyzed genes, the whale shark gene, annotated as “sparcl1,” was clustered with elephant shark sparcl1l1 (100% in the BI tree; Fig. 17.3). Elephant shark sparcl1l1 is characterized by a large exon, which encodes a highly acidic sequence and is flanked by a phase-0 intron at the 5′ border and a phase-1 intron at the 3′ border (Kawasaki et al. 2017). A similar large exon was also identified in this whale shark gene (649–1243 nucleotides of XM_020510428.1), but not known from any other SPARC family genes. Based on these findings, I consider this whale shark gene as the SPARCL1L1 ortholog. Unfortunately, exons 1, 2, 3, and the 5′ end of exon 4 of whale shark sparcl1l1 were not identified, and details of these exons remain to be elucidated.
Phylogenetic trees constructed by the ML and BI methods. Different colors show SPARC, SPARCL1, SPARCL1L1, and sparcb. Non-vertebrate SPARC genes are the common ancestor of SPARC, SPARCL1, and SPARCL1L1. Bootstrap values of >70% in the ML tree, and posterior probabilities of >80% in the BI tree are shown at the node. Vertical bars indicate genes in jawed and jawless vertebrates. Horizontal bars represent scales
Similar to our previous analysis (Kawasaki et al. 2017), SPARC genes in all jawed vertebrates formed a single cluster (95% and 100% in the ML and BI trees, respectively), whereas the phylogeny between SPARCL1L1 in cartilaginous vertebrates, SPARCL1L1 in bony vertebrates, and SPARCL1 in bony vertebrates was not resolved by significant statistical supports (Fig. 17.3). This result suggests a close phylogenetic relationship of SPARCL1 and SPARCL1L1.
In the present study, I used three jawless vertebrate genes, sparca and sparcb in lampreys and a hagfish gene. I tentatively call this hagfish gene sparc, because this gene encodes a small N-terminal acidic domain (71 residues), similar to SPARC in jawed vertebrates (Kawasaki et al. 2004). In the BI tree, SPARC, SPARCL1, and SPARCL1L1 in all jawed vertebrates formed a cluster (100%), and this cluster was most closely related to sparca (100%; Fig. 17.3), which is thought to be orthologous to SPARC (Kawasaki et al. 2017). However, the ML tree did not well support this relationship and did not resolve the phylogeny of these genes, sparcb, and hagfish sparc (80% or less; Fig. 17.3).
3.2 Arrangements of SCPP Genes and SPARC Family Genes in Vertebrate Genomes
In the genome of the elephant shark, gar, and coelacanth, g3bp2, uso1, and sparcl1l1 are clustered, and their order and directions are conserved (Fig. 17.1). The arrangement of these genes suggests that the last common ancestor of jawed vertebrates had a similar cluster. In the lamprey genome, sparcb is located adjacent to uso1, which reinforces our previous hypothesis that sparcb is co-orthologous to SPARCL1 and SPARCL1L1 (Fig. 17.1) (Kawasaki et al. 2017). Unlike SPARCL1 or SPARCL1L1, sparcb encodes a small N-terminal acidic domain, similar to SPARC (Kawasaki et al. 2017). This is presumably because the large N-terminal acidic domain evolved in the common ancestor of SPARCL1 and SPARCL1L1 in the jawed vertebrate lineage after the divergence of the lineage leading to modern jawless vertebrates (Fig. 17.2).
The phylogeny of sparcb as a co-ortholog of SPARCL1 and SPARCL1L1 is not consistent with the topology of the phylogenetic trees; sparcb, SPARC (including sparca), SPARCL1, and SPARCL1L1 are intermingled (Fig. 17.3). The size of the encoded N-terminal acidic domain is small in SPARC and sparcb but large in SPARCL1 and SPARCL1L1, suggesting an asymmetrical functional divergence of the common ancestor of SPARCL1 and SPARCL1L1. The functional divergence probably led to differential sequence changes, which may partly explain the low resolution of these genes in the phylogenetic analysis.
In the gar genome, sparcl1l1, 18 P/Q-rich SCPP genes, sparcl1, and six acidic SCPP genes are clustered on chromosome 4, while spp1, rchy1, 12 P/Q-rich SCPP genes, and sparcr1 form a cluster on chromosome 2 (Fig. 17.1). Adjacent locations of MEPE and SPP1 in the human and coelacanth genomes suggest that these two large SCPP gene clusters in the gar genome were originally connected to each other (double arrow in Fig. 17.1). The original cluster was presumably composed of sparcl1l1, P/Q-rich SCPP genes, sparcl1, acidic SCPP genes, rchy1, P/Q-rich SCPP genes, and sparcr1 in this order. The arrangement of the orthologs of these genes in the coelacanth genome suggests that the last common ancestor of bony vertebrates had a similar gene cluster, containing at least five acidic and three P/Q-rich SCPP genes (Fig. 17.1).
It was shown that two zebrafish SCPP gene clusters, located on chromosomes 5 and 10, originated by the teleost genome duplication (Braasch et al. 2016). The locations of spp1 and rchy1 in the gar and zebrafish genomes suggest that these two zebrafish SCPP gene clusters are co-orthologons of the SCPP gene cluster on gar chromosome 2 (Fig. 17.1). Twelve P/Q-rich SCPP genes are found on gar chromosome 2 and seven P/Q-rich SCPP genes on zebrafish chromosomes 5 and 10 (Fig. 17.1). By contrast, only scpppq8 (XR_322354.2) was identified in the syntenic region in the coelacanth genome and none in the tetrapod genomes (Fig. 17.1). Among these P/Q-rich SCPP genes, the only gene investigated to date is gsp37, which encodes a matrix protein of the surface layer of scales (Miyabe et al. 2012). Moreover, expression of ten P/Q-rich SCPP genes on gar chromosome 2 was detected in the skin that overlies scales but not in the teeth or bone (Kawasaki et al. 2017). Interestingly, both coelacanth scpppq8 and zebrafish gsp37 encode similar sequence elements, including a cluster of pSer residues, a Cys residue (rare in SCPP genes), and an RGD integrin-binding sequence. These findings suggest that one or more P/Q-rich SCPP genes involved primarily in scale formation arose in the common ancestor of bony vertebrates and that these scale SCPP genes were secondarily lost in tetrapods.
In summary, the present analysis is consistent with our previous hypothesis (Kawasaki et al. 2017): an SPP1-like structure originated in the 5′ half of SPARCL1L1 in a common ancestor of jawed vertebrates, roughly contemporaneous with the origin of skeletal mineralization. It is possible that early SCPP genes were secondarily lost in cartilaginous vertebrates, while common ancestors of bony vertebrates gained various acidic and P/Q-rich SCPP genes. Some of these P/Q-rich SCPP genes may have specifically involved in scale formation, but these scale genes were lost in tetrapods.
References
Bertrand S, Fuentealba J, Aze A, Hudson C, Yasuo H, Torrejon M et al (2013) A dynamic history of gene duplications and losses characterizes the evolution of the SPARC family in eumetazoans. Proc Biol Sci 280:20122963
Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J et al (2016) The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet 48:427–437
Bradshaw AD (2012) Diverse biological functions of the SPARC family of proteins. Int J Biochem Cell Biol 44:480–488
Donoghue PCJ, Sansom IJ (2002) Origin and early evolution of vertebrate skeletonization. Microsc Res Tech 59:352–372
Eames BF, Allen N, Young J, Kaplan A, Helms JA, Schneider RA (2007) Skeletogenesis in the swell shark Cephaloscyllium ventriosum. J Anat 210:542–554
Kawasaki K (2011) The SCPP gene family and the complexity of hard tissues in vertebrates. Cells Tissues Organs 194:108–112
Kawasaki K, Weiss KM (2003) Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster. Proc Natl Acad Sci U S A 100:4060–4065
Kawasaki K, Suzuki T, Weiss KM (2004) Genetic basis for the evolution of vertebrate mineralized tissue. Proc Natl Acad Sci U S A 101:11356–11361
Kawasaki K, Mikami M, Nakatomi M, Braasch I, Batzel P, Postlethwait JH et al (2017) SCPP genes and their relatives in gar: rapid expansion of mineralization genes in osteichthyans. J Exp Zool B Mol Dev Evol 328:645–665
Kuraku S, Meyer A, Kuratani S (2009) Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol 26:47–59
Miyabe K, Tokunaga H, Endo H, Inoue H, Suzuki M, Tsutsui N et al (2012) GSP-37, a novel goldfish scale matrix protein: identification, localization and functional analysis. Faraday Discuss 159:463–481
Ohno S (1970) Evolution by gene duplication. Springer, New York
Schultze HP (2016) Scales, enamel, cosmine, ganoine, and early osteichthyans. C R Palevol 15:83–102
Torres-Núñez E, Suarez-Bregua P, Cal L, Cal R, Cerdá-Reverter JM, Rotllant J (2015) Molecular cloning and characterization of the matricellular protein Sparc/osteonectin in flatfish, Scophthalmus maximus, and its developmental stage-dependent transcriptional regulation during metamorphosis. Gene 568:129–139
Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB et al (2014) Elephant shark genome provides unique insights into gnathostome evolution. Nature 505:174–179
Acknowledgments
I am grateful to Prof. Joan Richtsmeier at Penn State for encouragement and Prof. Hiromichi Nagasawa at the University of Tokyo for inviting me to BiominXIV. This work was made possible by the financial support from the Department of Anthropology at Penn State and from the National Institute of Health (P01HD078233).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2018 The Author(s)
About this paper
Cite this paper
Kawasaki, K. (2018). The Origin and Early Evolution of SCPP Genes and Tissue Mineralization in Vertebrates. In: Endo, K., Kogure, T., Nagasawa, H. (eds) Biomineralization. Springer, Singapore. https://doi.org/10.1007/978-981-13-1002-7_17
Download citation
DOI: https://doi.org/10.1007/978-981-13-1002-7_17
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-1001-0
Online ISBN: 978-981-13-1002-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)