Abstract
Background
Flooding due to global climate change is a serious problem that frequently decreases crop yields. Rice fields in flood-prone areas often experience full or partial submergence. Submergence has an adverse effect on internal oxygen availability, sugar status and survival. Complete submergence imposes severe pressure on plants, principally because the excess water in their surroundings deprives them of certain basic resources such as oxygen, carbon dioxide and light for photosynthesis. To better understand the mechanisms involved under different levels of flooding, it is necessary to further observe physiological responses and to identify the Rboh genes involved and determine how they are regulated during submergence.
Results
In this study, significant physiological changes were observed in plant height, leaf sheath elongation and chlorophyll a, b and total content under partial and full submergence treatments. Senescence-regulating genes were severely affected under full submergence. Additionally, intracellular oxidative homeostasis was disrupted by overproduction of H2O2 and O2 −, which affected cell viability and antioxidant enzyme activity, under different levels of submergence. Quantitative RT-PCR analyses revealed that complex regulation of Rboh genes is involved under different levels of submergence.
Conclusion
Our results demonstrated that the effect of physiological and the transcript levels of OsRboh genes were presented different responses to different levels of submergence in rice seedlings. There have different mechanism in intracellular to response different levels of submergence. Finally we discuss effects of the regulation of OsRboh expression and ROS production which was important to maintain homeostasis to help rice seedlings face different levels of submergence.
Similar content being viewed by others
Background
The effect of global climate change on crop yields is an urgent issue. Waterlogging is a serious problem that affects crop gas exchange in low-lying rain-fed areas. Approximately 30 % of the world’s rice (Oryza sativa) farmlands are at a low elevation and irrigated by rain (Bailey-Serres et al. 2010). Under limited oxygen availability, photosynthesis and respiration are restricted, leading to an energy crisis, toxic products from anaerobic respiration and the accumulation of reactive oxygen species (ROS) in plant cells (Licausi and Perata 2009; Pucciariello et al. 2012a; Yang and Hong 2015).
Several studies have focused on the important role of ROS during hypoxia signaling under full submergence conditions (Baxter et al. 2014; Fukao et al. 2011; Liu et al. 2015; Yang and Hong 2015 ). Plasma membrane NADPH oxidases in plants were discovered through their sequence similarity to mammalian respiratory burst NADPH oxidase subunit gp91phox and have been named respiratory burst oxidase homologs (Rbohs) (Sagi and Fluhr 2006). Rbohs participate in ROS production and are involved in the oxidative burst in response to pathogens (Lherminier et al. 2009; Sagi and Fluhr 2006). Some reports have indicated that Rboh proteins possess a resistance function, such as AtRboh D and AtRboh F, which may be involved in triggering cell death in inoculated plants (Pogany et al. 2009; Vellosillo et al. 2010). AtRboh D and AtRboh F are involved not only in the hypersensitive response but also the ABA- and ethylene-induced stomatal response and are associated with a very complicated interaction between the ABA-, ethylene-, JA- and SA-signaling pathways (Desikan et al. 2006; Maruta et al. 2011). Both ethylene and H2O2 were demonstrated promote aerenchyma formation in rice stems in a dose-dependent manner (Steffens et al. 2011). In a recent study, the NADPH oxidase Rboh D was found to be involved in primary hypoxia signaling and modulate down-stream gene expression (Yang and Hong 2015).
Floods are major constraints to crop production worldwide that affect oxygen availability in the environment and lead to serious physiological damage. Transient flash floods leading to partial or complete submergence can affect crop growth and yield (Kato et al. 2014). In recent years, many reports have indicated that complete submergence has an adverse effect on internal oxygen availability, sugar status and survival (Bailey-Serres and Voesenek 2010; Winkel et al. 2013). Complete submergence imposes severe pressure on plants, principally because excess water in their surroundings deprives them of certain basic resources such as oxygen, carbon dioxide and light for photosynthesis (Sakagami et al. 2013). Paddy field rice is generally tolerant to waterlogging and partial submergence compared with other economic crops such as maize and soybean. Lowland rice (Flood Resistant 13A; FR13A) can restrict its elongation growth, economizing its carbohydrate reserves to enable the development of new leaves upon desubmergence (Fukao et al. 2011).
So far, these results have mainly been obtained from complete submergence studies. To better understand the mechanisms involved under different levels of flooding, it is necessary to further identify the Rboh genes involved and to determine how they are regulated during submergence. In this study, we characterized the physiological responses and the transcript profiles of nine Rbohs under different levels of flooding treatments in rice (Oryza sativa L. japonica, Tai keng 9). Moreover, we examined the accumulation of H2O2 and O2 − using DAB and NBT staining and the activities of antioxidant enzymes. This study provides evidence of differential regulation of OsRboh expression and ROS accumulation under partial or full submergence in rice.
Results
Partial Submergence Leads to Faster Plant Height Increases and Leaf Sheath Elongation at a Relatively Early Stage
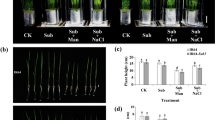
To assess several physiological mechanisms associated with partial and full submergence in rice, plant height and leaf sheath length were determined in 14 days-old rice seedlings under partial submergence (PS) or full submergence (FS) for 2, 4, 6 and 8 days. The leaves of the rice seedlings were yellowish, thin and long, and could not remain upright under FS for 8 days compared with regular growth (control check; CK) and PS conditions (Fig. 1a and b). After PS treatment, the yellowish color was only present in tissue that had been submerged (Fig. 1a and b). During the partial submergence treatment period, plant height and the length of the 3rd sheath were significantly increased after 2 days treatment. Subsequently, plant height and the length of the 3rd leaf sheath increased in both PS and FS conditions compared with the control (Fig. 1c and d). There were no significant differences in the lengths of the 1st and 2nd leaf sheaths after PS or FS treatment compared with the control (Additional file 1: Figure S1). These results indicated that the plant height increase of rice seedlings during submergence was mainly affected by elongation of the 3rd leaf sheath. Notably, partial submergence led to faster elongation at a relatively early stage compared with full submergence.
Characterization of rice (Oryza sativa) seedlings growth under different levels of submergence. a Photographs of 14 days-old rice seedlings after exposed different levels of submergence for 8 days. b The phenotypes of plant height after exposed different levels of submergence treatment for 8 days. (1st, 2nd and 3rd; length of the first, second and third leaf sheath). c Plant height of 14 days-old rice seedlings after treated submergence for 2, 4, 6 and 8 days. d The third leaves sheath length of 14 days-old rice seedlings after treated submergence for 2, 4, 6 and 8 days. Control check (CK), partial submergence (PS) and full submergence (FS). Bar = 3 cm. The data represent average values ± SD from 30 seedlings of each treatment obtained from six biologically independent experiments. Values with the different letters are significantly different at P < 0.05, according to post-hoc LSD test
The Chlorophyll Content and Senescence-regulating Genes are Severely Affected under Full Submergence
The photosynthetic apparatus and particularly PS II in rice seedlings is affected by submergence, which causes ROS damage to the photosystem and a decreased photosynthetic rate and chlorophyll content (Panda et al. 2008). The submergence tolerance regulator, SUBMERGENCE1A (SUB1A), is involved in acclimation responses during leaf senescence caused by prolonged darkness in rice (Fukao et al. 2012).
To dissect the influence of chlorophyll content and the expression of senescence-regulating genes under different levels of submergence, we determined the chlorophyll a, b and total content after PS or FS treatment for 2, 4, 6 and 8 d in 14 days-old rice seedlings. The results showed that the chlorophyll a, b and total contents were significantly decreased with submergence treatment, particularly in the FS treatment (Fig. 2). To evaluate whether senescence-associated gene (SAG) expression was affected by different levels of submergence, we used quantitative RT-PCR to determine the mRNA expression of Stay-Green (SGR), which is necessary for chlorophyll degradation in light-harvesting complex II (Hortensteiner 2009), Red Chlorophyll Catabolite Reductase (RCCR), which functions in chlorophyll degradation in chloroplasts (Pruzinska et al. 2007), and Osl85, which encodes isocitrate lyase, a marker gene that is upregulated during dark-induced and natural leaf senescence (Yamada et al. 2014). The quantitative RT-PCR results revealed that the transcripts of these senescence-regulating genes were highly induced by FS treatment compared with PS treatment (Fig. 3). These results demonstrated that FS conditions severely affect the chlorophyll content, which is accompanied by senescence induction, in rice seedlings more than PS conditions.
Different levels of submergence influence the content of chlorophyll in rice seedlings. The chlorophyll content of 14-days old seedlings after treated control (CK), partial submergence (PS) and full submergence (FS) conditions for 2, 4, 6 and 8 days. The content of chlorophyll a (a), chlorophyll b (b) and total chlorophyll (c) were determined. Values represent means ± standard deviation from six biologically independent experiments (n = 3). Values with the different letters are significantly different at P < 0.05, according to post-hoc LSD test
Transcript levels of gene associated with leaf senescence. Quantitative RT-PCR was used to quantify transcript levels of Stay-Green (SGR), Red Chlorophyll Catabolite Reductase (RCCR) and Isocitrate Lyase (Osl85) genes in 14 days-old rice seedlings after treated control (CK), partial submergence (PS) and full submergence (FS) conditions for 6 days. Relative amounts of transcripts were calculated and normalized to that of Ubiquitin mRNA. Values represent means ± standard deviation from five biologically independent experiment. Values with the different letters are significantly different at P < 0.05, according to post-hoc LSD test
Full Submergence Leads to H2O2 and O2 − Overproduction that Disrupts Intracellular Oxidative Homeostasis
Several studies have shown that ROS are common components of biochemical changes in chloroplasts, mitochondria and peroxisomes when plants are subjected to oxygen deprivation conditions (Blokhina et al. 2003; Damanik et al. 2010). To investigate ROS accumulation, H2O2 and O2 − accumulation under different levels of submergence was evaluated by DAB and NBT staining. The 3rd leaves in the submerged area appeared yellow after PS or FS treatment for 6 days (Fig. 4). As shown in Fig. 4b and c, H2O2 and O2 − were accumulated to higher levels in FS than in PS conditions, especially in the tips of leaves. We also determined the cell viability by Evans blue staining, and the results showed significant cell death in the tips and blades of leaves in FS conditions (Fig. 4d and e).
DAB, NBT and Evans blue staining for the detection of H2O2, O2 − and cell death in rice seedlings under different levels of submergence. a Phenotypes of 14 days-old rice seedlings after treated control (CK), partial submergence (PS) and full submergence (FS) for 6 days. b and c, DAB and NBT staining of H2O2 and O2 − in detached leaves of 14 days-old rice seedlings after treated control (CK), partial submergence (PS) and full submergence (FS) for 6 days. Same results were obtained in three independent experiments. Photograph shows results of three independent leaves. d and e, Evans blue staining and cell death analysis (see materials and methods for details). Bar = 1 cm. The data represent average values ± SD from five biologically independent experiments. Values with the different letters are significantly different at P < 0.05, according to post-hoc LSD test
To investigate the cellular scavenging capacity of the antioxidative enzymes in rice seedlings under different levels of submergence, the activities of antioxidant enzymes such as catalase (CAT), ascorbate peroxidase (APX), superoxide dismutase (SOD) and total peroxidase (POX) were evaluated. The CAT, SOD and APX activities were decreased under PS and FS treatment for 6 days and recovery for 1 day compared with the control plants. The APX activity was decreased more under FS treatment for 6 days than PS treatment. The POX activity was significantly increased under PS and FS treatment for 6 days and recovery for 1 day (Fig. 5). Thus, our results suggest that the overproduction of H2O2 and O2 − disrupts intracellular oxidative homeostasis, which affects cell viability and antioxidant enzyme activity under different levels of submergence.
The activities of CAT, APX, SOD and POX in rice seedlings under different levels of submergence. Enzyme activity was detected from detached shoots of 14 days-old rice seedlings after treated control (CK), partial submergence (PS) and full submergence (FS) for 6 days then recovery 1 day (R1D). The catalase (CAT), ascorbate peroxidase (APX), superoxide dismutase (SOD) and total peroxidase (POX) activity were determined, respectively. The data represent average values ± SD from six biologically independent experiments. Values with the different letters are significantly different at P < 0.05, according to post-hoc LSD test
Transcript Profiles of OsRboh Genes under Different Levels of Submergence
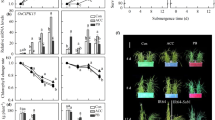
H2O2participates in hypoxia signal transduction to modulate the expression of a set of genes encoding heat-shock proteins and other groups of ROS-mediated proteins (Pucciariello et al. 2012b). NADPH oxidase enzymes are key players in H2O2 production. Previously, we showed that AtRboh genes are involved in hypoxia signaling, and revealed the different transcript profiles of Rboh genes under hypoxic stress in Arabidopsis. AtRboh D plays a major role at an early stage to modulate the expression of down-stream hypoxia-inducible genes under hypoxic stress (Yang and Hong 2015). Phylogenetic analysis of the corresponding corrected protein sequences of ten Arabidopsis and nine rice Rboh proteins were obtained from the TAIR (http://www.arabidopsis.org/) and Phytozome (https://phytozome.jgi.doe.gov/) databases (Additional file 2: Figure S2). To characterize the expression profiles of the nine Rboh genes in rice seedlings during submergence, their transcript levels were determined by quantitative RT-PCR under different levels of submergence. The transcript levels of the Rboh C (Os05g45210), D (Os05g38980), E (Os01g61880) and F (Os08g35210) genes increased under PS conditions, the transcript levels of Rboh H (Os12g35610) and I (Os11g33120) genes increased under FS conditions, the transcript levels of Rboh A (Os01g53294) decreased under PS treatment for 24 h and FS treatment for 24 or 48 h, the transcript levels of the Rboh B (Os01g25820) and G (Os09g26660) genes decreased under FS conditions (Fig. 6a and b). These results not only demonstrate that Rboh genes are involved in different levels of submergence, but also reveal the complex regulation of Rboh genes in hypoxia signaling in rice seedlings.
Transcript profiles of Rboh genes in rice seedlings under different levels of submergence. a quantitative RT-PCR analyses of transcript levels of Rbohs genes in 14 days-old rice seedlings after treated control (CK), partial submergence (PS) and full submergence (FS) conditions. b fold change were presented by different color. Total RNAs were isolated from shoots of 14-days-old seedlings after 24 and 48 h submergence treatment and levels of Rboh A–I mRNA were determined. Relative amounts of transcripts were calculated and normalized to that of ubiquitin mRNA. Values represent means standard deviation from five biologically independent experiments. Values with the different letters are significantly different at P < 0.05, according to post-hoc LSD test
Discussion
Rice fields in flood-prone areas often experience full or partial submergence. The many visible symptoms of injury caused by full or partial submergence include an initial phase of fast elongation by one or more leaves accompanied by yellowing of old leaves, and slow growth in the dry mass of roots and shoots (Jackson and Ram 2003; Kato et al. 2014; Sarkar and Bhattacharjee 2012). The survival strategies of flood-tolerant plants are characterized as low-O2 escape syndrome (LOES) and low-O2 quiescence syndrome (LOQS) (Voesenek and Bailey-Serres 2015). LOES facilitates escape from submergence stress and includes upward growth of leaves (hyponastic growth) and petiole/stem elongation initiated by ethylene accumulation (Jackson 2008). To further dissect the effects of different levels of flooding, we investigated the physiological and molecular responses of rice seedlings under partial and full submergence. In this study, our physiological experiment demonstrated that the plant height of rice seedlings was increased, which was mainly affected by elongation of the 3rd leaf sheath, during submergence. Furthermore, partial submergence led to faster elongation at a relatively early stage compared with full submergence (Fig. 1). Therefore, an early physiological response to partial submergence is the elongation of leaf sheaths, which implies sensing of different levels of submergence and different regulation mechanisms in rice seedlings.
A significant reduction of Rubisco activity, inhibition of the CO2 photosynthetic rate and ROS accumulation occurs when plants are exposed to flooding conditions for prolonged periods (Kamal and Komatsu 2015; Panda et al. 2008; Yang and Hong 2015; Yang 2014). The catabolism of chlorophyll is regulated by natural leaf senescence and environmental stress (Lim et al. 2007). Our results showed that the chlorophyll a, b and total contents were significant decreased by submergence treatment, particularly FS treatment (Fig. 2). The transcript accumulation of genes associated with senescence was highly induced by FS treatment compared with PS treatment (Fig. 3). These results reveal that different levels of submergence not only affect SAG expression, but also lead to a reduction of chlorophyll content. Notably, a characteristic enzyme gene of the glyoxylate cycle, Osl85 (isocitrate lyase), was significantly induced after FS treatment. This seems to indicate that the glyoxylate cycle is involved in the response of rice seedlings to different levels of submergence.
Under submergence, the O2 concentration of water is lower than that of air, leading to suppressed respiration in roots and enhanced fermentation (Nishiuchi et al. 2012; Yin et al. 2013). This metabolic shift can cause toxicity from ethanol accumulation. The most common group of toxic intermediates produced is ROS. We demonstrated that H2O2 and O2 − accumulation was highest in the tips of leaves by histochemical staining and observed significant cell death in FS conditions (Fig. 4). Antioxidative enzyme activity detection showed that POX activity was significantly increased under PS and FS treatment for 6 days and recovery for 1 day (Fig. 5). Thus, the overproduction of H2O2 and O2 − disrupts intracellular oxidative homeostasis, which affects cell viability and antioxidant enzyme activity under different levels of submergence.
Previously, we characterized the transcription profiles of ten Rbohs in Arabidopsis under hypoxic stress and found Rboh D is involved in primary hypoxia signaling to modulate the expression of hypoxia-inducible genes under hypoxic stress (Yang and Hong 2015). Here, we showed that the expression profiles of nine Rboh genes in rice seedlings differed under different levels of submergence (Fig. 6), indicating Rboh genes are involved in responses to different levels of submergence. As in Arabidopsis, our data reveal the complex regulation of Rboh genes in hypoxia signaling in rice seedlings.
Conclusions
Although flooding leads to chlorophyll content decreased and accompanied senescence induction, the transcript levels of OsRboh genes was presented different responses to different levels of submergence in rice seedlings. Our results demonstrate that have different mechanism in intracellular to response different levels of submergence. The regulation of OsRboh expression and ROS production was important to help rice seedlings face different levels of submergence.
Methods
Plant Materials, Growth Conditions and Stress Treatment
Rice (Oryza sativa, japonica.) Tai-keng 9 (TK9) was used in this study. Seeds were surface sterilized via immersion in a 3 % sodium hypochlorite solution for 30 min, and were then thoroughly rinsed with distilled water. Seeds were subsequently placed on wet filter paper for 3 day at 28 °C in a 16-h-light (236 μmolm−2s−1)/8-h-dark cycle in a growth chamber. The germinated seeds were transplanted onto iron grid in the beaker with Kimura B solution. For submergence treatment, 14-day old seedlings (2–3 leaf stage) were transferred into water box (W:L:H, 40 cm × 40 cm × 60 cm) filled with 23 cm high of water for partial submergence (PS) which the water level reached half of plants height or filled with 50 cm high of water for full submergence (FS). After each treatment, sampled tissue was immediately frozen in liquid nitrogen and stored in −80 °C until use.
Measurement of Plant Height, Length of Leaf Sheath and Chlorophyll Content
After 14-day old seedlings were treatments by PS or FS for 2, 4, 6 and 8 days, the length of each shoot and leaf sheath (at least 30 plants each) were recorded for indicated times. Experiments were repeated five times independently. For chlorophyll content assays, chlorophyll a, b and total contents were extracted from 50 mg of shoot tissue in 2 mL of sodium phosphate buffer (50 mM pH 6.8). Add 40 μL extracted solution in 960 μL 99 % ethanol then incubated for 30 min at RT in the dark with gentle shaking. After centrifugation at 4 °C for 15 min at 1000 g, the absorbance of the supernatant was measured at 665 and 649 nm with a spectrophotometer (Metertec SP8001).
Assay of O2 − and H2O2 Accumulation and Cell Viability by Histochemical Staining Method
Accumulation of O2 − in cell was visualized by the nitroblue tetrazolium (NBT) staining method. 14-day old seedlings after submergence treatment for 6 days, the 3 rd leaves were immersed into 1 mM NBT solution prepared in 10 mM phosphate buffer (pH 7.8) at RT under dark for 8 h. When blue spots appeared, the pigments were removed from leaves by boiling in 75 % ethanol for 5 min. Accumulation of H2O2 in cells was visualized by the 3, 3′ -diaminobenzidine (DAB) staining method as described in detail previously (Yang and Hong 2015). In the stained leaves, H2O2 is visualized as reddish-brown coloration. The leaves were photographed with a digital camera and the intensity of the colour in the NBT- or DAB-stained area was measured using Photoshop image software. Experiments were repeated three times independently. The stained intensity was measured at least 10 leaves for each biological repeat. Cell viability assay was visualized by the Evans blue staining. 14-day old seedlings after submergence treatment for 6 days, the 3 rd leaves were collected, weighed and immersed into 0.2 % Evans blue solution for 3 h with gentle shaking. The de-stained leaves (0.05 g/each) were homogenized with liquid nitrogen in 500 μL of 10 % SDS followed by the addition of 500 μL of de-ionized water. The solutions were centrifuged at 15,800 g for 30 min at 4 °C, the absorbance of the supernatant was measured at 590 nm with a spectrophotometer (Metertec SP8001). Experiments were repeated six times independently.
Determination of Antioxidative Enzyme Activity
The shoot samples (50 mg) were excised and immediately used for enzyme extraction. The shoot tissues were homogenized with 50 mM sodium phosphate buffer (pH 6.8) with liquid nitrogen. The homogenate was centrifuged at 12,000 g for 20 min and the resulted supernatant was used for determination of enzyme activity. For CAT activity was performed as described previously (Chao et al. 2012; Kato and Shimizu 1985). The decrease in H2O2 was determined as the decrease in absorbance at 240 nm with a spectrophotometer (Metertec SP8001). The activity was calculated by the extinction coefficient (40 mM−1 cm−1 at 240 nm) for H2O2. One unit of CAT was defined as the amount of enzyme that degraded 1 μmol H2O2 per min. For SOD activity was determined according to Chao et al. (2012). The reaction mixture contained 100 mM triethanolamine- diethanolamine buffer (pH 7.4), EDTA/MnCl2 (100 mM/50 mM, pH 7.4), 7.5 mM β-NADH, 10 mM 2-mercaptoethanol, and enzyme extract. The reaction was started by the addition of β-NADH and absorbance was measured at 340 nm for 1 min. One unit of SOD was defined as the amount of enzyme that inhibited by 50 % the rate of β-NADH oxidation. For APX activity, the decrease in ascorbic acid (AsA) concentration was determined as the decline in absorbance at 290 nm and activity was calculated by the extinction coefficient (2.8 mM−1 cm−1 at 290 nm) for AsA (Chao et al. 2012; Nakano and Asada 1981). For total peroxidase activity was performed as described previously (Lin and Kao 1999; MacAdam et al. 1992). The shoot tissues were homogenized with 50 mM potassium phosphate buffer (pH 5.8) containing 0.8 M KCl with liquid nitrogen to extract both soluble and ionically bound (‘total’) peroxidase. The reaction mixture contained 50 mM potassium phosphate buffer (pH 5.8), 21.6 mM guaiacol, 39 mM H2O2 and enzyme extract. The reaction was started by the addition of H2O2 and absorbance was measured at 470 nm for 1 min. The activity was calculated by the extinction coefficient (26.6 mM−1 cm−1 at 470 nm) for tetraguaiacol. One unit of peroxidase was defined as the amount of enzyme that caused the formation 1 μmol tetraguaiacol per min.
Quantitative RT-PCR Analyses
Shoot samples were collected from seedlings and frozen until analysis. RNA was isolated from frozen tissues by TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Total RNA samples from shoots were first treated with DNase I and then reverse transcribed into cDNA by Moloney murine leukemia virus reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed using an Illumina Eco Real-Time PCR system (Biogenesis Technology, San Diego, USA) with Power SYBR Green PCR Master Mix (Applied Biosystems), according to the manufacturer’s instructions. The relative expression level of each gene was quantified using the comparative threshold cycle method, as described in the manufacturer’s instructions for the Eco™ Real-Time PCR System User Guide. The ubiquitin (Os03g13170) gene was used as an internal control to normalize the cDNA levels. Data were analyzed using the EcoStudy software, version 4.0 (Biogenesis Technology). Amplification conditions were as follows: 94 °C for 5 min, and then 45 cycles at 94 °C for 15 s, 55 °C for 15 s and 72 °C for 30 s. The primers used for quantitative RT-PCR analyses were listed in Table 1. Quantitative RT-PCR experiments were repeated least six times independently in duplicate, and the data were averaged.
Abbreviations
- APX:
-
ascorbate peroxidase
- CAT:
-
catalase
- FS:
-
full submergence
- H2O2 :
-
hydrogen peroxide
- POX:
-
total peroxidase
- PS:
-
partial submergence
- Rbohs:
-
respiratory burst oxidase homologs
- ROS:
-
reactive oxygen species
- RT-PCR:
-
reverse transcription-polymerase chain reaction
- SOD:
-
superoxide dismutase
References
Bailey-Serres J, Voesenek LA (2010) Life in the balance: a signaling network controlling survival of flooding. Curr Opin Plant Biol 13(5):489–494
Bailey-Serres J, Fukao T, Ronald P, Ismail A, Heuer S, Mackill D (2010) Submergence tolerant rice: SUB1’s journey from Landrace to Modern Cultivar. Rice 3(2–3):138–147
Baxter A, Mittler R, Suzuki N (2014) ROS as key players in plant stress signalling. J Exp Bot 65(5):1229–1240
Blokhina O, Virolainen E, Fagerstedt KV (2003) Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann Bot 91(2):179–194
Chao YY, Chou TS, Kao CH (2012) Involvement of abscisic acid and hydrogen peroxide in regulating the activities of antioxidant enzymes in leaves of rice seedlings under magnesium deficiency. Plant Growth Regul 66(1):1–8
Damanik RI, Maziah M, Ismail MR, Ahmad S, Zain A (2010) Responses of the antioxidative enzymes in Malaysian rice (Oryza sativa L.) cultivars under submergence condition. Acta Physiol Plant 32(4):739–747
Desikan R, Last K, Harrett-Williams R, Tagliavia C, Harter K, Hooley R, Hancock JT, Neill SJ (2006) Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J 47(6):907–916
Fukao T, Yeung E, Bailey-Serres J (2011) The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23(1):412–427
Fukao T, Yeung E, Bailey-Serres J (2012) The submergence tolerance gene SUB1A delays leaf senescence under prolonged darkness through hormonal regulation in rice. Plant Physiol 160(4):1795–1807
Hortensteiner S (2009) Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci 14(3):155–162
Jackson MB (2008) Ethylene-promoted elongation: an adaptation to submergence stress. Ann Bot 101(2):229–248
Jackson MB, Ram PC (2003) Physiological and molecular basis of susceptibility and tolerance of rice plants to complete submergence. Ann Bot 91(2):227–241
Kamal AH, Komatsu S (2015) Involvement of reactive oxygen species and mitochondrial proteins in biophoton emission in roots of soybean plants under flooding stress. J Proteome Res 14(5):2219–2236
Kato M, Shimizu S (1985) Chlorophyll metabolism in higher plants VI. Involvement of peroxidase in chlorophyll degradation. Plant Cell Physiol 26(7):1291–1301
Kato Y, Collard BC, Septiningsih EM, Ismail AM (2014) Physiological analyses of traits associated with tolerance of long-term partial submergence in rice. AoB Plants 6:plus058. doi: 10.1093/aobpla/plu058
Lherminier J, Elmayan T, Fromentin J, Elaraqui KT, Vesa S, Morel J, Verrier JL, Cailleteau B, Blein JP, Simon-Plas F (2009) NADPH oxidase-mediated reactive oxygen species production: subcellular localization and reassessment of its role in plant defense. Mol Plant Microbe Interact 22(7):868–881
Licausi F, Perata P (2009) Low oxygen signaling and tolerance in plants. Adv Bot Res 50:139–198
Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58:115–136
Lin CC, Kao CH (1999) NaCl induced changes in ionically bound peroxidase activity in roots of rice seedlings. Plant and Soil 216:147–153
Liu B, Rennenberg H, Kreuzwieser J (2015) Hypoxia induces stem and leaf nitric oxide (NO) emission from poplar seedlings. Planta 241(3):579–589
MacAdam JW, Nelson CJ, Sharp RE (1992) Peroxidase activity in the leaf elongation zone of tall fescue. Plant Physiol 99(3):872–878
Maruta T, Inoue T, Tamoi M, Yabuta Y, Yoshimura K, Ishikawa T, Shigeoka S (2011) Arabidopsis NADPH oxidases, AtrbohD and AtrbohF, are essential for jasmonic acid-induced expression of genes regulated by MYC2 transcription factor. Plant Sci 180(4):655–660
Nakano Y, Asada K (1981) Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol 22(5):867–880
Nishiuchi S, Yamauchi T, Takahashi H, Kotula L, Nakazono M (2012) Mechanisms for coping with submergence and waterlogging in rice. Rice 5:2. doi:10.1186/1939-8433-5-2
Panda D, Sharma SG, Sarkar RK (2008) Chlorophyll fluorescence parameters, CO2 photosynthetic rate and regeneration capacity as a result of complete submergence and subsequent re-emergence in rice (Oryza sativa L.). Aquat Bot 88(2):127–133
Pogany M, von Rad U, Grun S, Dongo A, Pintye A, Simoneau P, Bahnweg G, Kiss L, Barna B, Durner J (2009) Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis-Alternaria pathosystem. Plant Physiol 151(3):1459–1475
Pruzinska A, Anders I, Aubry S, Schenk N, Tapernoux-Luthi E, Muller T, Krautler B, Hortensteiner S (2007) In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19(1):369–387
Pucciariello C, Banti V, Perata P (2012a) ROS signaling as common element in low oxygen and heat stresses. Plant Physiol Bioch 59:3–10
Pucciariello C, Parlanti S, Banti V, Novi G, Perata P (2012b) Reactive oxygen species-driven transcription in Arabidopsis under oxygen deprivation. Plant Physiol 159(1):184–196
Sagi M, Fluhr R (2006) Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol 141(2):336–340
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Sakagami JI, John Y, Sone C (2013) Complete submergence escape with shoot elongation ability by underwater photosynthesis in African rice, Oryza glaberrima Steud. Field Crop Res 152:17–26
Sarkar RK, Bhattacharjee B (2012) Rice genotypes with SUB1 QTL differ in submergence tolerance, elongation ability during submergence and re-generation growth at re-emergence. Rice 5:7
Steffens B, Geske T, Sauter M (2011) Aerenchyma formation in the rice stem and its promotion by H2O2. New Phytol 190(2):369–378
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Vellosillo T, Vicente J, Kulasekaran S, Hamberg M, Castresana C (2010) Emerging complexity in reactive oxygen species production and signaling during the response of plants to pathogens. Plant Physiol 154(2):444–448
Voesenek LA, Bailey-Serres J (2015) Flood adaptive traits and processes: an overview. New Phytol 206(1):57–73
Winkel A, Colmer TD, Ismail AM, Pedersen O (2013) Internal aeration of paddy field rice (Oryza sativa) during complete submergence---importance of light and floodwater O2. New Phytol 197(4):1193–1203
Yamada Y, Furusawa S, Nagasaka S, Shimomura K, Yamaguchi S, Umehara M (2014) Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 240(2):399–408
Yang CY (2014) Hydrogen peroxide controls transcriptional responses of ERF73/HRE1 and ADH1 via modulation of ethylene signaling during hypoxic stress. Planta 239(4):877–885
Yang CY, Hong CP (2015) The NADPH oxidase Rboh D is involved in primary hypoxia signalling and modulates expression of hypoxia-inducible genes under hypoxic stress. Environ Exp Bot 115:63–72
Yin D, Chen S, Chen F, Jiang J (2013) Ethylene promotes induction of aerenchyma formation and ethanolic fermentation in waterlogged roots of Dendranthema spp. Mol Biol Rep 40(7):4581–4590
Acknowledgment
The author thanks Dr. Jun-Yi Yang for instrument assistance of quantitative RT-PCR.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ Contributions
YSW carried out the experiments and analyzed the data. Dr. CYY defined the research theme, designed experiments and wrote the manuscript. Both authors read and approved the final manuscript.
Additional files
Additional file 1: Figure S1.
Characterization of rice (Oryza sativa) seedlings growth under different levels of submergence. The first (a) and second (b) leaves sheath length of 14 days-old rice seedlings after treated submergence for 2, 4, 6 and 8 days. Control check (CK), partial submergence (PS) and full submergence (FS). The data represent average values ± SD from 30 seedlings of each treatment obtained from six biologically independent experiments. Values with the different letters are significantly different at P < 0.05, according to post-hoc LSD test. (JPG 1543 kb)
Additional file 2: Figure S2.
Phylogenetic tree of members of Rbohs protein family. Phylogenetic tree of 10 Arabidopsis Rboh proteins and 9 rice (Oryza sativa) related Rboh proteins. The phylogenetic tree was constructed by the Neighbor Joining algorithm (Saitou and Nei 1987) implemented in the MEGA 6 software package (Tamura et al. 2013). (JPG 614 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wu, YS., Yang, CY. Physiological Responses and Expression Profile of NADPH Oxidase in Rice (Oryza Sativa) Seedlings under Different Levels of Submergence. Rice 9, 2 (2016). https://doi.org/10.1186/s12284-016-0074-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-016-0074-9