Abstract
Background
Migraine is a complex neurovascular disorder with a strong genetic component. There are rare monogenic forms of migraine, as well as more common polygenic forms; research into the genes involved in both types has provided insights into the many contributing genetic factors. This review summarises advances that have been made in the knowledge and understanding of the genes and genetic variations implicated in migraine etiology.
Findings
Migraine is characterised into two main types, migraine without aura (MO) and migraine with aura (MA). Hemiplegic migraine is a rare monogenic MA subtype caused by mutations in three main genes - CACNA1A, ATP1A2 and SCN1A - which encode ion channel and transport proteins. Functional studies in cellular and animal models show that, in general, mutations result in impaired glutamatergic neurotransmission and cortical hyperexcitability, which make the brain more susceptible to cortical spreading depression, a phenomenon thought to coincide with aura symptoms. Variants in other genes encoding ion channels and solute carriers, or with roles in regulating neurotransmitters at neuronal synapses, or in vascular function, can also cause monogenic migraine, hemiplegic migraine and related disorders with overlapping symptoms. Next-generation sequencing will accelerate the finding of new potentially causal variants and genes, with high-throughput bioinformatics analysis methods and functional analysis pipelines important in prioritising, confirming and understanding the mechanisms of disease-causing variants.
With respect to common migraine forms, large genome-wide association studies (GWAS) have greatly expanded our knowledge of the genes involved, emphasizing the role of both neuronal and vascular pathways. Dissecting the genetic architecture of migraine leads to greater understanding of what underpins relationships between subtypes and comorbid disorders, and may have utility in diagnosis or tailoring treatments. Further work is required to identify causal polymorphisms and the mechanism of their effect, and studies of gene expression and epigenetic factors will help bridge the genetics with migraine pathophysiology.
Conclusions
The complexity of migraine disorders is mirrored by their genetic complexity. A comprehensive knowledge of the genetic factors underpinning migraine will lead to improved understanding of molecular mechanisms and pathogenesis, to enable better diagnosis and treatments for migraine sufferers.
Similar content being viewed by others
Background
Migraine types and classification
Migraine is a common type of primary headache disorder, distinguished by recurrent attacks of moderate to severe unilateral throbbing pain, often accompanied by nausea and/or photophobia and phonophobia. It is classified into two major types: migraine without aura (MO) and migraine with aura (MA), with visual, sensory or other central nervous system (CNS) symptoms preceding the headache and associated migraine symptoms, in the latter [1]. Other subtypes or forms have been classified, including chronic migraine and episodic syndromes associated with migraine. Hemiplegic migraine (HM) is a rare, severe subtype of MA, in which migraine symptoms are accompanied by motor symptoms such as temporary numbness or weakness, affecting one side of the body (hemiparesis). Familial hemiplegic migraine (FHM) is a familial form of HM where it is usually inherited in an autosomal dominant manner. Investigating the genetic basis of FHM, as well as the common types of MO and MA, has greatly helped in our understanding of migraine pathophysiology through the discovery of the genes that contribute to the disorder.
Migraine phases and pathophysiology
Activation of the trigeminovascular system
Migraine is thought to be a complex brain network disorder that occurs when the brain loses control of its homeostasis, leading to the activation of the trigeminovascular system and a cascade of events [2]. Signals from activated nociceptors innervating the cranial blood vessels are transmitted to the trigeminal bipolar neurons, and further relayed to thalamic and cortical areas [3, 4]. The signal from the perivascular neurons is transmitted by endogenous mediators, including the vasoactive neuropeptides calcitonin gene-related peptide (CGRP), substance P, neurokinin A, and pituitary adenylate cyclase-activating peptide (PACAP), as well as release of vasoactive inflammatory mediators such as nitric oxide, coincident with inflammation in the meninges [2, 5]. Sensitization of pain relevant brainstem regions, including peripheral trigeminovascular neurons to dural stimuli, is thought to produce the characteristic sensation of throbbing pain in migraine [6, 7].
Migraine progression and mechanisms
During migraine, distinct areas of the brain are activated, each contributing to aspects of migraine pathophysiology, whether this is triggering the attack, generating the pain, or playing roles in some of the associated neurological symptoms that occur during an attack [2]. Migraine is characterised by multiple phases; trigeminal activation occurs in the headache phase, but these may be preceded by a premonitory phase, in which symptoms including fatigue, mood changes, food cravings, yawning, muscle tenderness, and photophobia may be experienced up to 3 days before the headache [8]. Some individuals also experience an aura phase, which may feature visual, sensory, speech/language, and motor disturbances, as well as disruption of higher cortical function, immediately preceding or concurrent with the headache [8]. Cortical spreading depression (CSD) is a slowly propagating wave of depolarization in neuronal and glial cell membranes accompanied by massive ion fluxes, which spreads across the brain cortex, followed by a suppression of activity [9]. It coincides with the initiation and progression of aura symptoms, but whether CSD is causally linked to the initiation of headache is still debated [10]. Evidence from experimental animals supports a pivotal role of CSD in aura, headache initiation and activation of trigeminal nociception [11,12,13]; CSD-associated opening of neuronal Panx1 mega-channels releases molecules that trigger an inflammatory cascade, which activates neighboring astrocytes and leads to sustained release of inflammatory mediators [13]. Most migraineurs, however, do not experience aura, and it is unlikely that CSD is involved in initiating the complete syndrome of migraine. Alternative triggers for trigeminovascular activation, such as cortical hyperexcitability and brain stem or hypothalamic dysfunction, may also be important [14].
Brain alterations in migraine
A variety of imaging techniques have revealed both structural and functional brain alterations in individuals that suffer migraine [14]. Furthermore, clinical and neurophysiological studies have found chronic hypersensitivity to sensory stimuli and or abnormal processing of sensory information in migraineurs [15,16,17], as well as cortical excitability which may make them more susceptible to CSD [17, 18]. While some of these changes may be the result of repetitive exposure to pain or stress, the brain biology of migraine sufferers appears to differ from healthy controls [2]. Migraine may be triggered by a range of external factors, including chemicals, lack of sleep, stress, and skipping meals. However, these triggers only lead to migraine in migraineurs. Some aspects of the altered brain biology are likely to be genetically predetermined.
A genetic basis for migraine
Family and twin studies have demonstrated that there are genetic factors that contribute to the susceptibility of an individual to migraine. This is clear for individuals with monogenic migraine disorders, such as FHM, in which a pathogenic variant in a single gene can lead to the disorder, with nearly complete penetrance. Family and twin studies also suggest that common migraine is also a heritable trait, with heritability estimated between 30 and 60% [19,20,21]. Common migraine forms, including MO and MA, are most likely due to the contribution of variants with small effect at many genetic loci, i.e. these are considered to be polygenic disorders. Different approaches have been used to identify and understand the function of the genes involved in monogenic and polygenic migraine. For the former, this has been achieved by linkage mapping of genetic markers and sequencing of candidate genes in family pedigrees featuring the disorder, followed with functional studies in cellular and animal models. In recent years Next-generation sequencing (NGS) techniques have accelerated the discovery of genes and variants linked to monogenic migraine-related disorders. With regards to polygenic forms, genome-wide association studies (GWAS) in large migraine case-control cohorts has greatly helped our understanding of the many genetic factors and pathways that contribute to common migraine, with subsequent transcriptomics and functional experiments required for further understanding of the causal mechanisms.
Main text
Genetics of monogenic migraine disorders
Valuable insights into how some of the underlying genetic factors contribute to the pathophysiology of migraine have been provided by a number of rare inherited migraine disorders, which can be caused by mutations in a single gene (Table 1). These include hemiplegic migraine (HM) and familial migraine (where migraine is inherited in a Mendelian manner), as well as a range of monogenic neurological and vascular disorders which can show symptomatic crossover. The latter include some types of episodic ataxias, paroxysmal movement disorders, and the stroke syndrome cerebral autosomal dominant arteriopathy with subcortical infarcts and leucoencephalopathy (CADASIL; Mendelian Inheritance in Man catalogue, MIM #125310), and commonly feature migraine and/or episodic attacks of associated symptoms such as motor weakness, vertigo and nausea, along with their other characteristic symptoms.
Hemiplegic migraine
Hemiplegic Migraine (HM) is a rare subtype of MA characterised by episodes of severe migraine and aura symptoms involving motor weakness or numbness, usually affecting one side of the body (hemiparesis), as well as visual, sensory, or speech disturbances [1, 22]. In some cases, patients experience additional neurological symptoms such as confusion, seizures, memory loss, and coma. Individuals usually fully recover between episodes, although some symptoms may persist for weeks or longer, and some patients can develop permanent ataxia (difficulty coordinating movements), which may worsen with time [23]. In rare cases HM can be fatal after a minor head trauma [24].
Familial hemiplegic migraine (FHM)
The prevalence of HM has been found to be up to 0.01% in European populations, with both familial and sporadic forms [23, 25, 26]. FHM is diagnosed when there is at least one 1st or 2nd degree relative in the family who also suffers HM attacks. FHM usually shows an autosomal dominant pattern of inheritance (with 70–90% penetrance) and is considered to be monogenic, but genetically heterogeneous. To date three main causative genes – CACNA1A, ATP1A2 and SCN1A – have been identified through linkage studies and mutational screening in FHM family pedigrees. FHM can be classified as FHM1 (MIM #141500), FHM2 (MIM #602481) and FHM3 (MIM #609634) according to whether patients have mutations in CACNA1A, ATP1A2 or SCN1A, respectively. Clinically these FHM sub-types are indistinguishable, as symptoms overlap, but there is wide variation in phenotypes, including between individuals with mutations in the same gene, or even family members with the same mutation [27,28,29]. This suggests that other genes or environmental factors can modify phenotype. It should be noted that the majority of cases (< 25%) do not appear to have mutations in the CACNA1A, ATP1A2 or SCN1A genes [30] and our results (under review). Nevertheless, identifying and studying the known FHM genes and mutations has greatly improved diagnostics as well as understanding of the underlying biology of HM. The three main HM genes encode ion channel or ion transport proteins, leading to the supposition that HM is a channelopathy [31].
FHM1 due to mutations in CACNA1A
CACNA1A on chromosome 19p13 was the first gene implicated in FHM (FHM1), identified via positional cloning and mutation analysis of candidate genes in multiple FHM family pedigrees [32]. The gene encodes the pore-forming α1 subunit of the neuronal voltage-gated Cav2.1 (P/Q-type) channels, which are predominantly localized at the presynaptic terminals of brain and cerebellar neurons, and play an important role in controlling neurotransmitter release [33]. > 25 pathogenic variants in CACNA1A have been reported for FHM1, which are inherited in an autosomal dominant pattern. CACNA1A deletions have been reported in FHM1 patients [34, 35], however the majority are missense variants, lying in significant functional domains of the calcium channel, i.e. the voltage sensor, pore, and pore-lining loops [36]. They usually have gain-of-function effects, leading to increased Ca2+ influx, which results in enhanced glutamatergic neurotransmission and neuronal hyperexcitability [32, 37, 38]. While a strict genotype-phenotype correlation does not exist [29, 39], symptoms and clinical severity may vary depending on the variant [40, 41]. Transgenic FHM1 knock-in (KI) mouse models have been generated: one, which expresses the milder R192Q CACNA1A mutation, shows no overt phenotype [42], while another with the severe S218 L mutation exhibits cerebellar ataxia and spontaneous seizures in accordance with severity of the clinical symptoms observed in patients (28). In both these mouse models the FHM1 mutations cause gain-of-function effects, leading to altered cortical excitatory-inhibitory balance, increased neurotransmission, and increased susceptibility to CSD action [42,43,44,45]. Additionally, increased trigeminal sensory firing [44, 46, 47], tissue anoxia attributing to prolonged aura [48], head pain when triggered [49], and altered CGRP-mediated trigeminal pain signalling and synaptic plasticity [4, 50], have been observed in FHM KI models.
What controls trigeminal sensory excitability in between FHM attacks remains unknown [44]; this, in conjunction with extreme clinical diversity and variability, suggests that a number of environmental factors and/or modifier genes may act independently on the function of neuronal P/Q calcium channels as compensatory mechanisms until a threshold is reached [29]. Screens for genetic modifiers in animal models are consistent with this. For example, genetic knockdown of Drosophila phospholipase C beta (PLCβ, which is involved in cardiovascular and neuronal signalling), or genetic variants affecting the receptors that gate intracellular calcium stores (e.g. inositol triphosphate [IP3] and Ryanodine receptors), partially alleviated some of the electrophysiological phenotypes of FHM1 mutations [51]. In another example, a large-scale functional RNAi screen in Caenorhabditis elegans for modifiers of unc-2, the worm orthologue of CACNA1A, identified genes in the TGF-β and Notch signalling pathways [52]. Interestingly, those pathways are relevant to both common migraine, as revealed by association studies [53], as well as other monogenic disorders such as CADASIL which has overlapping symptoms with FHM [54]. Studies in FHM1 transgenic mice have also demonstrated the role of female sex hormones in increased susceptibility to CSD (37), suggesting that hormones are also modifying factors, and may explain some of the variable expressivity and penetrance of FHM pathogenic variants and the female preponderance of migraine disorders (49).
Episodic Ataxia 2 and spinocerebellar Ataxia type 6 due to mutations in CACNA1A
In addition to FHM1, heterozygous mutations within CACNA1A can cause two other neurological disorders, episodic ataxia type 2 (EA2; MIM #108500) and spinocerebellar ataxia type 6 (SCA6; #MIM 183086) [32, 55]. EA2 is characterized by paroxysmal attacks of ataxia, vertigo, and nausea, while SCA6 is typified by adult-onset, slowly progressive cerebellar ataxia, dysarthria, and nystagmus. There can be overlapping clinical features between the three allelic disorders [56], e.g. ~ 50% EA2 patients also suffer migraine [57], and episodic headaches and nausea are also common in SCA6 [58]. EA2 mutations can be missense, truncating or cause aberrant splicing of CACNA1A [59]. However, unlike FHM mutations, they are usually loss-of-function and result in decreased Ca2+ influx [4]. SCA6 mutations are usually small expansions of a polyglutamine repeat in the COOH tail of CACNA1A [55] which leads to accumulation of mutant Cav2.1 channels and selective degeneration of cerebellar Purkinje cells due to a toxic gain-of-function effect [60].
FHM2 due to mutations in ATP1A2
In 2003, ATP1A2 at 1q23.2 was identified as second major FHM gene [61]. ATP1A2 encodes the α2 isoform of the catalytic subunit of the Na+/K+-ATPase ion transport pump, which is responsible for regulating electrochemical gradients across the cell membranes of the CNS, heart, skeletal and smooth muscle tissue [62]. The pump is mainly expressed on astrocytes at tripartite synapses in the CNS, and its function in the clearance of extracellular K+ and production of a Na+ gradient used in the reuptake of glutamate, is important to its role in HM [63]. ATP1A2 mutations (FHM2) are usually inherited in an autosomal dominant pattern, and patients have a wide clinical spectrum [62, 64], which includes neurological disorders such as alternating hemiplegia of childhood [65], epilepsy [66], seizures [67], and permanent mental retardation [68, 69], as well as neuromuscular periodic paralysis disorders [70] and recurrent coma and fever [71], secondary to recurrent FHM-like attacks. > 80 causal variants have been linked to FHM2, with ~ 25 diagnosed in sporadic cases, suggesting that de novo mutations are common at the ATP1A2 locus [62]. While CACNA1A mutations are reported as the most common in some HM cohorts [36, 72], using an NGS panel to screen the three main HM genes in an Australian patient cohort we found that ~two-thirds of the HM mutations identified were in ATP1A2 (under review).
The majority of FHM2 mutations are missense and cluster in the catalytic P domain, the transmembrane domain, or in the central region between these; small deletions, a mutation causing protein extension through stop codon alterations, and exonic duplication have also been reported [62, 73,74,75]. In vitro functional models have been used to determine the functional consequences of a number of ATP1A2 FHM2 mutations, with studies demonstrating significant protein dysfunction ranging from partial to complete loss [62]. ATP1A2 mutations have been found to: i) alter (increase or decrease) pump sensitivity to potassium [76, 77]; ii) reduce the sodium/potassium turnover rate [40]; or iii) generate non-functional proteins [78,79,80]. Homozygous Atp1a2 knock-out (KO) mice die immediately after birth [81], and recently biallelic loss of function variants in ATP1A2 have been reported in humans, resulting in death neonatally, with features of hydrops fetalis, microcephaly, arthrogryposis and extensive cortical malformations [82]. Heterozygote KO mice have altered behaviour and neurological defects [81], but also exhibit a low threshold for induction of CSD, faster propagation rate, and delayed recovery from mass depolarization compared to wild-type mice [83]. FHM2 KI mice carrying either the human W887R or G301R mutations, show altered CSD, with the former more susceptible to CSD due to a reduced rate of glutamate and K+ clearance by cortical astrocytes [84, 85], and the latter displaying a prolonged recovery phase following CSD [86]. Therefore, ATP1A2 mutations have been hypothesised to contribute to FHM pathophysiology by increasing the propensity for CSD action due to increased levels of synaptic K+ and glutamate as a result of dysfunctional Na+/K+ ATPase pump action [87, 88]. While many FHM2 ATP1A2 mutations abolish or greatly reduce pump activity, others cause more subtle effects, including shifts in voltage dependence, kinetics, or apparent cation affinities [62]. Nevertheless, they affect glutamatergic neurotransmission, causing the defective regulation of the balance of excitation and inhibition in the brain seen in migraine [89].
FHM3 due to mutations in SCN1A
SCN1A (chr 2q24.3) was identified as a third causative gene for FHM in 2005 [90]. FHM3 is rarer than FHM1 and 2 (up to ~ 10% of patients with a molecular diagnosis). SCN1A encodes the α1 subunit of the neuronal voltage-gated sodium channel Nav1.1, which mediates the voltage-dependent sodium ion permeability of excitable membranes (primarily the inhibitory gamma-Aminobutyric acid [GABA]-ergic interneurons) of the CNS [91]. SCN1A is commonly mutated in epilepsy syndromes with hundreds of heterozygous truncating and missense mutations reported [92]. Eleven FHM3 SCN1A mutations have been described to date, and are usually inherited in an autosomal dominant manner [93,94,95]. Mutations have been identified in both pure FHM families, and also in those with FHM and additional neurological disorders, including generalised tonic-clonic epilepsy, elicited repetitive transient daily blindness and childhood epilepsy [96,97,98].
Epileptic mutations mainly cause loss-of-function, resulting in reduced sodium currents and action potential firing in GABAergic inhibitory interneurons [99,100,101]; SCN1A KO mice suffer from ataxia and epileptic seizures [102, 103]. In FHM3, mutations in SCN1A are usually missense and cause gain-of-function effects on the channel, displaying increased threshold-near persistent current, delayed entry into inactivation, and a faster recovery and higher channel availability during repetitive stimulation [104,105,106,107]. This predicts increased firing of inhibitory GABAergic neurons, leading to higher extracellular potassium concentrations, enhanced glutamate release, and triggering of CSD [106, 108]. However, the mechanisms of SCN1A mutations in FHM3 can be complicated: some exhibit loss-of-function effects in heterologous cell systems [109]; a SCN1A T1174S mutation reported in a family with both epileptic and FHM phenotypes can act in both a gain- and loss-of-function manner [105]; and furthermore, the SCN1A L1670 W and L1649Q mutations induce folding and trafficking defects which, when rescued by incubation at lower temperatures, or when expressed in GABAergic cortical neurons, modifies the gating properties leading to an overall gain-of-function [110, 111]. KI mouse models of FHM3 mutations have not been reported to date, but would help further understanding of their mechanisms of pathogenesis.
Sporadic hemiplegic migraine (SHM)
Sporadic Hemiplegic Migraine (SHM) is diagnosed when there is no family history of HM, and estimates suggest in the general population approximately one-third of cases are sporadic [25]. SHM can be caused by pathogenic variants in the known FHM genes, including those that have arisen de novo, which may then become familial cases [41, 74, 112] Variants in ATP1A2 have been the most commonly found in SHM cases, possibly reflecting greater genetic heterogeneity, or more variable penetrance, in this gene [62]. SHM may result from less penetrant variants in the known FHM genes, mosaicism in the transmitting parent, pathogenic variants in other genes, and/or other modes of inheritance, e.g. compound recessive mutations and gene/environment interactions [23, 93]. Some SHM cases may also represent a phenotypic extreme of common migraine due to a combination of lower-risk genetic variants. For example, Pelzer et al. (2018) found that individuals with HM, but without mutations in CACNA1A, ATP1A2 or SCN1A, generally have a milder phenotype than those with mutations in those genes [41].
Hemiplegic migraine and disorders with overlapping symptoms caused by mutations in other genes
Although rare, pathogenic variants in other genes, including PRRT2, PNKD, SLC2A1, SLC1A3, SLC4A4, have been reported in HM. Mutations in PRRT2 and PNKD are more commonly associated with paroxysmal conditions, in particular movement disorders [113]. PNKD is the main causal gene for paroxysmal non-kinesigenic dyskinesia (PNKD; MIM #118800) [114, 115], while PRRT2 mutations can cause paroxysmal kinesigenic dyskinesia (PKD; MIM #128200) [116, 117], paroxysmal non-kinesigenic dyskinesia (PNKD) [118], paroxysmal exercise-induced dyskinesia (PED), and childhood epilepsy/seizure disorders [119, 120]. Some patients presenting with HM have been found to have mutations in PRRT2 [118, 121,122,123,124], leading to the suggestion that it is a fourth HM gene [121]. However, the relationship is complicated due to the clinical heterogeneity and pleiotropy of phenotypes, and it may mainly act in a modifying role [125]. PRRT2 encodes Proline Rich Transmembrane Protein 2 (PRRT2), a presynaptic transmembrane protein which interacts with members of the SNAP Receptor (SNARE) complex [126]. It is involved in synaptic vesicle fusion and regulation of voltage-gated calcium channels in glutamatergic neurons, and is important in the final steps of neurotransmitter release [127,128,129]. Heterozygous PRRT2 c.649dupC (p.Arg217Profs*8) or c.649delC (p.Arg217Glufs*12) loss-of-function truncating mutations are the most common in PRRT2-related conditions, including HM, and are likely to result in impaired interaction with the SNAP25/SNARE complex and increased presynaptic vesicle release, leading to a state of hyperexcitability [118].
Mutations in both PNKD, the main causal gene for PNKD, and SLC2A1, the glucose transporter protein type 1 (GLUT1 or EAAT2) gene implicated in PED and GLUT1 deficiency syndrome (MIM #606777), have also been found in HM patients [118, 130, 131]. They likely act via disruption of neurotransmitter regulation and impaired synaptic vesicle release [118]. Mutations in SLC1A3, the gene for the glial glutamate transporter EAAT1, can cause episodic ataxia, type 6, (EA6; MIM #612656), but have also been associated with HM [132, 133]. Similarly, mutations in SLC4A4, the gene for the sodium bicarbonate cotransporter NBCe1, which is usually involved in renal tubular acidosis syndromes (MIM #604278) are also found in some HM cases [134]. Analysis of whole exome sequencing (WES) data of HM patients without CACNA1A, ATP1A2 and SCN1A mutations suggests that mutations in all these genes are rare [41] and our results (under review), but should be considered in the molecular diagnosis of patients without mutations in the main HM genes.
Familial migraine with Aura and associated disorders
The majority of studies of migraine in family pedigrees with Mendelian inheritance have focussed on those with the HM phenotype. However, a few cases of familial MA have been reported, which have revealed other genes and molecular mechanisms involved in migraine biology.
Familial migraine with Aura caused by mutations in KCNK18 encoding the TRESK channel
A monogenic form of typical MA in a large multigenerational pedigree identified a frameshift mutation (F139Wfsx24) in the TWIK-related spinal cord potassium channel (TRESK, encoded by KCNK18), segregating with migraine [135]. TRESK is a member of the two pore domain potassium channel (K2P) family, which regulate the excitability of a variety of neurons involved in transducing pain stimuli, including the somatosensory neurons of the dorsal root ganglia (DRG) and trigeminal ganglia [136, 137]. KO mouse models suggest TRESK functions to modify certain forms of nociceptive afferentation [138, 139]. Functional analysis suggested a dominant negative effect of the TRESK F139Wfsx24 mutation on whole-cell TRESK currents resulting in hyperexcitability of trigeminal ganglion neurons [140]. However, another dominant negative TRESK mutation, C110R, which is not associated with migraine [141], does not trigger sensory neuron hyperexcitability, even though it reduces TRESK currents in sensory neurons [142]. A recent study by Royal et al. (2019) sheds light on this apparent contradiction and has revealed a novel mechanism by which frameshift mutations can alter the function of a gene [143]. Firstly, they found that TRESK can heterodimerise with two other K2P channels, TREK1 and TREK2, which when knocked out together in mice results in a migraine-like allodynia phenotype. The TRESK-C110R protein inhibits TRESK activity on dimerization, but does not affect TREK1 and TREK2, while TRESK-F139Wfsx24 inhibits activity of all three channels. Interestingly, the 2 bp frameshift puts an alternative start codon in frame, which results in translation of a second TRESK fragment. It is this that specifically downregulates TREK1 and TREK2 function, which appears to contribute to migraine induction. Furthermore, Royal et al. (2019) identified another TRESK frameshift mutation (Y121LfsX44) in a human exome sequence database, and which is associated with migraine in ClinVar, that appears to work via the same mechanism which they have termed frameshift mutation-induced alternative translation initiation [143]. Finally, this work suggests that TREK-related genes may also be involved in migraine.
Familial advanced sleep-phase syndrome (FASPS) and migraine caused mutations in CSNK1D
Casein kinase 1 delta (CKIδ) is a central component of the circadian clock. Mutations in the CKIδ gene, CSNK1D, were found to cause familial advanced sleep phase syndrome (FASPS) in two large independent pedigrees [144, 145]. FASPS patients show severe disruption of the sleep-wake-cycle and other circadian rhythms, but interestingly, the phenotype also co-segregated with MA in these pedigrees. Mice carrying a transgene with the human CKIδ-T44A mutation displayed sensitization to pain after triggering migraine with nitroglycerin, and a reduced threshold for CSD; cultured astrocytes showed increased spontaneous and induced calcium signalling [144, 145]. Further details of its role in migraine are to be elucidated, but CKIδ is a ubiquitous serine-threonine kinase that phosphorylates the circadian clock protein PER2, as well as other proteins involved in brain signalling [146]. CSNK1D is a notable exception to the ion channel and glutamatergic-related genes implicated in the majority of monogenic migraine, and the connection between migraine and FASPs is consistent with a likely role of the hypothalamus in regulating physiological stresses and migraine susceptibility [147,148,149].
ROSAH syndrome – retinal dystrophy, optic nerve edema, splenomegaly, anhidrosis and migraine headache – caused by mutations in ALPK1
ROSAH is a recently described distinct autosomal dominant ocular systemic disorder, which features migraine headache as one of the key clinical features. Exome and genome sequencing identified a heterozygous missense pathogenic variant in the ALPK1 gene (c.710C > T, p.[Thr237Met]) in five independent families [150]. ALPK1 encodes Alpha Kinase 1, which may play roles in inflammation and intracellular trafficking, although its function is poorly defined, and it is not yet understood the how mutations in the protein would contribute to migraine.
Monogenic vascular disorders which feature migraine
Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL)
There are a number of primarily vascular disorders caused by mutations in single genes, in which migraine is a common symptom. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL), is a cerebral small-vessel disease (SVD) characterised by vascular degeneration, recurrent subcortical ischaemic strokes, cognitive decline, dementia, and premature death [54]. It is the most common heritable cause of stroke and vascular dementia in adults, caused by toxic gain-mutations in NOTCH3, which are usually autosomal dominant. Migraine, in particular the MA subtype, is a common symptom accompanying CADASIL (in up to 75% cases) [151,152,153,154], often presenting decades before the onset of other symptoms [54, 155]. For example, a study of 300 symptomatic CADASIL patients found that three quarters had migraine (90% of which was MA), and in two-thirds of the patients it was the presenting symptom [153].
Retinal vasculopathy with cerebral leukodystrophy (RVCL) and COL4A1-related SVDs
Other SVDs that commonly feature migraine include syndromes such as retinal vasculopathy with cerebral leukodystrophy (RVCL; MIM #192315) caused by mutations in TREX1 [156, 157], and COL4A1 and COL4A2-related disorders [158,159,160]. The exact mechanism through which vascular disorders lead to an increased prevalence of migraine is unknown [154], but they indicate that some genes with roles in vascular function are also implicated in migraine, something which has also become apparent in polygenic migraine from both epidemiological studies and GWAS [161, 162].
Methods and applications for identifying disease-causing variants in monogenic migraine and related disorders
Next-generation sequencing for molecular testing of hemiplegic migraine
Until relatively recently HM genetic testing involved Sanger sequencing of selected exons in one, two or all three main HM causative genes (CACNA1A, ATP1A2 and SCN1A). This form of iterative testing was limited and could be costly and time consuming. The development of next-generation sequencing (NGS) technologies, in which millions of small fragments of DNA are sequenced in parallel, have revolutionised genomic research, allowing specific regions of interest to the entire genome to be sequenced concurrently. NGS applications include targeted gene panels, WES (in which all the coding regions of the genome are sequenced), and Whole Genome Sequencing (WGS), which also captures introns, regulatory regions and all other non-coding DNA. NGS has been applied clinically in genetic diagnostics, including for HM and overlapping disorders, facilitating the discovery of novel HM mutations [163,164,165]. Using a five gene panel designed for HM and overlapping disorders (EA2 and CADASIL), our laboratory has found that diagnostic success rates have increased considerably (~ 21%) when compared to that of previous Sanger sequencing testing methods (~ 9%), and have identified a number of novel causative variants for HM and related disorders [166, 167]. Clinicians also appreciate the option to test for overlapping neurological disorders when presented with complex cases with HM-related symptoms.
Discovering new genes in migraine-related disorders
Importantly, recent application of NGS sequencing techniques to screen HM patients have shown that the majority do not have exonic mutations in the main HM genes [30]. We find that > 75% patients sent for testing do not have likely pathogenic exonic variants in CACNA1A, ATP1A2 or SCN1A (under review). Furthermore, analysis of data from NGS panels or WES has revealed that likely pathogenic variants in other known familial migraine and migraine-related genes are also rare [41], [our results (under review)]. This low level of diagnostic success may largely be due to other causative genes or genetic factors, although no other major HM loci have been found so far [41]. In addition to the three main genes, HM may be highly genetically heterogeneous. From what is already known of the biology, other genes likely to be involved in HM may include ion channel and solute transporter genes, as well as genes involved in aspects of glutamatergic neurotransmission and vascular biology. Assigning causality for variants that are less dominant or penetrant than those in the known HM genes will be challenging. This is exemplified in a study by Klassen et al. (2011) comparing ion channel variant profiles of unaffected individuals to those with sporadic idiopathic epilepsy from targeted exome sequencing; rare missense variants were prevalent in both groups at similar complexity, demonstrating that even deleterious ion channel variants confer uncertain risk to an individual depending on the other variants with which they are combined [168]. In fact Hiekkala et al. have hypothesized that HM may not be a true monogenetic disease, but that it may reflect an extreme phenotype in the MA spectrum where rare and/or multiple common variants contribute to the disease outcome [30].
Assigning function to potential HM and migraine-causing variants
Determining the biological effect of variants on protein function is a major limitation in medical genetics. As NGS techniques reveal many more variants, particularly if HM is highly genetically heterogeneous, it will be necessary to improve functional testing pipelines to filter those likely to be pathogenic. Public databases which provide variant frequency (e.g. dbSNP, Genome Aggregation Database [169]) and previously reported pathogenicity information (e.g. ClinVar [170], Leiden Open Variation Databases), and in silico bioinformatics tools which predict functional consequences (e.g. SIFT [171], Polyphen2 [172], and MutationTaster) are useful in prioritising lists of candidate variants by providing first assessments of pathogenicity [173, 174, 175]. In silico methods to predict the impact of regulatory variants are also being developed [176, 177]. In addition to in silico analysis, functional assays are necessary to provide further evidence for pathogenicity, or otherwise, for prioritised variants, and to explore molecular mechanisms. Testing exogenous DNA constructs with engineered variants in cell and animal models can be complimented with genome-editing technologies, particularly the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system, which allows more refined and faster generation of knock-out or knock-in lines [178]. Coupled with induced pluripotent stem cells (iPSCs), which are able to be differentiated into various neuronal cell types [179, 180], as well as brain organoids [181], variants can be functionally tested in more relevant cell models, or generated from patients so they can be studied in the context of their genomic background. A range of approaches to scale up such assays are being developed [182], e.g. deep mutational scanning, which combines large scale generation of variants with deep sequencing, is a technique allowing the effect of a combination of variants to be tested at once [183], and high throughput electrophysiology platforms are available for testing ion channel variants [184].
Targeting treatment to genetic diagnosis in HM-related disorders
A molecular diagnosis is likely to improve management and treatment efficiencies for neurological disorders, even if symptoms may overlap, as the specific pathway or mechanism can be targeted. E.g. Glut1 deficiency caused by SLC2A1 mutations can be treated using a ketogenic diet and HM symptoms, if present, have been found to improve on a modified Atkins diet [131]. In HM cases with PRRT2 mutations, some benefit has been observed with carbamazepine, the most frequently used drug in treating PKD and PKD/IC patients [185]. A range of acute and prophylactic drugs are used for HM, and some may be more effective than others depending on the nature of the causative genetic mutation [22].
Genetics of common migraine
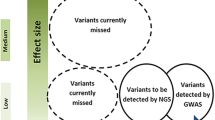
Monogenic migraine disorders have a large impact on the individuals and families involved, but they are rare. The majority of migraine is polygenic, i.e. it is a complex disorder in which multiple variants in genes contribute to the underlying risk, with each one usually having a relatively small effect. Disease susceptibility is further a result of interaction of these genetic variations with each other, and with environmental and lifestyle factors. Discovering loci and genes that contribute to common migraine requires different approaches to the Mendelian disorders, mainly based on finding differences in allele frequencies of genetic variants linked to genes, between cohorts of migraine cases and non-migraine controls, composed of unrelated individuals. Common genetic variation largely comprises of SNPs, small insertions or deletions, short tandem repeats, and copy number variants. Most effort in identifying variants that influence traits and disorders, including migraine, has been focussed on the SNPs that confer an increased or decreased risk of migraine. These studies are demanding as, although each variant may contribute to migraine susceptibility, it is neither necessary, nor sufficient, to cause it. Effect sizes for most loci are generally small (allelic odds ratio of 1.03–1.28), requiring genotyping of large numbers of individuals to robustly obtain results that pass significance thresholds [162]. Significant differences in allele frequencies of a SNP does not necessarily mean that the SNP is itself a susceptibility factor, but that a causal variant may be in linkage disequilibrium (LD) with it. Linking the associated polymorphism to the variant that elicits the effect, or even to the gene affected, is often challenging.
Association studies of polymorphisms in migraine candidate genes
For many years, association studies of SNPs in and around hypothesis-driven candidate genes was the main approach used to investigate genes thought to be involved in migraine. Studies generally genotyped either known functional variants, or tagging SNPs across gene loci selected from biological pathways thought to be relevant, e.g. neurological, vascular, hormonal, and inflammatory pathways [186]. Association studies of close to 200 polymorphisms in ~ 100 genes have been published for migraine [187], although subsequent and replication studies often reported conflicting results. The occurrence of false positive results in case-control study designs may be due to small sample sizes, lack of consideration for LD blocks, inadequate correction for multiple testing and phenotyping issues [40]. The C667T variant (rs1801133) in the 5,10-methylenetetrahydrofolate reductase gene (MTHFR), encoding a key enzyme in the folate pathway, results in an alanine to valine substitution in the catalytic domain, which reduces its activity by ~ 50% [188]. MTHFR C667T has been one of the most extensively studied polymorphisms in migraine; some meta-analyses report association of the T-allele with MA, but not MO [189,190,191,192], however, this has not been supported by other meta-analyses [193, 194]. Furthermore, a systematic re-evaluation of the most promising candidate gene SNPs, including MTHFR C667T, and others previously found to be positively associated with migraine, showed no clear evidence for involvement in migraine using International Headache Genetics Consortium (IGHC) GWAS data for 5175 clinic-based migraineurs and 13,972 controls [195]. Population stratification, where a significant association may be due to the underlying structure of the population irrespective of disease status, can contribute to biased or conflicting results in case-control studies [196]. Genetic background and population-specific risk factors may also lead to divergent findings. One MTHFR C667T meta-analysis reported association with migraine and MA of the T-allele, particularly in populations belonging to Asian ancestry [192].
Genome-wide association studies (GWAS) for migraine
Hypothesis-free GWAS present a more unbiased method to identify SNPs, and potentially genes, robustly involved in migraine to gain insights into its pathways and pathophysiology. SNP arrays have enabled the simultaneous genotyping of hundreds of thousands to millions of SNPs in a sample, essentially allowing the entire genome to be scanned. Genotyped SNPs serve as a proxy for any SNPs that are in strong LD, which are tested for association with the trait in question. A number of migraine GWAS have been performed, including five major studies [53, 197,198,199,200], with the most recent meta-analysis bringing the number of associated SNPs to 44 that mapped to 38 independent genomic loci [53]. Earlier GWAS identified migraine susceptibility SNPs nearby genes with mainly putative or known neuronal functions, including MTDH, PRDM16, TPRM8 and LRP1 [197, 198]. LRP1 has been shown to exert regulatory effects on a number of correlated cellular events including amyloid precursor protein metabolism, kinase dependent intracellular signalling, neuronal calcium signalling and modulation of synaptic transmission through the N-methyl-D-aspartate glutamate receptors via regulating the cellular distribution of GluA1 receptors on neurons [201,202,203]. TPRM8 encodes for a receptor-activated non-selective cation channel activated by cold environmental temperatures and is related to pain sensor channels [204]. PRDM16 plays roles in leukaemogenesis, palatogenesis, and brown fat cell differentiation from skeletal muscle [205], but also promotes stem cell maintenance in fetal hematopoietic and nervous systems and adult neural stem cell maintenance, neurogenesis, and ependymal cell differentiation, partly via modulating oxidative stress [206, 207].
A GWAS by Freilinger et al. (2012) had revealed that, in addition to genes involved in synapse and neuronal function and differentiation (MEF2D and ASTN2), genes with vascular functions (TGFBR2, PHACTR1) were also likely to be important in migraine susceptibility [199]. For example, TGFBR2 encodes part of the receptor complex which transduces TGF-β signalling and regulates both synaptic and endothelial functions [208, 209]. The GWAS meta-analyses of Antilla et al. (2013) and Gormley et al. (2016), with expanded sample sizes, reiterated this fact with the discovery of further loci near genes with neuronal functions, but also many more gene loci related to functions in vascular and smooth muscle tissues, underlining their contribution to migraine pathophysiology [53, 161]. The most recent meta-analysis by Gormley et al. (2016) combined 22 GWA studies from the International Headache Genetics Consortium (IGHC), comprised 59,674 migraine cases from clinic- and population-based collections, as well as samples obtained by partnerships with the commercial entities 23andMe and deCODE, and 316,078 controls [53]. This study brought the number of SNPs significantly associated with migraine to 44 independent SNPs at 38 distinct genomic loci, and included the majority of GWAS loci previously reported, as well as an additional 28 novel loci, including the first on the X chromosome (Near MED14-USP9X). Database annotations and relevant literature for the genes in LD with the SNPs have been reviewed by Gormley et al. (supplementary tables) [53] and Sutherland et al. (table) [93].
The meta-analysis by Gormley et al. confirmed the single most significant SNP as rs11172113 in the LRP1 gene locus, and that the genes prioritised as likely candidates at many of the loci have known or putative roles in vascular function (e.g. LRP1, PRDM16, ECM1, MEF2D, TGFBR2, ARHGEF26, REST, PHACTR1, NOTCH4, FHL5, GJA1, HEY2, NRP1, PLCE1, HTRA1, YAP1, FGF6, ZCCHC14, JAG1, and CCM2L) and the expression of many of these is highly enriched in vascular tissues [53, 162]. Furthermore, consistent with the mechanisms that have been elucidated from FHM, two of the loci are near ion channels genes, TPRM8 and KCNK5, the latter a member of the same family as KCNK18. Three additional loci are linked to the SLC24A3, ITPK1 and GJA1 genes, which all have a function in cellular ion homeostasis. More unexpectedly, many genes that contribute to migraine susceptibility are involved in metal ion homeostasis according to Gene Ontology (GO) terms (PRDM16, TGFBR2, REST, FHL5, NRP1, MMPED2, LRP1, ZCCHC14, RNF213, JAG1, SLC24A3) suggesting the importance of these pathways in migraine pathophysiology [162]. Metal ions (including Fe2+, Cu2+, Co2+, Mn2+, Ca2+, Na+, and Zn2+) are essential in many metabolic processes and their transport and storage into cellular compartments is highly regulated [210]. How these processes might be contribute to migraine remains to be fully elucidated, however, it is known for example, that synaptic zinc is a potent modulator of neurotransmission [211].
It should be noted that many of the loci have both neuronal and vascular functions, and/or roles in multiple pathways [53, 93, 162]. For example, NRP1 encodes neuropilin 1, a cell surface glycoprotein which mediates axon guidance and adhesion during GABAergic synapse formation in developing nervous system [212], but is also involved in vascular patterning and cardiovascular system development as a receptor for the vascular guidance molecule semaphoring 3d [213]. Furthermore, there is some overlap in pathways between monogenic migraine genes and GWAS loci. In common with the monogenic FHM and MA forms caused by ion channel gene mutations, some ion channel gene loci are implicated in polygenic migraine. Similarly, genes of the Notch signalling pathway are involved in both the monogenic migraine-related cerebrovascular disorder CADASIL (caused by pathogenic NOTCH3 variants) and common migraine, with GWAS loci identified near both the NOTCH4 receptor gene, and JAG1, which encodes Jagged1, a ligand of multiple Notch receptors.
Fine mapping and functional analysis of migraine associated SNPs
Analyses of the genes in the vicinity of GWAS loci has suggested the types of gene function and pathways that may be involved in migraine, however, it is important to remember that for the majority of loci, the gene that is actually influenced by the SNP remains unknown. SNPs affect the diversity of human traits/diseases via various mechanisms: changing encoded amino acids of a protein (non-synonymous) may affect its function or localisation; and SNPs that are either silent (synonymous), or more commonly, in noncoding regions, may affect gene expression levels via messenger RNA (mRNA) conformation and stability, subcellular localization, or its promoter/enhancer activity. Making the leap from associated SNPs to causal genes, and then to functional mechanisms, still presents a formidable task in the interpretation of GWAS.
Methods have been developed to fine-map GWAS loci, combining statistical and functional evidence [214, 215]. Firstly, association-test statistics can be combined with LD information to prioritise a credible set of SNPs likely to contain the causal disease-associated SNP. As susceptibility SNPs often lie in introns or intergenic regions, the next hurdle is to identify which gene is affected (not necessarily the nearest), by connecting the variants with genes by a range of methods and resources, complementing functional annotation with information from projects such as ENCyclopedia of DNA Elements (ENCODE), NIH Roadmap Epigenomics, and FANTOM5, which have characterized regulatory regions and expression quantitative trait loci (eQTL) [162, 214]. Once putative variants and genes have been pinpointed via in silico analysis, further functional experiments are required to confirm and understand molecular mechanisms. This process is illustrated by investigations into rs9349379 in intron 3 of the PHACTR1 gene, which has been identified as a causal susceptibility SNP in a range of vascular disorders including migraine [216]. From epigenomic data from human tissues, Gupta et al. (2017) identified an enhancer signature over rs9349379 in aorta suggesting a vascular regulatory function; then using CRISPR-edited stem cell-derived endothelial cells they demonstrated that the SNP actually regulates expression of the endothelin 1 gene (EDN1), located 600 kb upstream of PHACTR1 [216]. EDN1 encodes a 21 amino acid peptide that, along with its receptor, promotes vasoconstriction, vascular smooth muscle cell proliferation, extracellular matrix production, and fibrosis; these factors would contribute to the increased risk of coronary artery disease and decreased risk of cervical artery dissection, fibromuscular dysplasia and migraine, conferred by the SNP [216]. This work underlines the importance of functional assays in cellular and animal models in further characterisation of migraine GWAS signals.
In another effort to refine GWAS loci, Hannon et al. applied summary-data-based Mendelian randomization (SMR) to large DNA methylation quantitative trait locus (mQTL) datasets generated from blood and fetal brain to prioritize genes for > 40 complex traits with well-powered GWAS data, including migraine [217]. Using this approach they showed that, with respect to the HEY2-NOCA7 GWAS signal identified by Gormley et al. [53], whole blood and fetal brain have a mQTL profile highly comparable to that of the migraine GWAS, which implicated HEY2 in migraine. These results are consistent with genetic signals influencing DNA methylation in both tissues and migraine, and shows utility of this approach in prioritizing specific genes within genomic regions identified by GWAS [217]. The expansion of resources with gene expression and epigenetic data in tissues relevant to migraine-related pathophysiology will be critical to advancing these types of studies. Recent studies have used gene expression datasets (including single cell analysis) to begin to link genetic loci to their expression in migraine-relevant brain tissues and cell types [218,219,220].
Migraine susceptibility loci in migraine sub-types
There has been some discussion about whether MO and MA are different entities or part of a disease spectrum [221,222,223]. Subtype analysis in high-powered GWAS with large samples sizes may reveal whether particular genes may contribute to phenotypic consequences. Most of the migraine loci identified by Gormley et al., (2016) were implicated in both MO and MA, although seven genomic loci (near TSPAN2, TRPM8, PHACTR1, FHL5, ASTN2, near FGF6 and LRP1) were significantly associated with the MO subtype [53]. None were significant for MA, likely reflecting the smaller sample size. Some genetic loci may be selectively associated with particular features (e.g. pain character, duration, frequency, nausea, photophobia and triggers) of the migraine attack [224, 225]. Menstrual migraine affects a subset of female MO sufferers; replication of migraine GWAS loci in a menstrual migraine case-control cohort suggested a particular role for NRP1 in this subgroup [226]. However, the small sample sizes often make it difficult to obtain robust associations for such specific phenotypes. Nevertheless, it will be interesting to identify genes that might be involved in specific aspects of migraine.
Shared genetic factors with other disorders
A wider view is also informative and can be used to explore the etiology of related and comorbid traits. A GWAS of broadly defined headache using the UK Biobank data found significant associations at 28 loci, of which 14 overlapped with migraine, including the rs11172113 in the LRP1 as the top SNP [227]. Some migraine-associated genes and SNPs have more systemic effects and are involved in a wide range of disorders. A large analysis of shared heritability between common brain disorders found that while most psychiatric and neurologic disorders share relatively little common genetic risk, suggesting largely independent etiological pathways, migraine appears to share some genetic architecture with psychiatric disorders, including attention deficit hyperactivity disorder (ADHD), Tourette’s syndrome, and major depressive disorder [228]. This, together with genetic correlations with other neurological (epilepsy) and vascular disorders (stroke, coronary artery disease), is consistent with comorbidities that have been documented for migraine and suggests they are underpinned by shared genetic factors [228,229,230,231,232,233]. Similarly, the monogenic migraine disorders show comorbidity with epilepsy, depression, vascular and sleep disorders [54, 145, 234, 235]. Understanding these relationships can impact the management and treatment of conditions with overlapping etiologies [235, 236].
Migraine susceptibility loci in migraine in specific populations
As the large migraine GWAS have been performed in predominantly Caucasian populations of European heritage, questions remain as to whether the genes and SNPs identified are relevant to other ethnicities, and if there are population-specific genes and polymorphisms. One way to address the former is to test whether there is replication of association of the GWAS SNPs in a particular population. A number of studies have taken this approach, both in specific European cohorts, as well as North Indian and Han Chinese. For example, association of the minor C allele for the PRDM16 polymorphism rs2651899 was replicated in Swedish [237], Spanish [238] and Han Chinese cohorts [239, 240], while rs2651899 and LRP1 rs11172113 showed a protective effect on migraine susceptibility in a North Indian population [241]. Polymorphisms rs4379368 (Succinyl-CoA:Glutarate-CoA Transferase gene locus, C7orf10) and rs13208321 (FHL5) showed some replication in a cohort of the Chinese She people [242]. However, GWAS conducted in specific ethnic populations will determine whether the genetic contributions to migraine vary, and identify migraine susceptibility loci which may be particular to different groups. While still limited, and with relatively small sample sizes, GWAS have been performed in Norfolk Islander, Taiwanese Han Chinese and African American pediatric cohorts [243,244,245]. The Norfolk Island genetic isolate is a unique admixed Polynesian-Caucasian population with a high prevalence of migraine (25%). A GWAS for migraine revealed a number of loci of suggestive significance near neurotransmitter-related genes [245]. A GWAS in Taiwanese Han Chinese identified two novel migraine susceptibility SNPs: rs655484 in DLG2, a gene involved in glutamatergic neurotransmission; and rs3781545 in GFRA1, which encodes a receptor for glial cell line-derived neurotrophic factor (GDNF) in trigeminal neurons [243]. The GWAS in American African children found association of migraine with SNPs, including rs72793414, which were strongly correlated with the mRNA expression levels of NMUR2, encoding the G protein-coupled receptor of the CNS neuropeptide neuromedin-U [244].
Genetic risk scores (GRS) and applications for migraine
Due to low effect sizes that the majority of variants have on associated traits, the genotype at an individual SNP does not have particular diagnostic or prognostic value in common migraine. However, calculating a genetic risk score (GRS) or polygenic risk score (PRS), which assesses the additive effect of many associated SNPs from sufficiently powered studies, may have utility in disease prediction [246]. With the availability of increasingly large GWAS data sets for migraine, GRS may be applied to: investigating migraine subtypes and endophenotypes, understanding migraine pleiotropy and co-morbidites, disease and phenotype prediction, and for assessing pharmocogenetic effects for personalised medicine [247]. Higher GRS have been correlated with migraine diagnosis in specific cohorts [226, 248], as well as migraine severity, and in cases where migraine is aggregated in families suggesting this results from a higher common variant burden [225, 249]. One particular use of GRS may be in understanding drug reactions and efficacy of therapies. Studies to predict response and efficacy of treatment with triptans in migraineurs have used this approach [250, 251]. While sensitivity and specificity are still relatively low, the diagnostic value of GRS will improve with the discovery of more SNPs. With respect to drug and treatment responses, this would include variants that affect the genes targeted by drugs, but also those involved in drug transport and metabolism [252, 253].
Powering up GWAS and genomic sequencing
It is likely that common variants will not completely explain common migraine, but that rare private variants (with small to medium effects) will contribute as well. This has been demonstrated by the well-studied trait of adult human height, which has a strong genetic component (estimated heritability up to 80%). Meta-analysis of multiple GWAS with a combined sample size of > 250,000 individuals has yielded ~ 700 common SNPs clustered in 423 independent loci that contribute to height [254]. These, however, still only capture ~ 20% of the heritability. Compound heterozygote-like SNP interactions may further contribute to phenotypic variance [255]. Furthermore, using ExomeChips, Marouli et al. identified a further 83 coding variants with lower minor-allele frequencies (in the range of 0.1–4.8%) associated with height [256]. However, in addition to further scaling up of sample sizes, ultimately WGS will be required to truly discover all of the DNA sequence contribution to the trait. For migraine, sample sizes are still relatively small compared to the studies that have been done for traits like height and obesity, i.e. > 500,000 individuals including 170,000 Japanese [257, 258]. It is likely that more migraine-related loci will be discovered as sample numbers increase in migraine GWAS using SNP-chips (including from various ethnicities), and the effect of rare variants identified from exonic and genomic sequencing becomes clearer. Integrating genetic and other genomic information, such as transcriptional and epigenetic data, will deepen understanding of the important tissues and pathways in migraine [218, 259].
Conclusions
Migraine is a multifactorial disorder with genetics playing an important role in the susceptibility, and symptomology, as well as comorbidity with other traits and conditions. Investigation of the genetic factors involved in migraine have used family studies for the rare, Mendelian forms of migraine, as well as GWAS in case-control cohorts for the common polygenic form of migraine, for gene discovery and further understanding of the pathways and basic biology of the disorder (Fig. 1). For monogenic migraine, mapping of loci in family pedigrees, coupled with genomic sequencing to find variants, led to the discovery of the main FHM genes, CACNA1A, ATP1A2 and SCN1A. Knowledge of their roles as ion channels and in ion transport, along with functional experiments in cellular and animal models, has contributed to uncovering how their dysfunction may lead to cortical hyperexcitability and migraine. Mutations in other genes can also cause HM, and it is likely that pathogenic variants in more genes will be discovered, with NGS technologies (WES and WGS) accelerating this research. With respect to the common polygenic forms of migraine, GWAS analyses using high-throughput SNP genotyping arrays has revealed many variants around genes with roles in neurological and vascular pathways in migraine. With increasing sample sizes more susceptibility loci are likely to be found, some of which may contribute to specific migraine subtypes or symptoms. Moving from finding a risk SNP, to the gene, to the molecular mechanism, still remains challenging, but developments around methods for functional studies, including iPSC models and genome-editing, will facilitate such research.
Genetics has further emphasized the complexity of migraine disorders, but it is an exciting time to be working in the field of migraine biology, with the end game – to better diagnose, manage and treat migraine sufferers.
Availability of data and materials
Not applicable.
Abbreviations
- BFIE:
-
Benign familial infantile epilepsy
- CADASIL:
-
Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy
- CGRP:
-
Calcitonin gene-related peptide
- CNS:
-
Central nervous system
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- dbSNP:
-
Single Nucleotide Polymorphism Database
- DMRs:
-
Differentially methylated regions
- EA:
-
Episodic ataxia
- ENCODE:
-
ENCyclopedia of DNA Elements
- ExAC:
-
Exome Aggregation Consortium
- FANTOM:
-
Functional Annotation of the Mammalian Genome
- FASPS:
-
Familial advanced sleep phase syndrome
- FHM:
-
Familial hemiplegic migraine
- GABA:
-
Gamma-Aminobutyric acid
- GDNF:
-
Glial cell line-derived neurotrophic factor
- gnomAD:
-
Genome Aggregation Database
- GRS:
-
Genetic risk score
- GTex:
-
Gene-tissue expression project
- GWAS:
-
Genome-wide association study
- HM:
-
Hemiplegic migraine
- ICCA:
-
Infantile convulsions and choreoarthetosis
- IHGC:
-
International Headache Genetics Consortium
- iPSCs:
-
Induced pluripotent stem cells
- KI:
-
Knock-in
- KO:
-
Knock-out
- LD:
-
Linkage disequilibrium
- LOVD:
-
Leiden Open Variation Databases
- MA:
-
Migraine with aura
- MIM:
-
Mendelian Inheritance in Man
- MO:
-
Migraine without aura
- mQTL:
-
Methylation quantitative trait locus
- mRNA:
-
Messenger RNA
- NGS:
-
Next-generation sequencing
- NIH:
-
National Institute of Health
- PACAP:
-
Pituitary adenylate cyclase-activating peptide
- PED:
-
Paroxysmal exercise-induced dyskinesia
- PKD:
-
Paroxysmal kinesigenic dyskinesia
- PNKD:
-
Paroxysmal nonkinesigenic dyskinesia
- PRS:
-
Polygenic risk score
- ROSAH:
-
Retinal dystrophy, optic nerve edema, splenomegaly, anhidrosis and migraine headache
- RTA:
-
Renal tubular acidosis
- SHM:
-
Sporadic hemiplegic migraine
- SMR:
-
Summary-data-based Mendelian randomization
- SNARE:
-
SNAP Receptor
- WES:
-
Whole exome sequencing
- WGS:
-
Whole genome sequencing
References
(IHS) HCCotIHS (2018) The international classification of headache disorders, 3rd edition. Cephalalgia 38(1):1–211
Goadsby PJ, Holland PR, Martins-Oliveira M, Hoffmann J, Schankin C, Akerman S (2017) Pathophysiology of migraine: a disorder of sensory processing. Physiol Rev 97(2):553–622
Noseda R, Burstein R (2013) Migraine pathophysiology: anatomy of the trigeminovascular pathway and associated neurological symptoms, cortical spreading depression, sensitization, and modulation of pain. Pain 154(Suppl 1):S44–S53
Ferrari MD, Klever RR, Terwindt GM, Ayata C, van den Maagdenberg AM (2015) Migraine pathophysiology: lessons from mouse models and human genetics. Lancet Neurol 14(1):65–80
Messlinger K, Fischer MJ, Lennerz JK (2011) Neuropeptide effects in the trigeminal system: pathophysiology and clinical relevance in migraine. Keio J Med 60(3):82–89
Levy D (2012) Endogenous mechanisms underlying the activation and sensitization of meningeal nociceptors: the role of immuno-vascular interactions and cortical spreading depression. Curr Pain Headache Rep 16(3):270–277
Burstein R, Jakubowski M, Rauch SD (2011) The science of migraine. J Vestib Res 21(6):305–314
Dodick DW (2018) A phase-by-phase review of migraine pathophysiology. Headache 58(Suppl 1):4–16
Kramer DR, Fujii T, Ohiorhenuan I, Liu CY (2016) Cortical spreading depolarization: pathophysiology, implications, and future directions. J Clin Neurosci 24:22–27
Goadsby PJ (2001) Migraine, aura, and cortical spreading depression: why are we still talking about it? Ann Neurol 49(1):4–6
Bolay H, Reuter U, Dunn AK, Huang Z, Boas DA, Moskowitz MA (2002) Intrinsic brain activity triggers trigeminal meningeal afferents in a migraine model. Nat Med 8(2):136–142
Zhang X, Levy D, Kainz V, Noseda R, Jakubowski M, Burstein R (2011) Activation of central trigeminovascular neurons by cortical spreading depression. Ann Neurol 69(5):855–865
Karatas H, Erdener SE, Gursoy-Ozdemir Y, Lule S, Eren-Kocak E, Sen ZD et al (2013) Spreading depression triggers headache by activating neuronal Panx1 channels. Science 339(6123):1092–1095
Burstein R, Noseda R, Borsook D (2015) Migraine: multiple processes, complex pathophysiology. J Neurosci 35(17):6619–6629
Stankewitz A, Aderjan D, Eippert F, May A (2011) Trigeminal nociceptive transmission in migraineurs predicts migraine attacks. J Neurosci 31(6):1937–1943
Lang E, Kaltenhauser M, Neundorfer B, Seidler S (2004) Hyperexcitability of the primary somatosensory cortex in migraine--a magnetoencephalographic study. Brain 127(Pt 11):2459–2469
Aurora SK, Barrodale PM, Tipton RL, Khodavirdi A (2007) Brainstem dysfunction in chronic migraine as evidenced by neurophysiological and positron emission tomography studies. Headache 47(7):996–1003 discussion 4-7
Vecchia D, Pietrobon D (2012) Migraine: a disorder of brain excitatory-inhibitory balance? Trends Neurosci 35(8):507–520
Honkasalo ML, Kaprio J, Winter T, Heikkila K, Sillanpaa M, Koskenvuo M (1995) Migraine and concomitant symptoms among 8167 adult twin pairs. Headache 35(2):70–78
Mulder EJ, Van Baal C, Gaist D, Kallela M, Kaprio J, Svensson DA et al (2003) Genetic and environmental influences on migraine: a twin study across six countries. Twin Res 6(5):422–431
Polderman TJ, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM et al (2015) Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 47(7):702–709
Pelzer N, Stam AH, Haan J, Ferrari MD, Terwindt GM (2013) Familial and sporadic hemiplegic migraine: diagnosis and treatment. Curr Treat Options Neurol 15(1):13–27
Russell MB, Ducros A (2011) Sporadic and familial hemiplegic migraine: pathophysiological mechanisms, clinical characteristics, diagnosis, and management. Lancet Neurol 10(5):457–470
Kors EE, Terwindt GM, Vermeulen FL, Fitzsimons RB, Jardine PE, Heywood P et al (2001) Delayed cerebral edema and fatal coma after minor head trauma: role of the CACNA1A calcium channel subunit gene and relationship with familial hemiplegic migraine. Ann Neurol 49(6):753–760
Lykke Thomsen L, Kirchmann Eriksen M, Faerch Romer S, Andersen I, Ostergaard E, Keiding N et al (2002) An epidemiological survey of hemiplegic migraine. Cephalalgia 22(5):361–375
Barros J, Ruano L, Domingos J, Tuna A, Damasio J, Alonso I et al (2014) The prevalence of familial hemiplegic migraine with cerebellar ataxia and spinocerebellar ataxia type 6 in Portugal. Headache 54(5):911–915
Terwindt GM, Ophoff RA, Haan J, Vergouwe MN, van Eijk R, Frants RR et al (1998) Variable clinical expression of mutations in the P/Q-type calcium channel gene in familial hemiplegic migraine. Dutch Migraine Genetics Research Group. Neurology 50(4):1105–1110
Angelini C, Van Gils J, Bigourdan A, Jouk PS, Lacombe D, Menegon P et al (2018) Major intra-familial phenotypic heterogeneity and incomplete penetrance due to a CACNA1A pathogenic variant. Eur J Med Genet. https://doi.org/10.1016/j.ejmg.2018.08.011. [Epub ahead of print]
Kors EE, Haan J, Giffin NJ, Pazdera L, Schnittger C, Lennox GG et al (2003) Expanding the phenotypic spectrum of the CACNA1A gene T666M mutation: a description of 5 families with familial hemiplegic migraine. Arch Neurol 60(5):684–688
Hiekkala ME, Vuola P, Artto V, Happola P, Happola E, Vepsalainen S et al (2018) The contribution of CACNA1A, ATP1A2 and SCN1A mutations in hemiplegic migraine: a clinical and genetic study in Finnish migraine families. Cephalalgia 38(12):1849–1863
Cregg R, Momin A, Rugiero F, Wood JN, Zhao J (2010) Pain channelopathies. J Physiol 588(Pt 11):1897–1904
Ophoff RA, Terwindt GM, Vergouwe MN, van Eijk R, Oefner PJ, Hoffman SM et al (1996) Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell 87(3):543–552
Catterall WA (1998) Structure and function of neuronal Ca2+ channels and their role in neurotransmitter release. Cell Calcium 24(5–6):307–323
Labrum RW, Rajakulendran S, Graves TD, Eunson LH, Bevan R, Sweeney MG et al (2009) Large scale calcium channel gene rearrangements in episodic ataxia and hemiplegic migraine: implications for diagnostic testing. J Med Genet 46(11):786–791
Grieco GS, Gagliardi S, Ricca I, Pansarasa O, Neri M, Gualandi F et al (2018) New CACNA1A deletions are associated to migraine phenotypes. J Headache Pain 19(1):75
Ducros A, Denier C, Joutel A, Cecillon M, Lescoat C, Vahedi K et al (2001) The clinical spectrum of familial hemiplegic migraine associated with mutations in a neuronal calcium channel. N Engl J Med 345(1):17–24
Di Lorenzo C, Grieco GS, Santorelli FM (2012) Migraine headache: a review of the molecular genetics of a common disorder. J Headache Pain 13(7):571–580
Tottene A, Fellin T, Pagnutti S, Luvisetto S, Striessnig J, Fletcher C et al (2002) Familial hemiplegic migraine mutations increase ca (2+) influx through single human CaV2.1 channels and decrease maximal CaV2.1 current density in neurons. Proc Natl Acad Sci U S A 99(20):13284–13289
Tournier-Lasserve E (1999) CACNA1A mutations: hemiplegic migraine, episodic ataxia type 2, and the others. Neurology 53(1):3–4
de Vries B, Frants RR, Ferrari MD, van den Maagdenberg AM (2009) Molecular genetics of migraine. Hum Genet 126(1):115–132
Pelzer N, Haan J, Stam AH, Vijfhuizen LS, Koelewijn SC, Smagge A et al (2018) Clinical spectrum of hemiplegic migraine and chances of finding a pathogenic mutation. Neurology 90(7):e575–ee82
van den Maagdenberg AM, Pietrobon D, Pizzorusso T, Kaja S, Broos LA, Cesetti T et al (2004) A Cacna1a knockin migraine mouse model with increased susceptibility to cortical spreading depression. Neuron 41(5):701–710
Eikermann-Haerter K, Dilekoz E, Kudo C, Savitz SI, Waeber C, Baum MJ et al (2009) Genetic and hormonal factors modulate spreading depression and transient hemiparesis in mouse models of familial hemiplegic migraine type 1. J Clin Invest 119(1):99–109
Eroli F, Vilotti S, van den Maagdenberg A, Nistri A (2017) Hyperpolarization-activated current Ih in mouse trigeminal sensory neurons in a transgenic mouse model of familial hemiplegic migraine type-1. Neuroscience 351:47–64
Tottene A, Conti R, Fabbro A, Vecchia D, Shapovalova M, Santello M et al (2009) Enhanced excitatory transmission at cortical synapses as the basis for facilitated spreading depression in ca(v)2.1 knockin migraine mice. Neuron 61(5):762–773
Hullugundi SK, Ansuini A, Ferrari MD, van den Maagdenberg AM, Nistri A (2014) A hyperexcitability phenotype in mouse trigeminal sensory neurons expressing the R192Q Cacna1a missense mutation of familial hemiplegic migraine type-1. Neuroscience 266:244–254
Marchenkova A, van den Maagdenberg AM, Nistri A (2016) Loss of inhibition by brain natriuretic peptide over P2X3 receptors contributes to enhanced spike firing of trigeminal ganglion neurons in a mouse model of familial hemiplegic migraine type-1. Neuroscience 331:197–205
Khennouf L, Gesslein B, Lind BL, van den Maagdenberg AM, Lauritzen M (2016) Activity-dependent calcium, oxygen, and vascular responses in a mouse model of familial hemiplegic migraine type 1. Ann Neurol 80(2):219–232
Chanda ML, Tuttle AH, Baran I, Atlin C, Guindi D, Hathaway G et al (2013) Behavioral evidence for photophobia and stress-related ipsilateral head pain in transgenic Cacna1a mutant mice. Pain 154(8):1254–1262
Chen SP, Tolner EA, Eikermann-Haerter K (2016) Animal models of monogenic migraine. Cephalalgia 36(7):704–721
Brusich DJ, Spring AM, James TD, Yeates CJ, Helms TH, Frank CA (2018) Drosophila CaV2 channels harboring human migraine mutations cause synapse hyperexcitability that can be suppressed by inhibition of a Ca2+ store release pathway. PLoS Genet 14(8):e1007577
Pereira Mda C, Morais S, Sequeiros J, Alonso I (2016) Large-Scale Functional RNAi Screen in C. elegans Identifies TGF-beta and Notch Signaling Pathways as Modifiers of CACNA1A. ASN Neuro 8(2):1-10
Gormley P, Anttila V, Winsvold BS, Palta P, Esko T, Pers TH et al (2016) Meta-analysis of 375,000 individuals identifies 38 susceptibility loci for migraine. Nat Genet 48(8):856–866
Chabriat H, Joutel A, Dichgans M, Tournier-Lasserve E, Bousser MG (2009) Cadasil. Lancet Neurol 8(7):643–653
Zhuchenko O, Bailey J, Bonnen P, Ashizawa T, Stockton DW, Amos C et al (1997) Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat Genet 15(1):62–69
Blumenfeld AE, Victorio MC, Berenson FR (2016) Complicated migraines. Semin Pediatr Neurol 23(1):18–22
Jen J, Kim GW, Baloh RW (2004) Clinical spectrum of episodic ataxia type 2. Neurology 62(1):17–22
Sinke RJ, Ippel EF, Diepstraten CM, Beemer FA, Wokke JH, van Hilten BJ et al (2001) Clinical and molecular correlations in spinocerebellar ataxia type 6: a study of 24 Dutch families. Arch Neurol 58(11):1839–1844
Mantuano E, Romano S, Veneziano L, Gellera C, Castellotti B, Caimi S et al (2010) Identification of novel and recurrent CACNA1A gene mutations in fifteen patients with episodic ataxia type 2. J Neurol Sci 291(1–2):30–36
Watase K, Barrett CF, Miyazaki T, Ishiguro T, Ishikawa K, Hu Y et al (2008) Spinocerebellar ataxia type 6 knockin mice develop a progressive neuronal dysfunction with age-dependent accumulation of mutant CaV2.1 channels. Proc Natl Acad Sci U S A 105(33):11987–11992
De Fusco M, Marconi R, Silvestri L, Atorino L, Rampoldi L, Morgante L et al (2003) Haploinsufficiency of ATP1A2 encoding the Na+/K+ pump alpha2 subunit associated with familial hemiplegic migraine type 2. Nat Genet 33(2):192–196
Friedrich T, Tavraz NN, Junghans C (2016) ATP1A2 mutations in migraine: seeing through the facets of an ion pump onto the neurobiology of disease. Front Physiol 7:239
Benarroch EE (2010) Glutamate transporters: diversity, function, and involvement in neurologic disease. Neurology 74(3):259–264
Jurkat-Rott K, Freilinger T, Dreier JP, Herzog J, Gobel H, Petzold GC et al (2004) Variability of familial hemiplegic migraine with novel A1A2 Na+/K+-ATPase variants. Neurology 62(10):1857–1861
Bassi MT, Bresolin N, Tonelli A, Nazos K, Crippa F, Baschirotto C et al (2004) A novel mutation in the ATP1A2 gene causes alternating hemiplegia of childhood. J Med Genet 41(8):621–628
Deprez L, Weckhuysen S, Peeters K, Deconinck T, Claeys KG, Claes LR et al (2008) Epilepsy as part of the phenotype associated with ATP1A2 mutations. Epilepsia 49(3):500–508
Al-Bulushi B, Al-Hashem A, Tabarki B (2014) A wide clinical phenotype spectrum in patients with ATP1A2 mutations. J Child Neurol 29(2):265–268
Vanmolkot KR, Stroink H, Koenderink JB, Kors EE, van den Heuvel JJ, van den Boogerd EH et al (2006) Severe episodic neurological deficits and permanent mental retardation in a child with a novel FHM2 ATP1A2 mutation. Ann Neurol 59(2):310–314
Roth C, Freilinger T, Kirovski G, Dunkel J, Shah Y, Wilken B et al (2014) Clinical spectrum in three families with familial hemiplegic migraine type 2 including a novel mutation in the ATP1A2 gene. Cephalalgia 34(3):183–190
Sampedro Castaneda M, Zanoteli E, Scalco RS, Scaramuzzi V, Marques Caldas V, Conti Reed U et al (2018) A novel ATP1A2 mutation in a patient with hypokalaemic periodic paralysis and CNS symptoms. Brain 141(12):3308–3318
Pelzer N, Blom DE, Stam AH, Vijfhuizen LS, Hageman A, van Vliet JA et al (2017) Recurrent coma and fever in familial hemiplegic migraine type 2. A prospective 15-year follow-up of a large family with a novel ATP1A2 mutation. Cephalalgia 37(8):737–755
Carreno O, Corominas R, Serra SA, Sintas C, Fernandez-Castillo N, Vila-Pueyo M et al (2013) Screening of CACNA1A and ATP1A2 genes in hemiplegic migraine: clinical, genetic, and functional studies. Mol Genet Genomic Med 1(4):206–222
Riant F, De Fusco M, Aridon P, Ducros A, Ploton C, Marchelli F et al (2005) ATP1A2 mutations in 11 families with familial hemiplegic migraine. Hum Mutat 26(3):281
Riant F, Ducros A, Ploton C, Barbance C, Depienne C, Tournier-Lasserve E (2010) De novo mutations in ATP1A2 and CACNA1A are frequent in early-onset sporadic hemiplegic migraine. Neurology 75(11):967–972
Gagliardi S, Grieco GS, Gualandi F, Caniatti LM, Groppo E, Valente M et al (2017) De novo exonic duplication of ATP1A2 in Italian patient with hemiplegic migraine: a case report. J Headache Pain 18(1):63
Segall L, Mezzetti A, Scanzano R, Gargus JJ, Purisima E, Blostein R (2005) Alterations in the alpha2 isoform of Na,K-ATPase associated with familial hemiplegic migraine type 2. Proc Natl Acad Sci U S A 102(31):11106–11111
Tavraz NN, Friedrich T, Durr KL, Koenderink JB, Bamberg E, Freilinger T et al (2008) Diverse functional consequences of mutations in the Na+/K+-ATPase alpha2-subunit causing familial hemiplegic migraine type 2. J Biol Chem 283(45):31097–31106
Capendeguy O, Horisberger JD (2004) Functional effects of Na+,K+-ATPase gene mutations linked to familial hemiplegic migraine. NeuroMolecular Med 6(2–3):105–116
Koenderink JB, Zifarelli G, Qiu LY, Schwarz W, De Pont JJ, Bamberg E et al (2005) Na,K-ATPase mutations in familial hemiplegic migraine lead to functional inactivation. Biochim Biophys Acta 1669(1):61–68
Tavraz NN, Durr KL, Koenderink JB, Freilinger T, Bamberg E, Dichgans M et al (2009) Impaired plasma membrane targeting or protein stability by certain ATP1A2 mutations identified in sporadic or familial hemiplegic migraine. Channels (Austin) 3(2):82–87
Ikeda K, Onaka T, Yamakado M, Nakai J, Ishikawa TO, Taketo MM et al (2003) Degeneration of the amygdala/piriform cortex and enhanced fear/anxiety behaviors in sodium pump alpha2 subunit (Atp1a2)-deficient mice. J Neurosci 23(11):4667–4676
Monteiro FP, Curry CJ, Hevner R, Elliott S, Fisher JH, Turocy J et al (2019) Biallelic loss of function variants in ATP1A2 cause hydrops fetalis, microcephaly, arthrogryposis and extensive cortical malformations. Eur J Med Genet. https://doi.org/10.1016/j.ejmg.2019.01.014. [Epub ahead of print]
Unekawa M, Ikeda K, Tomita Y, Kawakami K, Suzuki N (2018) Enhanced susceptibility to cortical spreading depression in two types of Na(+),K(+)-ATPase alpha2 subunit-deficient mice as a model of familial hemiplegic migraine 2. Cephalalgia 38(9):1515–1524
Leo L, Gherardini L, Barone V, De Fusco M, Pietrobon D, Pizzorusso T et al (2011) Increased susceptibility to cortical spreading depression in the mouse model of familial hemiplegic migraine type 2. PLoS Genet 7(6):e1002129
Capuani C, Melone M, Tottene A, Bragina L, Crivellaro G, Santello M et al (2016) Defective glutamate and K+ clearance by cortical astrocytes in familial hemiplegic migraine type 2. EMBO Mol Med 8(8):967–986
Bottger P, Glerup S, Gesslein B, Illarionova NB, Isaksen TJ, Heuck A et al (2016) Glutamate-system defects behind psychiatric manifestations in a familial hemiplegic migraine type 2 disease-mutation mouse model. Sci Rep 6:22047
Gritz SM, Radcliffe RA (2013) Genetic effects of ATP1A2 in familial hemiplegic migraine type II and animal models. Hum Genomics 7:8
Isaksen TJ, Lykke-Hartmann K (2016) Insights into the pathology of the alpha2-Na(+)/K(+)-ATPase in neurological disorders; lessons from animal models. Front Physiol 7:161
Pietrobon D, Moskowitz MA (2013) Pathophysiology of migraine. Annu Rev Physiol 75:365–391
Dichgans M, Freilinger T, Eckstein G, Babini E, Lorenz-Depiereux B, Biskup S et al (2005) Mutation in the neuronal voltage-gated sodium channel SCN1A in familial hemiplegic migraine. Lancet 366(9483):371–377
Catterall WA (2000) From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron 26(1):13–25
Meng H, Xu HQ, Yu L, Lin GW, He N, Su T et al (2015) The SCN1A mutation database: updating information and analysis of the relationships among genotype, functional alteration, and phenotype. Hum Mutat 36(6):573–580
Sutherland HG, Griffiths LR (2017) Genetics of migraine: insights into the molecular basis of migraine disorders. Headache 57(4):537–569
Zhang Y, Chen N, Zhou M, Guo J, Guo J, He L (2017) A novel SCN1A mutation identified in a Chinese family with familial hemiplegic migraine: a case report. Cephalalgia 37(13):1294–1298
Mantegazza M, Cestele S (2018) Pathophysiological mechanisms of migraine and epilepsy: similarities and differences. Neurosci Lett 667:92–102
Le Fort D, Safran AB, Picard F, Bouchardy I, Morris MA (2004) Elicited repetitive daily blindness: a new familial disorder related to migraine and epilepsy. Neurology 63(2):348–350
Castro MJ, Stam AH, Lemos C, de Vries B, Vanmolkot KR, Barros J et al (2009) First mutation in the voltage-gated Nav1.1 subunit gene SCN1A with co-occurring familial hemiplegic migraine and epilepsy. Cephalalgia 29(3):308–313
Vanmolkot KR, Babini E, de Vries B, Stam AH, Freilinger T, Terwindt GM et al (2007) The novel p.L1649Q mutation in the SCN1A epilepsy gene is associated with familial hemiplegic migraine: genetic and functional studies. Mutation in brief #957. Online. Hum Mutat 28(5):522
Catterall WA (2018) Dravet syndrome: a Sodium Channel Interneuronopathy. Curr Opin Physiol 2:42–50
Escayg A, Goldin AL (2010) Sodium channel SCN1A and epilepsy: mutations and mechanisms. Epilepsia 51(9):1650–1658
Marini C, Scheffer IE, Nabbout R, Suls A, De Jonghe P, Zara F et al (2011) The genetics of Dravet syndrome. Epilepsia 52(Suppl 2):24–29
Ogiwara I, Miyamoto H, Morita N, Atapour N, Mazaki E, Inoue I et al (2007) Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci 27(22):5903–5914
Yu FH, Mantegazza M, Westenbroek RE, Robbins CA, Kalume F, Burton KA et al (2006) Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci 9(9):1142–1149
Cestele S, Scalmani P, Rusconi R, Terragni B, Franceschetti S, Mantegazza M (2008) Self-limited hyperexcitability: functional effect of a familial hemiplegic migraine mutation of the Nav1.1 (SCN1A) Na+ channel. J Neurosci 28(29):7273–7283
Cestele S, Labate A, Rusconi R, Tarantino P, Mumoli L, Franceschetti S et al (2013) Divergent effects of the T1174S SCN1A mutation associated with seizures and hemiplegic migraine. Epilepsia 54(5):927–935
Fan C, Wolking S, Lehmann-Horn F, Hedrich UB, Freilinger T, Lerche H et al (2016) Early-onset familial hemiplegic migraine due to a novel SCN1A mutation. Cephalalgia 36(13):1238-1247
Bertelli S, Barbieri R, Pusch M, Gavazzo P (2018) Gain of function of sporadic/familial hemiplegic migraine-causing SCN1A mutations: use of an optimized cDNA. Cephalalgia 39(4):477-488
Pellacani S, Sicca F, Di Lorenzo C, Grieco GS, Valvo G, Cereda C et al (2016) The revolution in migraine genetics: from aching channels disorders to a next-generation medicine. Front Cell Neurosci 10:156
Kahlig KM, Rhodes TH, Pusch M, Freilinger T, Pereira-Monteiro JM, Ferrari MD et al (2008) Divergent sodium channel defects in familial hemiplegic migraine. Proc Natl Acad Sci U S A 105(28):9799–9804
Cestele S, Schiavon E, Rusconi R, Franceschetti S, Mantegazza M (2013) Nonfunctional NaV1.1 familial hemiplegic migraine mutant transformed into gain of function by partial rescue of folding defects. Proc Natl Acad Sci U S A 110(43):17546–17551
Dhifallah S, Lancaster E, Merrill S, Leroudier N, Mantegazza M, Cestele S (2018) Gain of function for the SCN1A/hNav1.1-L1670W mutation responsible for familial hemiplegic migraine. Front Mol Neurosci 11:232
de Vries B, Freilinger T, Vanmolkot KR, Koenderink JB, Stam AH, Terwindt GM et al (2007) Systematic analysis of three FHM genes in 39 sporadic patients with hemiplegic migraine. Neurology 69(23):2170–2176
Silveira-Moriyama L, Kovac S, Kurian MA, Houlden H, Lees AJ, Walker MC et al (2018) Phenotypes, genotypes, and the management of paroxysmal movement disorders. Dev Med Child Neurol 60(6):559–565
Lee HY, Xu Y, Huang Y, Ahn AH, Auburger GW, Pandolfo M et al (2004) The gene for paroxysmal non-kinesigenic dyskinesia encodes an enzyme in a stress response pathway. Hum Mol Genet 13(24):3161–3170
Rainier S, Thomas D, Tokarz D, Ming L, Bui M, Plein E et al (2004) Myofibrillogenesis regulator 1 gene mutations cause paroxysmal dystonic choreoathetosis. Arch Neurol 61(7):1025–1029
Chen W-J, Lin Y, Xiong Z-Q, Wei W, Ni W, Tan G-H et al (2011) Exome sequencing identifies truncating mutations in PRRT2 that cause paroxysmal kinesigenic dyskinesia. Nat Genet 43:1252
Wang J-L, Cao L, Li X-H, Hu Z-M, Li J-D, Zhang J-G et al (2011) Identification of PRRT2 as the causative gene of paroxysmal kinesigenic dyskinesias. Brain 134(12):3493–3501
Gardiner AR, Jaffer F, Dale RC, Labrum R, Erro R, Meyer E et al (2015) The clinical and genetic heterogeneity of paroxysmal dyskinesias. Brain 138(Pt 12):3567–3580
Heron SE, Grinton BE, Kivity S, Afawi Z, Zuberi SM, Hughes JN et al (2012) PRRT2 mutations cause benign familial infantile epilepsy and infantile convulsions with choreoathetosis syndrome. Am J Hum Genet 90(1):152–160
Ono S, Yoshiura K, Kinoshita A, Kikuchi T, Nakane Y, Kato N et al (2012) Mutations in PRRT2 responsible for paroxysmal kinesigenic dyskinesias also cause benign familial infantile convulsions. J Hum Genet 57(5):338–341
Riant F, Roze E, Barbance C, Meneret A, Guyant-Marechal L, Lucas C et al (2012) PRRT2 mutations cause hemiplegic migraine. Neurology 79(21):2122–2124
Marini C, Conti V, Mei D, Battaglia D, Lettori D, Losito E et al (2012) PRRT2 mutations in familial infantile seizures, paroxysmal dyskinesia, and hemiplegic migraine. Neurology 79(21):2109–2114
Dale RC, Gardiner A, Antony J, Houlden H (2012) Familial PRRT2 mutation with heterogeneous paroxysmal disorders including paroxysmal torticollis and hemiplegic migraine. Dev Med Child Neurol 54(10):958–960
Ebrahimi-Fakhari D, Saffari A, Westenberger A, Klein C (2015) The evolving spectrum of PRRT2-associated paroxysmal diseases. Brain 138(Pt 12):3476–3495
Pelzer N, de Vries B, Kamphorst JT, Vijfhuizen LS, Ferrari MD, Haan J et al (2014) PRRT2 and hemiplegic migraine: a complex association. Neurology 83(3):288–290
Lee HY, Huang Y, Bruneau N, Roll P, Roberson ED, Hermann M et al (2012) Mutations in the gene PRRT2 cause paroxysmal kinesigenic dyskinesia with infantile convulsions. Cell Rep 1(1):2–12
Valente P, Castroflorio E, Rossi P, Fadda M, Sterlini B, Cervigni RI et al (2016) PRRT2 is a key component of the ca (2+)-dependent neurotransmitter release machinery. Cell Rep 15(1):117–131
Fruscione F, Valente P, Sterlini B, Romei A, Baldassari S, Fadda M et al (2018) PRRT2 controls neuronal excitability by negatively modulating Na+ channel 1.2/1.6 activity. Brain 141(4):1000–1016
Condliffe SB, Corradini I, Pozzi D, Verderio C, Matteoli M (2010) Endogenous SNAP-25 regulates native voltage-gated calcium channels in glutamatergic neurons. J Biol Chem 285(32):24968–24976
Weller CM, Leen WG, Neville BG, Duncan JS, de Vries B, Geilenkirchen MA et al (2015) A novel SLC2A1 mutation linking hemiplegic migraine with alternating hemiplegia of childhood. Cephalalgia 35(1):10–15
Mohammad SS, Coman D, Calvert S (2014) Glucose transporter 1 deficiency syndrome and hemiplegic migraines as a dominant presenting clinical feature. J Paediatr Child Health 50(12):1025–1026
Jen JC, Wan J, Palos TP, Howard BD, Baloh RW (2005) Mutation in the glutamate transporter EAAT1 causes episodic ataxia, hemiplegia, and seizures. Neurology 65(4):529–534
Kovermann P, Hessel M, Kortzak D, Jen JC, Koch J, Fahlke C et al (2017) Impaired K(+) binding to glial glutamate transporter EAAT1 in migraine. Sci Rep 7(1):13913
Suzuki M, Van Paesschen W, Stalmans I, Horita S, Yamada H, Bergmans BA et al (2010) Defective membrane expression of the Na(+)-HCO (3)(−) cotransporter NBCe1 is associated with familial migraine. Proc Natl Acad Sci U S A 107(36):15963–15968
Lafreniere RG, Cader MZ, Poulin JF, Andres-Enguix I, Simoneau M, Gupta N et al (2010) A dominant-negative mutation in the TRESK potassium channel is linked to familial migraine with aura. Nat Med 16(10):1157–1160
Kang D, Kim D (2006) TREK-2 (K2P10.1) and TRESK (K2P18.1) are major background K+ channels in dorsal root ganglion neurons. Am J Phys Cell Physiol 291(1):C138–C146
Enyedi P, Czirjak G (2015) Properties, regulation, pharmacology, and functions of the K (2) p channel, TRESK. Pflugers Arch 467(5):945–958
Dobler T, Springauf A, Tovornik S, Weber M, Schmitt A, Sedlmeier R et al (2007) TRESK two-pore-domain K+ channels constitute a significant component of background potassium currents in murine dorsal root ganglion neurones. J Physiol 585(Pt 3):867–879
Chae YJ, Zhang J, Au P, Sabbadini M, Xie GX, Yost CS (2010) Discrete change in volatile anesthetic sensitivity in mice with inactivated tandem pore potassium ion channel TRESK. Anesthesiology 113(6):1326–1337
Liu P, Xiao Z, Ren F, Guo Z, Chen Z, Zhao H et al (2013) Functional analysis of a migraine-associated TRESK K+ channel mutation. J Neurosci 33(31):12810–12824
Guo Z, Liu P, Ren F, Cao YQ (2014) Nonmigraine-associated TRESK K+ channel variant C110R does not increase the excitability of trigeminal ganglion neurons. J Neurophysiol 112(3):568–579
Andres-Enguix I, Shang L, Stansfeld PJ, Morahan JM, Sansom MS, Lafreniere RG et al (2012) Functional analysis of missense variants in the TRESK (KCNK18) K channel. Sci Rep 2:237
Royal P, Andres-Bilbe A, Avalos Prado P, Verkest C, Wdziekonski B, Schaub S et al (2019) Migraine-associated TRESK mutations increase neuronal excitability through alternative translation initiation and inhibition of TREK. Neuron 101(2):232–45 e6
Brennan KC, Bates EA, Shapiro RE, Zyuzin J, Hallows WC, Huang Y et al (2013) Casein kinase idelta mutations in familial migraine and advanced sleep phase. Sci Transl Med 5(183):183ra56, 1–183ra56,11
Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N et al (2005) Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature 434(7033):640–644
Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptacek LJ (2007) Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell 128(1):59–70
Alstadhaug KB (2009) Migraine and the hypothalamus. Cephalalgia 29(8):809–817
Ahn AH, Goadsby PJ (2013) Migraine and sleep: new connections. Cerebrum 2013:15
Maniyar FH, Sprenger T, Monteith T, Schankin C, Goadsby PJ (2014) Brain activations in the premonitory phase of nitroglycerin-triggered migraine attacks. Brain 137(Pt 1):232–241
Williams LB, Javed A, Sabri A, Morgan DJ, Huff CD, Grigg JR et al (2019) ALPK1 missense pathogenic variant in five families leads to ROSAH syndrome, an ocular multisystem autosomal dominant disorder. Genet Med. https://doi.org/10.1038/s41436-019-0476-3. [Epub ahead of print]
Dichgans M, Mayer M, Uttner I, Bruning R, Muller-Hocker J, Rungger G et al (1998) The phenotypic spectrum of CADASIL: clinical findings in 102 cases. Ann Neurol 44(5):731–739
Guey S, Mawet J, Herve D, Duering M, Godin O, Jouvent E et al (2016) Prevalence and characteristics of migraine in CADASIL. Cephalalgia 36(11):1038–1047
Tan RY, Markus HS (2016) CADASIL: migraine, encephalopathy, stroke and their inter-relationships. PLoS One 11(6):e0157613
Liem MK, Oberstein SA, van der Grond J, Ferrari MD, Haan JCADASIL (2010) Migraine: a narrative review. Cephalalgia 30(11):1284–1289
Rossi G, Shambhu S (2018) Hemiplegic migraine as the initial presentation of biopsy positive cerebral autosomal dominant Arteriopathy with subcortical infarcts and leukoencephalopathy. Cureus 10(5):e2631
Rice GI, Rodero MP, Crow YJ (2015) Human disease phenotypes associated with mutations in TREX1. J Clin Immunol 35(3):235–243
Stam AH, Kothari PH, Shaikh A, Gschwendter A, Jen JC, Hodgkinson S et al (2016) Retinal vasculopathy with cerebral leukoencephalopathy and systemic manifestations. Brain 139(11):2909-2922
Stam AH, Haan J, van den Maagdenberg AM, Ferrari MD, Terwindt GM (2009) Migraine and genetic and acquired vasculopathies. Cephalalgia 29(9):1006–1017
Breedveld G, de Coo IF, Lequin MH, Arts WF, Heutink P, Gould DB et al (2006) Novel mutations in three families confirm a major role of COL4A1 in hereditary porencephaly. J Med Genet 43(6):490–495
Sondergaard CB, Nielsen JE, Hansen CK, Christensen H (2017) Hereditary cerebral small vessel disease and stroke. Clin Neurol Neurosurg 155:45–57
Gormley P, Winsvold BS, Nyholt DR, Kallela M, Chasman DI, Palotie A (2016) Migraine genetics: from genome-wide association studies to translational insights. Genome Med 8(1):86
van den Maagdenberg A, Nyholt DR, Anttila V (2019) Novel hypotheses emerging from GWAS in migraine? J Headache Pain 20(1):5
Hansen RD, Christensen AF, Olesen J (2017) Family studies to find rare high risk variants in migraine. J Headache Pain 18(1):32
Hu Y, Jiang H, Wang Q, Xie Z, Pan S (2013) Identification of a novel nonsense mutation p.Tyr1957Ter of CACNA1A in a Chinese family with episodic ataxia 2. PLoS One 8(2):e56362
Oh SK, Baek JI, Weigand KM, Venselaar H, Swarts HG, Park SH et al (2015) A missense variant of the ATP1A2 gene is associated with a novel phenotype of progressive sensorineural hearing loss associated with migraine. Eur J Hum Genet 23(5):639–645
Maksemous N, Roy B, Smith RA, Griffiths LR (2016) Next-generation sequencing identifies novel CACNA1A gene mutations in episodic ataxia type 2. Mol Genet Genomic Med 4(2):211–222
Maksemous N, Smith RA, Haupt LM, Griffiths LR (2016) Targeted next generation sequencing identifies novel NOTCH3 gene mutations in CADASIL diagnostics patients. Hum Genomics 10(1):38
Klassen T, Davis C, Goldman A, Burgess D, Chen T, Wheeler D et al (2011) Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell 145(7):1036–1048
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T et al (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616):285–291
Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S et al (2016) ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44(D1):D862–D868
Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC (2012) SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 40(Web Server issue):W452–W457
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P et al (2010) A method and server for predicting damaging missense mutations. Nat Methods 7(4):248–249
Karchin R (2009) Next generation tools for the annotation of human SNPs. Brief Bioinform 10(1):35–52
Frousios K, Iliopoulos CS, Schlitt T, Simpson MA (2013) Predicting the functional consequences of non-synonymous DNA sequence variants--evaluation of bioinformatics tools and development of a consensus strategy. Genomics 102(4):223–228
Schwarz JM, Rodelsperger C, Schuelke M, Seelow D (2010) MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 7(8):575–576
Lee D, Gorkin DU, Baker M, Strober BJ, Asoni AL, McCallion AS et al (2015) A method to predict the impact of regulatory variants from DNA sequence. Nat Genet 47(8):955–961
Rojano E, Seoane P, Ranea JAG, Perkins JR (2018) Regulatory variants: from detection to predicting impact. Brief Bioinform. https://doi.org/10.1093/bib/bby039. [Epub ahead of print]
Sander JD, Joung JK (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32(4):347–355
Nemeth AH, Kwasniewska AC, Lise S, Parolin Schnekenberg R, Becker EB, Bera KD et al (2013) Next generation sequencing for molecular diagnosis of neurological disorders using ataxias as a model. Brain 136(Pt 10):3106–3118
Handel AE, Chintawar S, Lalic T, Whiteley E, Vowles J, Giustacchini A et al (2016) Assessing similarity to primary tissue and cortical layer identity in induced pluripotent stem cell-derived cortical neurons through single-cell transcriptomics. Hum Mol Genet 25(5):989–1000
Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME et al (2013) Cerebral organoids model human brain development and microcephaly. Nature 501(7467):373–379
Ipe J, Swart M, Burgess KS, Skaar TC (2017) High-throughput assays to assess the functional impact of genetic variants: a road towards genomic-driven medicine. Clin Transl Sci 10(2):67–77
Fowler DM, Stephany JJ, Fields S (2014) Measuring the activity of protein variants on a large scale using deep mutational scanning. Nat Protoc 9(9):2267–2284
Li T, Lu G, Chiang EY, Chernov-Rogan T, Grogan JL, Chen J (2017) High-throughput electrophysiological assays for voltage gated ion channels using SyncroPatch 768PE. PLoS One 12(7):e0180154
Dale RC, Gardiner A, Branson JA, Houlden H (2014) Benefit of carbamazepine in a patient with hemiplegic migraine associated with PRRT2 mutation. Dev Med Child Neurol 56(9):910
Maher BH, Griffiths LR (2011) Identification of molecular genetic factors that influence migraine. Mol Gen Genomics 285(6):433–446
Kondratieva N, Azimova J, Skorobogatykh K, Sergeev A, Naumova E, Kokaeva Z et al (2016) Biomarkers of migraine: part 1 - genetic markers. J Neurol Sci 369:63–76
Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG et al (1995) A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet 10(1):111–113
Rubino E, Ferrero M, Rainero I, Binello E, Vaula G, Pinessi L (2009) Association of the C677T polymorphism in the MTHFR gene with migraine: a meta-analysis. Cephalalgia 29(8):818–825
Schurks M, Rist PM, Kurth T (2010) MTHFR 677C>T and ACE D/I polymorphisms in migraine: a systematic review and meta-analysis. Headache 50(4):588–599
Samaan Z, Gaysina D, Cohen-Woods S, Craddock N, Jones L, Korszun A et al (2011) Methylenetetrahydrofolate reductase gene variant (MTHFR C677T) and migraine: a case control study and meta-analysis. BMC Neurol 11:66
Liu R, Geng P, Ma M, Yu S, Yang M, He M et al (2014) MTHFR C677T polymorphism and migraine risk: a meta-analysis. J Neurol Sci 336(1–2):68–73
Kaunisto MA, Kallela M, Hamalainen E, Kilpikari R, Havanka H, Harno H et al (2006) Testing of variants of the MTHFR and ESR1 genes in 1798 Finnish individuals fails to confirm the association with migraine with aura. Cephalalgia 26(12):1462–1472
Todt U, Freudenberg J, Goebel I, Netzer C, Heinze A, Heinze-Kuhn K et al (2006) MTHFR C677T polymorphism and migraine with aura. Ann Neurol 60(5):621–622 author reply 2-3
de Vries B, Anttila V, Freilinger T, Wessman M, Kaunisto MA, Kallela M et al (2016) Systematic re-evaluation of genes from candidate gene association studies in migraine using a large genome-wide association data set. Cephalalgia 36(7):604–614
Cardon LR, Palmer LJ (2003) Population stratification and spurious allelic association. Lancet 361(9357):598–604
Anttila V, Stefansson H, Kallela M, Todt U, Terwindt GM, Calafato MS et al (2010) Genome-wide association study of migraine implicates a common susceptibility variant on 8q22. Nat Genet 42(10):869–873
Chasman DI, Schurks M, Anttila V, de Vries B, Schminke U, Launer LJ et al (2011) Genome-wide association study reveals three susceptibility loci for common migraine in the general population. Nat Genet 43(7):695–698
Freilinger T, Anttila V, de Vries B, Malik R, Kallela M, Terwindt GM et al (2012) Genome-wide association analysis identifies susceptibility loci for migraine without aura. Nat Genet 44(7):777–782
Anttila V, Winsvold BS, Gormley P, Kurth T, Bettella F, McMahon G et al (2013) Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat Genet 45(8):912–917
Gan M, Jiang P, McLean P, Kanekiyo T, Bu G (2014) Low-density lipoprotein receptor-related protein 1 (LRP1) regulates the stability and function of GluA1 alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptor in neurons. PLoS One 9(12):e113237
Spuch C, Ortolano S, Navarro C (2012) LRP-1 and LRP-2 receptors function in the membrane neuron. Trafficking mechanisms and proteolytic processing in Alzheimer's disease. Front Physiol 3:269
Nakajima C, Kulik A, Frotscher M, Herz J, Schafer M, Bock HH et al (2013) Low density lipoprotein receptor-related protein 1 (LRP1) modulates N-methyl-D-aspartate (NMDA) receptor-dependent intracellular signaling and NMDA-induced regulation of postsynaptic protein complexes. J Biol Chem 288(30):21909–21923
Dussor G, Cao YQ (2016) TRPM8 and migraine. Headache 56(9):1406–1417
Chi J, Cohen P (2016) The multifaceted roles of PRDM16: adipose biology and beyond. Trends Endocrinol Metab 27(1):11–23
Chuikov S, Levi BP, Smith ML, Morrison SJ (2010) Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat Cell Biol 12(10):999–1006
Shimada IS, Acar M, Burgess RJ, Zhao Z, Morrison SJ (2017) Prdm16 is required for the maintenance of neural stem cells in the postnatal forebrain and their differentiation into ependymal cells. Genes Dev 31(11):1134–1146
Nguyen HL, Lee YJ, Shin J, Lee E, Park SO, McCarty JH et al (2011) TGF-beta signaling in endothelial cells, but not neuroepithelial cells, is essential for cerebral vascular development. Lab Investig 91(11):1554–1563
Luo SX, Timbang L, Kim JI, Shang Y, Sandoval K, Tang AA et al (2016) TGF-beta signaling in dopaminergic neurons regulates dendritic growth, excitatory-inhibitory synaptic balance, and reversal learning. Cell Rep 17(12):3233–3245
Nelson N (1999) Metal ion transporters and homeostasis. EMBO J 18(16):4361–4371
McAllister BB, Dyck RH (2017) Zinc transporter 3 (ZnT3) and vesicular zinc in central nervous system function. Neurosci Biobehav Rev 80:329–350
Telley L, Cadilhac C, Cioni JM, Saywell V, Jahannault-Talignani C, Huettl RE et al (2016) Dual function of NRP1 in axon guidance and subcellular target recognition in cerebellum. Neuron 91(6):1276–1291
Aghajanian H, Cho YK, Manderfield LJ, Herling MR, Gupta M, Ho VC et al (2016) Coronary vasculature patterning requires a novel endothelial ErbB2 holoreceptor. Nat Commun 7:12038
Spain SL, Barrett JC (2015) Strategies for fine-mapping complex traits. Hum Mol Genet 24(R1):R111–R119
Wellcome Trust Case Control C, Maller JB, McVean G, Byrnes J, Vukcevic D, Palin K et al (2012) Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet 44(12):1294–1301
Gupta RM, Hadaya J, Trehan A, Zekavat SM, Roselli C, Klarin D et al (2017) A genetic variant associated with five vascular diseases is a distal regulator of Endothelin-1 gene expression. Cell 170(3):522–33 e15
Hannon E, Weedon M, Bray N, O'Donovan M, Mill J (2017) Pleiotropic effects of trait-associated genetic variation on DNA methylation: utility for refining GWAS loci. Am J Hum Genet 100(6):954–959
Eising E, Huisman SM, Mahfouz A, Vijfhuizen LS, Anttila V, Winsvold BS et al (2016) Gene co-expression analysis identifies brain regions and cell types involved in migraine pathophysiology: a GWAS-based study using the Allen human brain atlas. Hum Genet 135(4):425–439
Renthal W (2018) Localization of migraine susceptibility genes in human brain by single-cell RNA sequencing. Cephalalgia 38(13):1976–1983
LaPaglia DM, Sapio MR, Burbelo PD, Thierry-Mieg J, Thierry-Mieg D, Raithel SJ et al (2018) RNA-Seq investigations of human post-mortem trigeminal ganglia. Cephalalgia 38(5):912–932
Russell MB, Ulrich V, Gervil M, Olesen J (2002) Migraine without aura and migraine with aura are distinct disorders. A population-based twin survey. Headache 42(5):332–336
Nyholt DR, Gillespie NG, Heath AC, Merikangas KR, Duffy DL, Martin NG (2004) Latent class and genetic analysis does not support migraine with aura and migraine without aura as separate entities. Genet Epidemiol 26(3):231–244
Zhao H, Eising E, de Vries B, Vijfhuizen LS (2016) International headache genetics C, Anttila V, et al. gene-based pleiotropy across migraine with aura and migraine without aura patient groups. Cephalalgia 36(7):648–657
Chasman DI, Anttila V, Buring JE, Ridker PM, Schurks M, Kurth T et al (2014) Selectivity in genetic association with sub-classified migraine in women. PLoS Genet 10(5):e1004366
Esserlind AL, Christensen AF, Steinberg S, Grarup N, Pedersen O, Hansen T et al (2016) The association between candidate migraine susceptibility loci and severe migraine phenotype in a clinical sample. Cephalalgia 36(7):615–623
Pollock CE, Sutherland HG, Maher BH, Lea RA, Haupt LM, Frith A et al (2018) The NRP1 migraine risk variant shows evidence of association with menstrual migraine. J Headache Pain 19(1):31
Meng W, Adams MJ, Hebert HL, Deary IJ, McIntosh AM, Smith BH (2018) A genome-wide association study finds genetic associations with broadly-defined headache in UK biobank (N=223,773). EBioMedicine 28:180–186
Brainstorm Consortium, Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J et al (2018) Analysis of shared heritability in common disorders of the brain. Science 360(6395):eaap8757
Yang Y, Zhao H, Boomsma DI, Ligthart L, Belin AC, Smith GD et al (2018) Molecular genetic overlap between migraine and major depressive disorder. Eur J Hum Genet 26(8):1202–1216
Keezer MR, Bauer PR, Ferrari MD, Sander JW (2015) The comorbid relationship between migraine and epilepsy: a systematic review and meta-analysis. Eur J Neurol 22(7):1038–1047
Malik R, Freilinger T, Winsvold BS, Anttila V, Vander Heiden J, Traylor M et al (2015) Shared genetic basis for migraine and ischemic stroke: a genome-wide analysis of common variants. Neurology 84(21):2132–2145
Winsvold BS, Nelson CP, Malik R, Gormley P, Anttila V, Vander Heiden J et al (2015) Genetic analysis for a shared biological basis between migraine and coronary artery disease. Neurol Genet 1(1):e10
Kutuk MO, Tufan AE, Guler G, Yalin OO, Altintas E, Bag HG et al (2018) Migraine and associated comorbidities are three times more frequent in children with ADHD and their mothers. Brain and Development 40(10):857–864
Prontera P, Sarchielli P, Caproni S, Bedetti C, Cupini LM, Calabresi P et al (2018) Epilepsy in hemiplegic migraine: genetic mutations and clinical implications. Cephalalgia 38(2):361–373
Louter MA, Pelzer N, de Boer I, Kuijvenhoven BE, van Oosterhout WP, van Zwet EW et al (2016) Prevalence of lifetime depression in a large hemiplegic migraine cohort. Neurology 87(22):2370–2374
Linde M, Mulleners WM, Chronicle EP, McCrory DC (2013) Valproate (valproic acid or sodium valproate or a combination of the two) for the prophylaxis of episodic migraine in adults. Cochrane Database Syst Rev 24(6):CD010611
Ran C, Graae L, Magnusson PK, Pedersen NL, Olson L, Belin AC (2014) A replication study of GWAS findings in migraine identifies association in a Swedish case-control sample. BMC Med Genet 15:38
Sintas C, Fernandez-Morales J, Vila-Pueyo M, Narberhaus B, Arenas C, Pozo-Rosich P et al (2015) Replication study of previous migraine genome-wide association study findings in a Spanish sample of migraine with aura. Cephalalgia 35(9):776–782
An XK, Ma QL, Lin Q, Zhang XR, Lu CX, Qu HL (2013) PRDM16 rs2651899 variant is a risk factor for Chinese common migraine patients. Headache 53(10):1595–1601
Fan X, Wang J, Fan W, Chen L, Gui B, Tan G et al (2014) Replication of migraine GWAS susceptibility loci in Chinese Han population. Headache 54(4):709–715
Ghosh J, Pradhan S, Mittal B (2013) Genome-wide-associated variants in migraine susceptibility: a replication study from North India. Headache 53(10):1583–1594
Lin QF, Fu XG, Yao LT, Yang J, Cao LY, Xin YT et al (2015) Association of genetic loci for migraine susceptibility in the she people of China. J Headache Pain 16:553
Chen SP, Fuh JL, Chung MY, Lin YC, Liao YC, Wang YF et al (2018) Genome-wide association study identifies novel susceptibility loci for migraine in Han Chinese resided in Taiwan. Cephalalgia 38(3):466–475
Chang X, Pellegrino R, Garifallou J, March M, Snyder J, Mentch F et al (2018) Common variants at 5q33.1 predispose to migraine in African-American children. J Med Genet 55(12):831–836
Cox HC, Lea RA, Bellis C, Carless M, Dyer TD, Curran J et al (2012) A genome-wide analysis of 'Bounty' descendants implicates several novel variants in migraine susceptibility. Neurogenetics 13(3):261–266
Dudbridge F (2013) Power and predictive accuracy of polygenic risk scores. PLoS Genet 9(3):e1003348
Chalmer MA, Esserlind AL, Olesen J, Hansen TF (2018) Polygenic risk score: use in migraine research. J Headache Pain 19(1):29
Rodriguez-Acevedo AJ, Ferreira MA, Benton MC, Carless MA, Goring HH, Curran JE et al (2015) Common polygenic variation contributes to risk of migraine in the Norfolk Island population. Hum Genet 134(10):1079–1087
Gormley P, Kurki MI, Hiekkala ME, Veerapen K, Happola P, Mitchell AA et al (2018) Common variant burden contributes to the familial aggregation of migraine in 1,589 families. Neuron 98(4):743–53 e4
Christensen AF, Esserlind AL, Werge T, Stefansson H, Stefansson K, Olesen J (2016) The influence of genetic constitution on migraine drug responses. Cephalalgia 36(7):624–639
Cargnin S, Viana M, Sances G, Cantello R, Tassorelli C, Terrazzino S (2019) Using a genetic risk score approach to predict headache response to Triptans in migraine without Aura. J Clin Pharmacol 59(2):288–294
Schurks M, Kurth T, Stude P, Rimmbach C, de Jesus J, Jonjic M et al (2007) G protein beta3 polymorphism and triptan response in cluster headache. Clin Pharmacol Ther 82(4):396–401
Gentile G, Missori S, Borro M, Sebastianelli A, Simmaco M, Martelletti P (2010) Frequencies of genetic polymorphisms related to triptans metabolism in chronic migraine. J Headache Pain 11(2):151–156
Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S et al (2014) Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 46(11):1173–1186
Zhong K, Zhu G, Jing X, Hendriks AEJ, Drop SLS, Ikram MA et al (2017) Genome-wide compound heterozygote analysis highlights alleles associated with adult height in Europeans. Hum Genet 136(11–12):1407–1417
Marouli E, Graff M, Medina-Gomez C, Lo KS, Wood AR, Kjaer TR et al (2017) Rare and low-frequency coding variants alter human adult height. Nature 542(7640):186–190
Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR et al (2015) Genetic studies of body mass index yield new insights for obesity biology. Nature 518(7538):197–206
Akiyama M, Okada Y, Kanai M, Takahashi A, Momozawa Y, Ikeda M et al (2017) Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat Genet 49(10):1458–1467
Gerring ZF, McRae AF, Montgomery GW, Nyholt DR (2018) Genome-wide DNA methylation profiling in whole blood reveals epigenetic signatures associated with migraine. BMC Genomics 19(1):69
Acknowledgements
Not applicable.
Funding
Our research group has previously received funding for migraine genetics research from the Brain Foundation, the Migraine Research Foundation and the National Health and Medical Research Council, Australia (Project grant APP1122387). No funding was obtained for this review.
The APCs (article processing charges) for the articles in this thematic series ‘The Changing faces of migraine’ were made possible through independent educational sponsorship by Eli Lilly. Eli Lilly provided the funds through an educational grant which included enduring materials within the context of a symposium at the 12th European Headache Federation Congress in September 2018, chaired by Paolo Martelletti. This grant was provided to Springer Healthcare IME who organized the symposium and all of the enduring materials. Three of the articles in this thematic series were developed from content presented at the symposium. Eli Lilly were not involved in the planning of the thematic series, the selection process for topics, nor in any peer review or decision-making processes.
The articles have undergone the journal’s standard peer review process overseen by the Editor-in-Chief. For articles where the Editor-in-Chief is an author, the peer review process was overseen by one of the other Editors responsible for this thematic series.
Author information
Authors and Affiliations
Contributions
All authors contributed to the writing and revision of the manuscript. The final manuscript was read and approved by all authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Sutherland, H.G., Albury, C.L. & Griffiths, L.R. Advances in genetics of migraine. J Headache Pain 20, 72 (2019). https://doi.org/10.1186/s10194-019-1017-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s10194-019-1017-9