Abstract
Background
Klebsiella pneumoniae is a frequent nosocomial pathogen, with the multidrug-resistant (MDR) K. pneumoniae being a major public health concern, frequently causing difficult-to-treat infections worldwide. The aim of this study was to investigate the molecular characterization of clinical MDR Klebsiella pneumoniae isolates.
Methods
A total of 27 non-duplicate MDR K. pneumoniae isolates with a CTX-CIP-AK resistance pattern were investigated for the prevalence of antimicrobial resistance genes including extended spectrum β-lactamase genes (ESBLs), plasmid-mediated quinolone resistance (PMQR) genes, 16S rRNA methylase (16S-RMTase) genes, and integrons by polymerase chain reaction (PCR) amplification and DNA sequencing. Plasmid replicons were typed by PCR-based replicon typing (PBRT). Multi-locus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) were carried out to characterize the strain relatedness.
Results
All the isolates co-harbored 3 or more resistance determinants. OqxAB, CTX-M-type ESBLs and RmtB were the most frequent determinants, distributed among19 (70.4%),18 (66.7%) and 8 (29.6%) strains. Fourteen isolates harbored class 1 integrons, with orfD-aacA4 being the most frequent gene cassette array. Class 3 integrons were less frequently identified and contained the gene cassette array of blaGES-1-blaOXA-10-aac(6′)-Ib. IncFII replicon was most commonly found in this collection. One cluster was observed with ≥80% similarity among profiles obtained by PFGE, and one sequence type (ST) by MLST, namely ST11, was observed in the cluster.
Conclusion
K. pneumoniae carbapenemase (KPC)–producing ST11 was the main clone detected. Of particular concern was the high prevalence of multiple resistance determinants, classs I integrons and IncFII plasmid replicon among these MDR strains, which provide advantages for the rapid development of MDR strains.
Similar content being viewed by others
Background
Klebsiella pneumoniae is an opportunistic pathogen associated with both community-acquired and nosocomial infections, including pneumonia, urinary tract infections, septicemia and wound infections, with the increasingly multidrug-resistant (MDR) K. pneumoniae being a major public health concern.
The prevailing hypothesis is that these bacteria acquire multidrug resistance through horizontal transfer of antimicrobial resistance genes mediated by mobile genetic elements such as integrons [1]. Several genes that are frequently involved in multidrug resistance to commonly used antimicrobial agents include plasmid-mediated quinolone resistance (PMQR) genes [2], exogenously acquired 16S rRNA methyltransferase (16S-RMTase) genes [3], and extended-spectrum β-lactamases (ESBLs) encoding genes [4]. Furthermore, carbapenem-hydrolyzing β-lactamases (CHβLs) has rapidly emerged in recent years, with K. pneumoniae being the most common organism associated with K. pneumoniae carbapenemase (KPC) resistance determinants [5].
Although the high prevalence of these resistance determinants has been reported among MDR K. pneumoniae strains [6, 7], there is little information available on the distribution of integrons and plasmid replicons among these strains, and limited data on the genetic relationship between these strains.
The aim of this study therefore was to investigate the prevalence of frequently reported antimicrobial resistance determinants among the 27 clinical MDR K. pneumoniae isolates, to explore the distribution of integrons and plasmid replicons, as well as to analyze the genetic clonality by pulse field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST).
Method
Bacterial isolates
A total of 27 non-duplicate MDR K. pneumoniae isolates simultaneously displaying resistance to cefotaxime, amikacin and levofloxacin were collected from our hospitals from March to July 2011(Susceptibility to antimicrobial agents was determined using Kirby–Bauer’s disc diffusion method and the results were analyzed and interpreted according to CLSI guidelines [8]). Among them, 12 strains were resistant to imipenem. The clinical sources of these specimens include sputum (n = 13), urine (n = 4), blood (n = 7), and wound secretions (n = 3). Among these 27 isolates, 2 strains originated from community-acquired infections, defined as the pathogen being isolated within 48 h after hospital admittance. The other 25 ones were from nosocomial infections, where the pathogens were isolated more than 48 h after being admitted to a hospital.
Detection of antimicrobial resistance determinants
DNA templates were prepared by the boiling method. All the isolates were analyzed for the presence of bla ESBLs (bla CTX, bla TEM, bla SHV, bla VEB, and bla OXA) [9], PMQRs (qnrA, qnrB, qnrC, qnrD, qnrS, aac (6′)-Ib-cr, and qepA) [10], and 16S-RMTases (armA, npmA, rmtA, rmtB, rmtC, rmtD, and rmtE) [11]. The 12 strains resistant to imipenem were further analyzed for CHβLs endocing genes (bla KPC, bla OXA-48, bla IMP, bla VIM, bla NDM, bla DIM, bla SPM, and bla SIM) [12]. All genes were screened by multiplex PCR followed by single PCR for gene confirmation. Positive products were further purified with a DNA purification kit and then sent to the Majorbio Company (Shanghai, China) for sequencing. Sequences were analyzed by using the Chromas-Pro application and BLAST (http://www.ncbi.nlm.nih.gov/BLAST).
Detection of integrons
Integrons were detected by PCR amplification of class 1, 2, and 3 integrase-specific int-1, int-2, and int-3 genes, as described previously [13]. To reveal the gene cassettes of the variable region of integrons, the variable region of class 1 and 3 integrons were further amplified and purified, and then sequenced to determine their gene cassette composition. The resulting nucleotide sequences and deduced protein sequences were analyzed with the BLAST and FASTA programs of the National Center for Biotechnology Information (http://www.ncbi.nlm.nhi.gov).
PCR-based replicon typing
Main plasmid incompatibility groups including F, FIA, FIB, FIC, HI1, HI2, I1-Ic, L/M, N, P, W, T, A/C, K, B/O, X, Y, and FII were determined using the PCR-based replicon typing (PBRT) scheme as described by Carattoli [14].
Pulsed-field gel electrophoresis
Clonal relatedness of 27 K. pneumoniae isolates was analyzed by pulsed-field electrophoresis (PFGE) as described [15]. Prepared genomic DNA was digested using the restriction endonuclease Xba I (Fermentas, ABI, Germany), and DNA fragments were separated in a PFGE CHEF-DR III system (Bio-Rad Laboratories, Hercules, CA) in 0.5× Tris-borate-EDTA buffer at 120 V for 19 h, with pulse times ranging from 2.2 s to 54.2 s. The banding patterns were analyzed by the BioNumerics software (Applied Math, Sint-Maten-Latem, Belgium). Cutoff lines at 65% and 80% were used to analyze genetic relatedness.
Multi-locus sequence typing
Genotyping for the 27 MDR strains was further determined by multi-locus sequence typing (MLST) analysis. MLST with 7 genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) was performed according to Diancourt et al. [16]. Alleles and sequence types (STs) were assigned by using the MLST database (http://www.pasteur.fr/mlst/Kpneumoniae.html).
Results
Wide distribution of resistance determinants
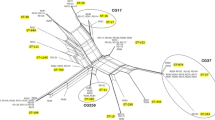
Diverse resistance determinants were found among the MDR strains (Figure 1). β-Lactamse including CTX-M-type ESBLs, sulfhydryl variable (SHV) variants, TEM-1b, KPC, and OXA, were carried by 18 (66.7%), 15 (55.6%), 13 (48.1%), 15 (55.6%), 12 (44.4%), and 10 (37.0%) of the isolates, respectively. PMQRs were detected in 21 isolates. Of these, 19 (70.4%) belonged to OqxAB, 11 (40.7%) to Qnr (7 QnrS1, 1 QnrB2, and 4 QnrB4), 10 (37.0%) to AAC (6′)-Ib-cr, and 1 to QepA. Determinants conferring resistance to amikacin included 15 (55.6%) 16S-RMTase determinants, with RmtB being present in 29.6% (8/27), ArmA in 25.9% (7/27), AAC(6′)-Ib-cr in 10 (37.0%), and AAC(6′)-Ib in 10 (37.0%) (encoded by 10 aacA4 located within class 1 integron).
Furthermore, all the isolates co-harbored 3 or more resistance determinants. The PMQRs were present in 19 out of the 22 ESBL-carrying K. pneumoniae isolates. Of the 11 KPC-positive MDR K. pneumoniae isolates, 7 co-carried rmtB or armA.
Prevalence of integrons
Integron analysis showed that 14 isolates carried class 1 integrons; 6 carried class 3 integrons; and 2 contained both of class 1 and 3 integrons (Figure 1). No class 2 integrons were found. Sequencing analysis for intI1-positive strains revealed that the cassette arrays of class 1 integron include orfD-aacA4 (n = 10 strains) and aad5-dfrA17 (n = 4 strains). Moreover, the variable regions in class 3 integrons of all the 6 isolates were the same as those comprising blaGES-1-blaOXA-10-aac(6′)-Ib, which has been identified in an earlier study [17].
Distribution of plasmid replicons
PBRT revealed that IncFII type was the predominant plasmid replicon among the MDR K. pneumoniae strains (Figure 1), IncFII (18/27, 66.7%), IncA/C (6/27, 22.2%) and IncI1 (2/27, 7.4%) were found alone or in combination; IncN(4/27, 14.8%), IncFIC (2/27, 7.4%), IncY(1/27, 3.7%), IncFIB (1/27, 3.7%) and IncB/O (1/27, 3.7%) were found in combination with IncFII.
Clonal relatedness
According to the PFGE patterns of the isolates (Figure 1), 27 different clonal patterns were observed among the 27 MDR K. pneumoniae isolates with ≥65% similarity.
One cluster of 7 closely related isolates was found that exhibited ≥80% similarities, these isolates were identified to be ST11 clones.
Sequence types for the K. pneumoniae isolates
MLST was conducted to determine the extent of genotypic diversity among the K. pneumoniae isolates. Fifteen different STs were identified. The most dominant ST was ST11 (29.6%, 8/27), followed by ST686 (14.8%, 4/27), ST1 (7.4%, 2/27), ST756 (7.4%, 2/27). These 4 STs accounted for 59.3% (16/27) of the total isolates. Among the STs identified, ST415, ST186, ST276, and ST866 have previously not been identified to be MDR strains.
Discussion
MDR K. pneumoniae strains have caused major therapeutic problems worldwide. The increasing prevalence of clinical MDR isolates has been associated with higher morbidity and mortality rates, posing a considerable threat to public health. In this study, we provide the current data on the molecular characterization of MDR K. pneumoniae isolates isolated from different clinical samples of hospitalized patients.
Our study revealed a wide distribution of diverse resistance determinants among the MDR K. pneumoniae strains. The most prevalent determinants were oqxAB genes, which have been reported to be the most frequent PMQRs in K. pneumoniae isolates [18]. CTX-M has been found to be widely disseminated among clinical Enterobacteriaceae such as Escherichia coli and K. pneumoniae[19], the even higher prevalence of bla CTX-M among our strains corresponds with an earlier report showing that CTX-M–producing strains are generally MDR ones [19]. Furthermore, in line with the previous investigation [20], KPC is highly prevalent among our strains, which also demonstrated that the production of KPC-type carbapenemases was the most prevalent carbapenem resistance mechanism in K. pneumoniae isolates. In addition, the widely distributed 16S-RMTase encoding genes in our MDR strains have been previously reported in China [21], together with AAC (6′)-Ib-cr and AAC (6′)-Ib accounting for the amikacin resistance. Recently, co-production of 16S rRNA methylases (ArmA and RmtB) and KPC were frequently reported in Enterobacteriaceae[22], which was also observed in our study, leading to few choices for antimicrobial treatment. Notably, there seems to be an intimate association between the occurrence of oqxAB and ESBLs; a similar phenomenon was noted for 16S-RMTase and β-lactamase. Such a frequent co-existence of 16S-RMTase with PMQRs and β-lactamase among MDR K. pneumoniae isolates in our study suggests a horizontal dissemination of these determinants amongst clinical MDR K. pneumoniae isolates. These MDR strains co-carrying diverse and numerous multiple resistance determinants may impose limitations in the therapeutic options available for the treatment of infections.
Epidemic resistance plasmids including IncFII, IncA/C, IncL/M, IncN and IncI1 plasmids has been worldwide detected in Enterobacteriaceae of different origin and sources [23]. In our study, the wide distribution of IncFII plasmid replicon is in accordance with the previous studies showing that IncFII plasmid replicons were most frequently detected in CTX-M-producing enterobacterial isolates in China and in Europe [24, 25]. Furthermore, other plasmid replicons such as IncN, IncI1, IncY and IncA/C distributed among our strains have also been previously reported [26, 27]. Additionally, the widely prevalent IncFII plasmid replicons and class I integrons identified in our study indicate that they might be playing an important role in attributing MDR to the clinical K. pneumoniae isolates. It is noteworthy that class 3 integrons were also detected among our strains. which is also reported earlier by Qi et al. in 2011 [28], showing the presence of class 3 integrons in MDR K. pneumoniae isolates from clinical settings in China, since class 3 integrons have also been found and characterized in Europe [29].
The genetic diversity of our isolates revealed that most isolates were of different strains, indicating the ease of transmission of these resistance determinants between bacterial species by mobile elements. This has been partially confirmed by the high prevalence of integrons and plasmid replicons in our study. However, it seems that the epidemic dissemination of the major clone ST11 producing bla KPC also played an additional role in our study, this corresponds with the report that ST11 is a truly international sequence type and associated with KPC [30]. Which is partly different from the KPC-producing K. pneumoniae clonal complex 258 (ST258, ST512, and ST101) in Europe and USA [31, 32]. Of note, the other STs such as ST415, ST186, ST276, and ST866 have not been previously identified to be MDR clones. Such a high heterogeneity of ST clones observed may further indicate the role of plasmids and integrons in the development of clinical MDR isolates.
Conclusion
In summary, our study showed that clinical MDR K. pneumoniae isolates may result mainly from the horizontal dissemination of multiple resistance determinants and the clonal dissemination of MDR ST11 strains–producing KPC-2, which alerts us the emergency and necessity to vigorously implement the infection control practice to prevent the dissemination of these MDR isolates in the healthcare settings.
Abbreviations
- MDR:
-
Multidrug-resistant
- MLST:
-
Multi-locus sequence typing
- PFGE:
-
Pulsed-field gel electrophoresis
- KPC:
-
K. pneumoniae carbapenemase
- PMQRs:
-
Plasmid-mediated quinolone resistance genes
- 16S-RMTase:
-
16S rRNA methyltransferase
- CHβL:
-
Carbapenem-hydrolyzing beta-lactamase
- ESBL:
-
Extended spectrum β-lactamase
- ST:
-
Sequence type
- PBRT:
-
PCR-based replicon typing.
References
Stalder T, Barraud O, Casellas M, Dagot C, Ploy MC: Integron involvement in environmental spread of antibiotic resistance. Front Microbiol 2012, 3: 119.
Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A: Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev 2009, 22: 664-689. 10.1128/CMR.00016-09
Wachino J, Arakawa Y: Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic gram-negative bacteria: an update. Drug Resist Updat 2012, 15: 133-148. 10.1016/j.drup.2012.05.001
Naseer U, Sundsfjord A: The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb Drug Resist 2011, 17: 83-97. 10.1089/mdr.2010.0132
Nordmann P, Naas T, Poirel L: Global spread of carbapenemase-producing Enterobacteriaceae . Emerg Infect Dis 2011, 17: 1791-1798. 10.3201/eid1710.110655
Luo Y, Yang J, Zhang Y, Ye L, Wang L, Guo L: Prevalence of beta-lactamases and 16S rRNA methylase genes amongst clinical Klebsiella pneumoniae isolates carrying plasmid-mediated quinolone resistance determinants. Int J Antimicrob Agents 2011, 37: 352-355. 10.1016/j.ijantimicag.2010.12.018
Elgorriaga-Islas E, Guggiana-Nilo P, Domínguez-Yévenes M, González-Rocha G, Mella-Montecinos S, Labarca-Labarca J, García-Cañete P, Bello-Toledo H: Prevalence of plasmid-mediated quinolone resistance determinant aac(6′)-Ib-cr among ESBL producing enterobacteria isolates from chilean hospitals. Enferm Infecc Microbiol Clin 2012, 30: 466-468. 10.1016/j.eimc.2012.01.024
Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing. Wayne, PA USA: Twenty-Second Informational Supplement M100-S22; 2012.
Dallenne C, Da Costa A, Decré D, Favier C, Arlet G: Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae . J Antimicrob Chemother 2010, 65: 490-495. 10.1093/jac/dkp498
Park KS, Kim MH, Park TS, Nam YS, Lee HJ, Suh JT: Prevalence of the plasmid-mediated quinolone resistance genes, aac(6′)-Ib-cr , qepA , and oqxAB in clinical isolates of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae in Korea. Ann Clin Lab Sci 2012, 42: 191-197.
Doi Y, Arakawa Y: 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis 2007, 45: 88-94. 10.1086/518605
Poirel L, Walsh TR, Cuvillier V, Nordmann P: Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 2011, 70: 119-123. 10.1016/j.diagmicrobio.2010.12.002
Mazel D, Dychinco B, Webb VA, Davies J: Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob Agents Chemother 2000, 44: 1568-1574. 10.1128/AAC.44.6.1568-1574.2000
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ: Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 2005, 63: 219-228. 10.1016/j.mimet.2005.03.018
Carrër A, Lassel L, Fortineau N, Mansouri M, Anguel N, Richard C, Nordmann P: Outbreak of CTX-M-15-producing Klebsiella pneumoniae in the intensive care unit of a french hospital. Microb Drug Resist 2009, 15: 47-54. 10.1089/mdr.2009.0868
Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S: Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 2005, 43: 4178-4182. 10.1128/JCM.43.8.4178-4182.2005
Correia M, Boavida F, Grosso F, Salgado MJ, Lito LM, Cristino JM, Mendo S, Duarte A: Molecular characterization of a new class 3 integron in Klebsiella pneumoniae . Antimicrob Agents Chemother 2003, 47: 2838-2843. 10.1128/AAC.47.9.2838-2843.2003
Zhao J, Chen Z, Chen S, Deng Y, Liu Y, Tian W, Huang X, Wu C, Sun Y, Sun Y, Zeng Z, Liu JH: Prevalence and dissemination of oqxAB in Escherichia coli isolates from animals, farmworkers, and the environment. Antimicrob Agents Chemother 2010, 54: 4219-4224. 10.1128/AAC.00139-10
Canton R, Gonzalez-Alba JM, Galan JC: CTX-M enzymes: origin and diffusion. Front Microbiol 2012, 3: 110.
El Salabi A, Walsh TR, Chouchani C: Extended spectrum beta-lactamases, carbapenemases and mobile genetic elements responsible for antibiotics resistance in gram-negative bacteria. Crit Rev Microbiol 2013, 39: 113-122. 10.3109/1040841X.2012.691870
Zhang Y, Yang J, Ye L, Luo Y, Wang W, Zhou W, Cui Z, Han L: Characterization of clinical multidrug-resistant Escherichia coli and Klebsiella pneumoniae isolates, 2007–2009, China. Microb Drug Resist 2012, 18: 465-470. 10.1089/mdr.2012.0016
Li JJ, Sheng ZK, Deng M, Bi S, Hu FS, Miao HF, Ji ZK, Sheng JF, Li LJ: Epidemic of Klebsiella pneumoniae ST11 clone coproducing KPC-2 and 16S rRNA methylase RmtB in a chinese university hospital. BMC Infect Dis 2012, 12: 373. 10.1186/1471-2334-12-373
Carattoli A: Plasmids in gram negatives: molecular typing of resistance plasmids. Int J Med Microbiol 2011, 301: 654-658. 10.1016/j.ijmm.2011.09.003
Ho PL, Lo WU, Yeung MK, Li Z, Chan J, Chow KH, Yam WC, Tong AH, Bao JY, Lin CH, Lok S, Chiu SS: Dissemination of pHK01-like incompatibility group IncFII plasmids encoding CTX-M-14 in Escherichia coli from human and animal sources. Vet Microbiol 2012, 158: 172-179. 10.1016/j.vetmic.2012.02.004
Habeeb MA, Haque A, Nematzadeh S, Iversen A, Giske CG: High prevalence of 16S rRNA methylase RmtB among CTX-M extended-spectrum β-lactamase-producing Klebsiella pneumoniae from Islamabad, Pakistan. Int J Antimicrob Agents 2013, 41: 524-526. 10.1016/j.ijantimicag.2013.02.017
Carattoli A: Resistance plasmid families in Enterobacteriaceae . Antimicrob Agents Chemother 2009, 53: 2227-2238. 10.1128/AAC.01707-08
Wang XR1, Chen JC, Kang Y, Jiang N, An SC, Gao ZC: Prevalence and characterization of plasmid-mediated bla ESBLwith their genetic environment in Escherichia coli and Klebsiella pneumoniae in patients with pneumonia. Chin Med J (Engl) 2012, 125: 894-900.
Qi Y1, Wei Z, Ji S, Du X, Shen P, Yu Y: ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother 2011, 66: 307-312. 10.1093/jac/dkq431
Barraud O, Casellas M, Dagot C, Ploy MC: An antibiotic-resistant class 3 integron in an Enterobacter cloacae isolate from hospital effluent. Clin Microbiol Infect 2013, 19: E306-E308. 10.1111/1469-0691.12186
Cuzon G, Naas T, Truong H, Villegas MV, Wisell KT, Carmeli Y, Gales AC, Venezia SN, Quinn JP, Nordmann P: Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. Emerg Infect Dis 2010, 16: 1349-1356. 10.3201/eid1609.091389
Woodford N, Turton JF, Livermore DM: Multiresistant gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 2011, 35: 736-755. 10.1111/j.1574-6976.2011.00268.x
Castanheira M, Farrell SE, Wanger A, Rolston KV, Jones RN, Mendes RE: Rapid expansion of KPC-2-producing Klebsiella pneumoniae isolates in two Texas hospitals due to clonal spread of ST258 and ST307 lineages. Microb Drug Resist 2013, 19: 295-297. 10.1089/mdr.2012.0238
Acknowledgment
This study is supported by Nan Jing Medical Science and Technique Development Foundation (grant no. QRX11191). We sincerely thank the EU Reference Laboratory for antimicrobial resistance of National Food Institute of Technical University of Denmark for providing the positive controls for our experiments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XC performed experimental work and drafted manuscript; XX and ZZ analyzed the study data; JC and HS provided interpretation of data; KZ conceived the study and provided data interpretation. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Cao, X., Xu, X., Zhang, Z. et al. Molecular characterization of clinical multidrug-resistant Klebsiella pneumoniae isolates. Ann Clin Microbiol Antimicrob 13, 16 (2014). https://doi.org/10.1186/1476-0711-13-16
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-0711-13-16